Abstract
Recently, image fusion has become one of the most promising fields in image processing since it plays an essential role in different applications, such as medical diagnosis and clarification of medical images. Multimodal Medical Image Fusion (MMIF) enhances the quality of medical images by combining two or more medical images from different modalities to obtain an improved fused image that is clearer than the original ones. Choosing the best MMIF technique which produces the best quality is one of the important problems in the assessment of image fusion techniques. In this paper, a complete survey on MMIF techniques is presented, along with medical imaging modalities, medical image fusion steps and levels, and the assessment methodology of MMIF. There are several image modalities, such as Computed Tomography (CT), Positron Emission Tomography (PET), Magnetic Resonance Imaging (MRI), and Single Photon Emission Computed Tomography (SPECT). Medical image fusion techniques are categorized into six main categories: spatial domain, transform fusion, fuzzy logic, morphological methods, and sparse representation methods. The MMIF levels are pixel-level, feature-level, and decision-level. The fusion quality evaluation metrics can be categorized as subjective/qualitative and objective/quantitative assessment methods. Furthermore, a detailed comparison between obtained results for significant MMIF techniques is also presented to highlight the pros and cons of each fusion technique.
1. Introduction
Image Fusion is concerned with combining features from multiple input images into a single image. The produced single image is more informative and accurate than any single source image since it contains all the necessary information. The purpose of image fusion is not only to decrease the quantity of data but also to construct images that are more appropriate and more clear for human and machine perception [1]. Multi-sensor image fusion is an application of image fusion that is the process of combining relevant information from two or more source images into one output image. Image fusion is one of the most exciting fields in image processing since it deploys different methodologies to produce a clear image that can be used for examination and diagnosis [2]. There are five basic steps to perform image fusion, which can be summarized as image registration, image decomposition, fusion rules, image reconstruction, and evaluation methods.
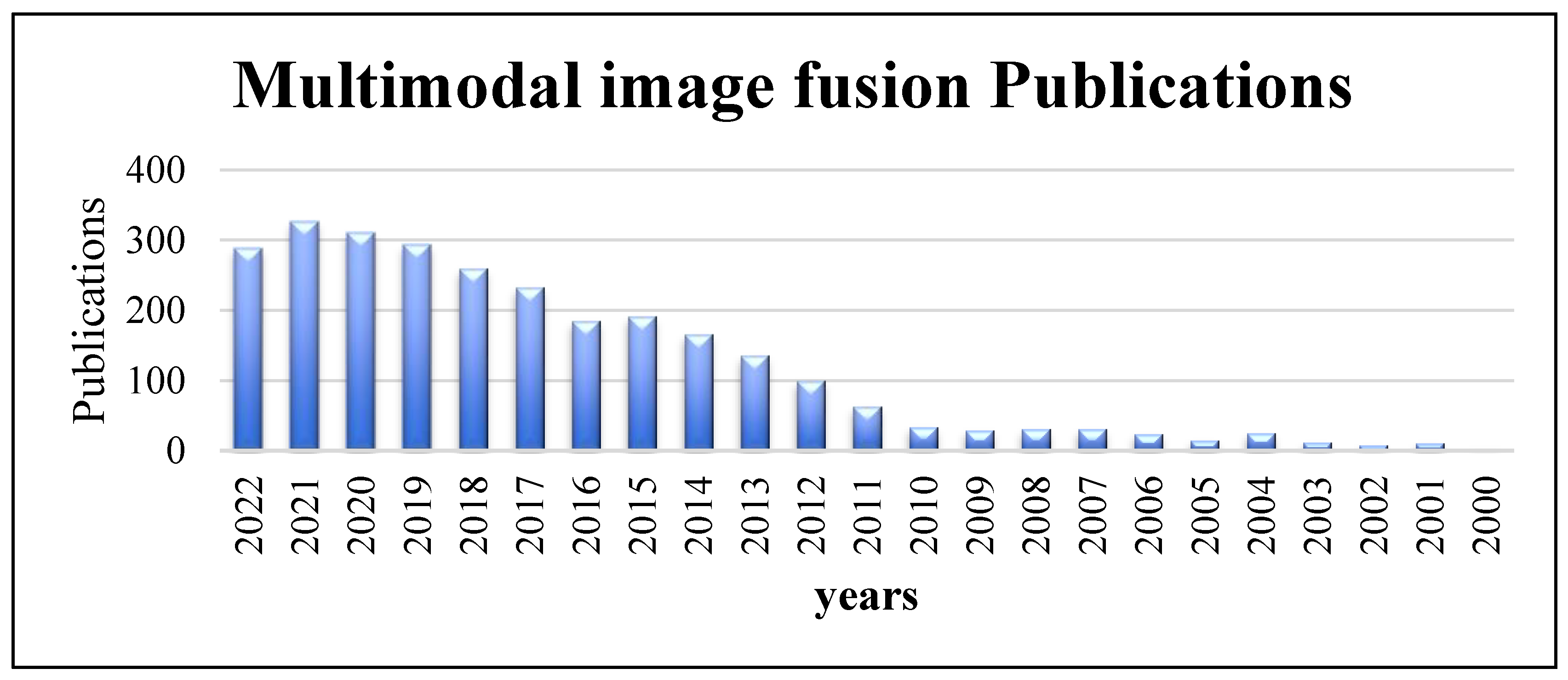
The many types of medical image modalities that exist led to the need to use images for improving medical therapy and treatment. MMIF is a medical imaging technique for combining data from different medical image modalities [3]. It is an effective approach for obtaining useful features and necessary information to understand accurately the status of human organs being diagnosed, such as the condition of the bones, metabolic rate, etc. The output of image fusion is a new image that is clearer for human and machine perception, which improves the accuracy and reliability of systems that rely on image fusion. It is generally utilized in illness diagnosis, treatment planning, and follow-up investigations. It aids in the accurate location and delineation of lesions. Because each medical imaging modality has its own strengths and limits, information from more than one imaging modality would be necessary for enhancing the diagnosis and treatment [4]. The increased number of scientific papers about multimodal image fusion shows the popularity and importance of MMIF. Figure 1 shows the number of publications in multimodal image fusion per year obtained from an online resource for biomedical subjects, PubMed, from 2000 to the third-quarter year of 2022 [5]. As can be seen from the column chart, there has been a huge number of fusion techniques in recent years that have been developed to meet the standards of MMIF [6].
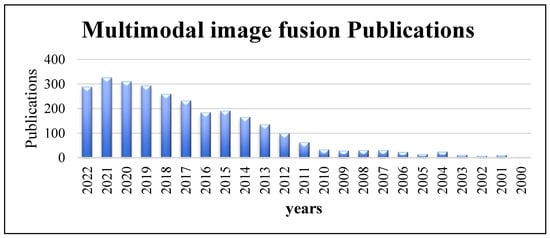
Figure 1.
Multimodal image fusion publications per year obtained from PubMed (2000 to third-quarter year, 2022).
Medical image fusion enhances the clinical readability of medical images for a better understanding of the image’s content in evaluation and diagnosis. In order to achieve that, the complementary information supplied in two or more images of different modalities is captured in the fusion result. The medical image fusion modalities can be categorized as Magnetic Resonance Imaging (MRI), Magnetic Resonance Angiogram (MRA), Positron Emission Tomography (PET), Structural Positron Emission Tomography (SPET), Computerized Tomography (CT), and Single Photon Emission Computed Tomography (SPECT). Researchers proposed many algorithms for combining data from several imaging modalities [7]. Each algorithm has advantages and disadvantages. The medical image fusion field is constantly evolving to meet the new challenges and demands that emerge. The fusion techniques utilize medical images of human organs of concern, such as the brain, breast, and lung [8].
Image fusion can be classified according to the dataset, such as multi-focus, multi-sensor, multi-scale, multi-spectral, and multi-temporal [9]. In multi-focus techniques for image fusion, several input images with different focuses are combined together into a single image that preserves all necessary information [10]. In multi-sensor techniques for image fusion, medical diagnosis applications combine information obtained from different sources to create a clearer image with all the necessary information. Actually, this information may not be visible to the visible human system [11]. The location of abnormalities can be accurately determined from the information extracted from merged images [8]. In multi-scale techniques for image fusion, this technique is suited to integrate more differently exposed Low Dynamic Range (LDR) images into a more informational LDR image. In multi-spectral techniques for image fusion, the corresponding information and spatiotemporal correlation are used to obtain an understandable image by merging the features in the spectrum domain [12]. In multi-temporal techniques for image fusion, the risk of information failure is reduced, and details of all clinical variables are obtained [13].
Image fusion techniques can be further divided into three levels: pixel-level, feature-level, and decision-level techniques to produce a fused image that is more informative for visual perception [14]. Pixel-level image fusion techniques are the simplest of all in which the original information from the source images or their multi-resolution transformations are combined. At this level, image fusion is executed at the lowest level. The feature-level image fusion techniques are the middle level between image fusion levels in which the important properties of the source image are extracted, such as length, form, segments, edges, and orientations. These properties are combined to produce more significant features which provide better descriptive and thorough images. Decision-level image fusion techniques, also called higher-level techniques, are used to identify real targets by combining the output from several techniques to obtain a final fusion judgment [10].
The traditional medical image fusion techniques can also be divided into the spatial domain, transform domain, and Hybrid transform. Many research papers focused on spatial domain medical image fusion [15]. Principal Component Analysis (PCA) and Intensity–Hue–Saturation (IHS) are two methodologies in spatial domain technology. Because of spectral and spatial distortion in the fused image by using the spatial domain, researchers use the transform domain (multi-scale-based transform) to improve fusion effects. The main process to fuse images in the transform domain is to transform the source image into the transform domain, performing the fusion based on transform coefficients, then applying an inverse transform to reconstruct the merged image. Discrete Wavelet Transforms (DWT), contour transformations, and pyramid transforms are examples of transform domains that are widely used in image fusion. The transform domain-based technique offers several advantages, such as clear structure and little amount of distortion, but the main disadvantage is that it always produces noise during the fusion process. Therefore, denoising is also a challenge for image fusion [16]. When reviewing the articles published in the last two years, it can be noticed that most of the proposed fusion techniques are not using spatial domain alone. Some Researchers combine the spatial and transform domains to perform fusion in the hybrid domain [17]. However, several modern approaches, such as PCA-DWT, integrates spatial domain and transform domain methodologies.
The performance evaluation of image fusion techniques is performed according to several qualitative and quantitative metrics. There are two types of evaluation approaches, namely, subjective and objective evaluation methods. The subjective method performs a visual examination to compare the source images with the final fused image. However, the subjective evaluation metrics are costly, troublesome, and time-consuming in several fusion applications [18]. The most prominent method is an objective evaluation method that tests the quality of the fused image compared with a reference image if it exists.
There are many challenges related to image fusion that will be discussed in subsequent sections. However, appropriate, accurate, and reliable image fusion techniques are required for the various types of images and for different types of problems. Furthermore, several transform domain fusion techniques may be combined at the decision level to obtain better results. Furthermore, image fusion techniques must be robust against uncontrollable acquisition conditions. They should also be inexpensive in terms of computation time in real-time systems. The problem of misregistration is a major error found while fusing images. The main objective of this paper is to summarize research progress in the field of medical image fusion and the future trends in this field. It focuses on multimodal medical image fusion, which combines several images of the same area of the human body using various imaging techniques.
The rest of the paper is organized as follows: Section 2 introduces a review of the medical imaging modalities. Section 3 presents the image fusion steps. Section 4 classifies MMIF levels. Section 5 classifies image fusion techniques. Section 6 provides the evaluation methods used to ensure the quality of fused images. Finally, Section 7 provides the conclusions.
2. Medical Imaging Modalities
This section presents the different medical imaging modalities. Each imaging modality has unique characteristics and properties which facilitate the study of specific human organs, illness, diagnosis, patient, and follow-up therapies. Microscopy, 3D reconstruction, visible photography light, radiology, and printed signals (waves) are some examples of imaging modalities [19]. The advancement of medical diagnosis is thanks to the recent progress in medical imaging capturing and enhancement. Medical imaging can be divided into structural systems and functional systems. All these types can be used to determine the location of the lesion. Functional and structural data from medical imaging can be combined to generate more meaningful information. In the treatment of the same human organ, medical image fusion is crucial since it enables more accurate disease monitoring and analysis. A brief overview of various medical imaging modalities is shown in Table 1.

Table 1.
A brief overview of various medical imaging modalities.
2.1. Structural Systems
Structural images such as MRI, CT, X-rays, and Ultrasound (US) provide high-resolution images with anatomical information. Specifically, CT can better distinguish tissues with different densities, such as bones and blood vessels. On the other hand, MRI can clearly show different soft tissues rather than bones.
2.1.1. Magnetic Resonance Image (MRI)
Magnetic field and radio transform signals are used in the medical imaging procedure, which is known as magnetic resonance imaging. It’s used to produce images of the nearest location to the illness, various organic functions, and anatomical structures. The essential feature of MRI is that it uses magnetic signals to create slices that mimic the human body and provides details on diseased soft tissues.
2.1.2. Computed Tomography (CT)
CT is one of the main modalities included for making image fusion. A thin cross-segment can be seen using the CT approach, which is based on detecting X-ray weakening. CT is one of the major non-invasive diagnostic methods in contemporary medicine. The advantages of CT images are they are introduced with fewer warps and provide details concerning dense construction, such as bones and show a higher ability to see little differences in tissue construction.
2.1.3. X-rays
X-rays were utilized to create “shadow grams” of the human body. Radiography is the use of X-rays to visualize inside organs. Today, radiation is not typically recorded, but its force is measured and then converted to an image. As a result, the image’s more subdued features become more obvious. The main task of X-rays is to identify bone anomalies and fractures in the human body. Mammography is a breast cancer assessment technique that uses X-rays as its primary imaging source. Ultrasound-X rays and Vibroacoustography with X-ray mammography are two examples of current modalities that combine X-ray information with some of them.
2.1.4. Ultrasound (US)
Ultrasound imaging relies on low-recurrence vibrations induced in the body. Because of the radiation power of Vibroacoustography (VA), a different imaging technique made possible by ultrasound-animated acoustic emission is used in conjunction with mammography to improve the detection of breast cancer. In order to study retrieved human tissues such as the liver, breast, prostate, and arteries, ultrasound imaging is being tested as a non-invasive imaging tool. By determining changes in the mechanical response to vibration in an excited state, the depth and thickness of the objects are not recorded by X-ray mammography. However, tissue thickness does not interfere with ultrasound imaging. Applications for ultrasound images include improving mass lesions and breast development analysis. More auxiliary and demonstrative data may be observed in ultrasound and X-ray mammography by combining images from two distinct imaging modalities, ultrasound and X-ray, using either pixel-based or color-based fusion techniques.
2.2. Functional Systems
Functional images such as PET and SPECT provide functional information with low spatial resolution.
2.2.1. Positron Emission Tomography (PET)
Low-recurrence vibrations are necessary for ultrasound imaging. A crucial component of atomic drug imaging is PET. It’s a non-invasive imaging technique that provides a representation and evaluation of a preselected tracer’s digestion. The essential characteristics of PET images are that they provide important information about the human brain, allow for the recording of changes in the solid cerebrum’s movement, and provide signs of various disorders. For imaging any area of the body, such as for whole-body cancer diagnoses, PET has emerged as one of the most frequently utilized clinical technologies. The PET image’s exceptional sensitivity is the greatest benefit. PET-CT, MRI-PET, MRI-CT-PET, and MRI-SPECTPET are some of the contemporary modalities that use image fusion techniques that integrate PET information in fusion.
2.2.2. Single Photon Emission Computed Tomography (SPECT)
Gamma rays are commonly utilized in the atomic pharmaceutical tomographic imaging technique known as SPECT to evaluate blood flow to tissues and organs. The internal organs’ functionality is examined using SPECT scan. It provides actual 3D data. Slices through the body are frequently used to display this data. One of the most popular scans for tissues outside the brain where tissue placement is very varied is the SPECT scan. SPECT-CT and MR-SPECT are two examples of modalities that use SPECT information infusion in conjunction with some of the existing image fusion techniques.
3. Fusion Steps
The multimodal image fusion methods attempt to increase quality and accuracy without changing the complementary information of the images by integrating many images from one or various imaging modalities. Several medical image modalities exist, such as MRI, CT, PET, US, and SPECT. MRI, US, and CT modalities deliver images with anatomical information about the body with high spatial resolution. PET and SPECT provide images with functional information about the body, such as blood flow and movement of soft tissue, although having low spatial resolution. The functional image will be merged with the structural image to yield a multimodal image containing better information for health specialists to diagnose diseases.
Image fusion is the process that aims to produce a more representative image by merging the input images with each other. Two images are geometrically aligned using medical image registration, and then an image fusion technique is used to combine the two input source images to create a new image with additional and complementary information. During the image fusion process, two requirements must be satisfied: (1) the fused image must have all relevant medical information that was present in the input images, and (2) it must not contain any additional information that was not present in the input images. Fusion can be applied to multimodal images widely used in medicine, multi-focus images that are usually obtained from the same modality, and multi-sensor images taken from various sources of imaging modalities. The multimodal fusion Steps and process, as well as the MMIF approaches and their different parts [20,21], are described in this section.
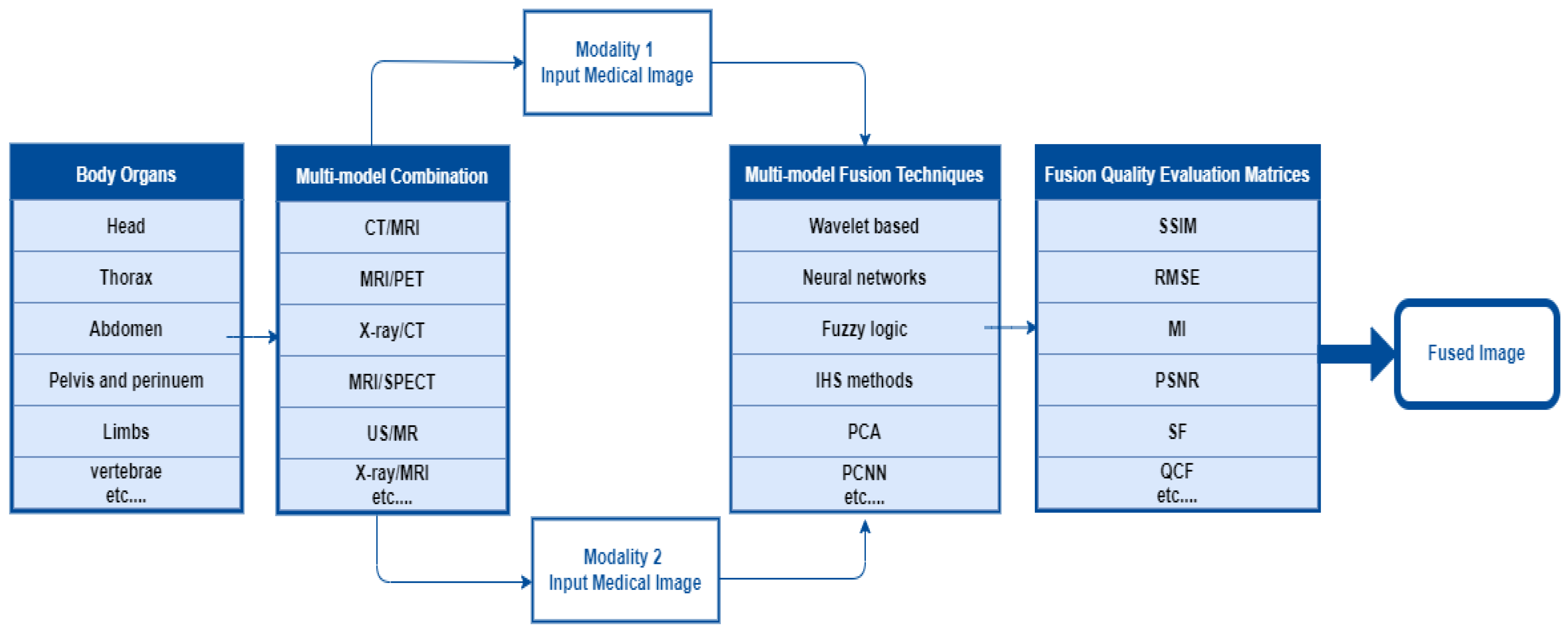
In the multimodal fusion process, first, the researcher determines the body organ of interest. Second, select two or more imaging modalities to perform image fusion using the appropriate fusion algorithm. It requires performance metrics to validate the fusion algorithm. In the final step, the fused image contains more information than the input images about the scanned area of the body organ. The overall MMIF procedure is shown in Figure 2. These steps are mostly used in the spatial domain.
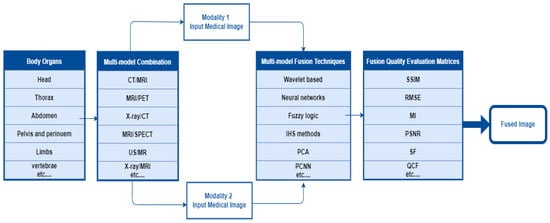
Figure 2.
The Overall procedure of multimodal medical image fusion.
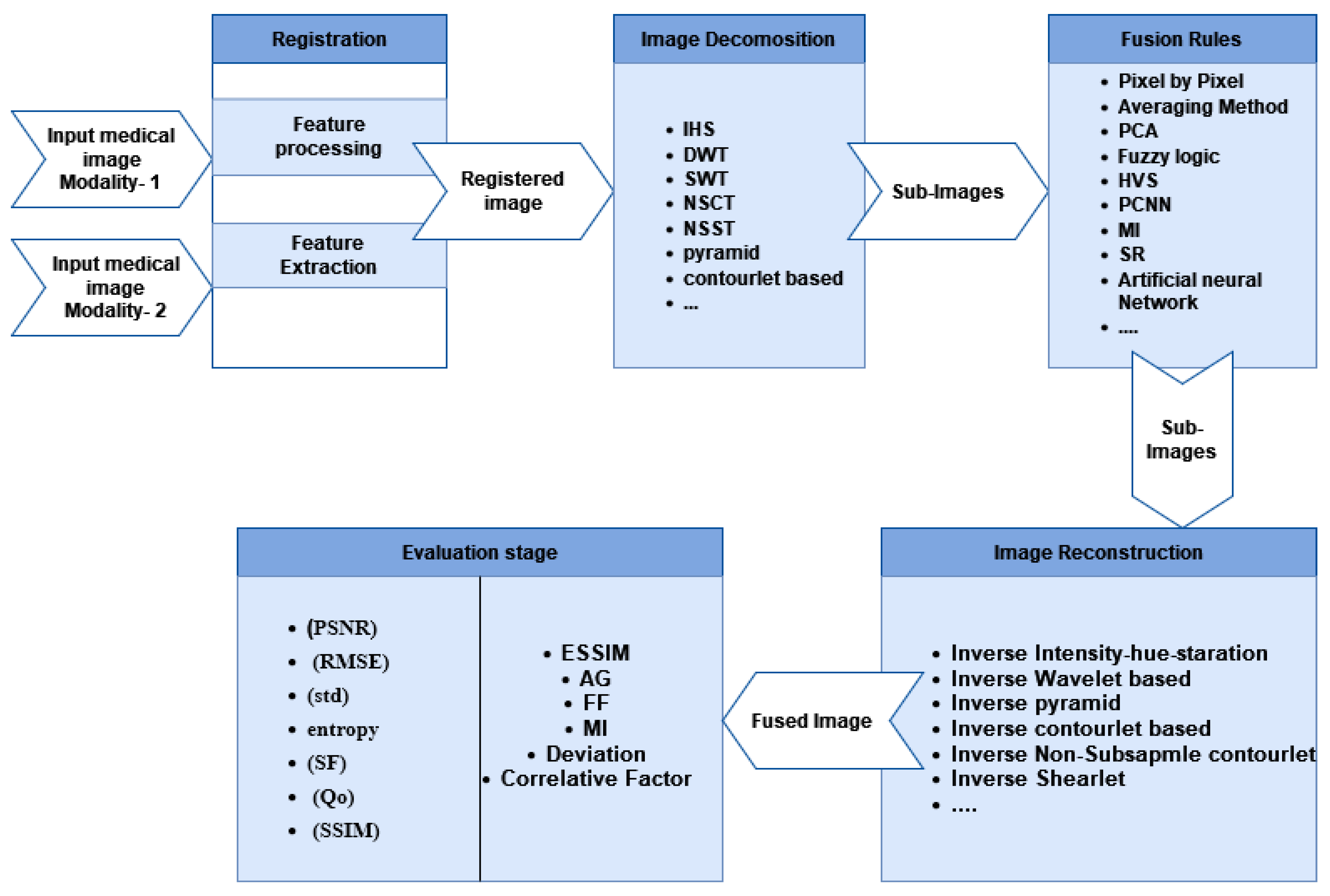
In the transform domain method, medical image fusion step-by-step is illustrated in Figure 3. There are five steps as follows: (1) Image Registration: the input source image is registered by mapping it with the reference image to match the corresponding images, (2) Image Decomposition: the source images are initially divided into smaller images using decomposition algorithms, and fusion algorithms such as Intensity–Hue–Saturation (IHS), pyramid, distinctive wavelet, Non-Subsampled Contourlet Transform (NSCT), shearlet transform, Sparse Representation (SR), and others are applied to merge multiple features from these sub-images [22]. (3) Fusion Rules: fusion algorithms such as fuzzy logic, Human Visual System (HVS) fusion, Artificial Neural Networks (ANNs), Principal Component Analysis (PCA), Mutual Information (MI) fusion, and Pulse-Coupled Neural Networks (PCNNs) are used to extract critical information and several features from sub-images which helpful in the following processes, (4) Image Reconstruction: in this step, the fused image is reconstructed. Image construction is the process of assembling the sub-images using an inverse algorithm, and (5) Image Quality Evaluation Methods: the image quality assessment is the last step in evaluating the quality of the fusion result by using both subjective and objective assessment measures. The radiologists are asked to assess the fusion outcome subjectively [23].
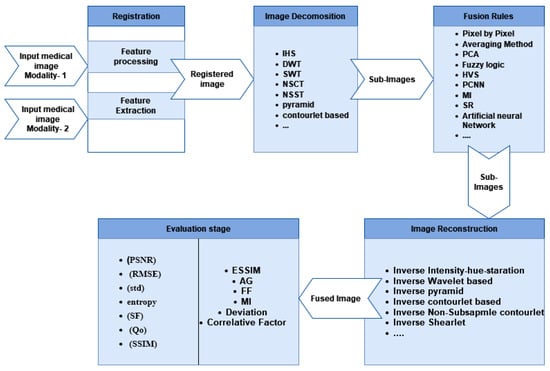
Figure 3.
Medical image fusion step-by-step process.
4. MMIF Levels
To help with understanding classification, we start by giving an overview of pixel-level, feature-level, and decision-level image fusion. The three levels of image fusion are pixel-level, feature-level, and decision-level techniques. In order to produce a final image that is more informative for visual perception, pixel-level image fusion involves directly combining the original information from the source images or their multi-resolution transformations. The goal of feature-level fusion is to extract important properties such as form, length, edges, segments, and orientations from the source Image. The characteristics that were taken from the input photographs are combined to create more significant features, which provide better descriptive and thorough images. A high level of fusion that identifies the real target is called decision-level fusion. It combines the output from many algorithms to obtain a final fusion judgment.
4.1. Pixel-Level Fusion
In pixel-level-based fusion techniques, images are effectively combined using individual pixels to determine the fusion decision [24]. It is further divided into a spatial domain and a transform domain [25].
4.2. Feature-Level Fusion
At the feature level, fusion use segmentation techniques to extract objects of interest for several image modalities. Then, factual approaches are used to combine comparison components (such as areas) from various modalities of images.
Fei et al. [26] proposed MMIF based on a decision map and sparse representation to simultaneously deal with these problems. The raw photos are first segmented into patches, and then the patches are sorted into vectors based on where they were in the original images. In the second step, a decision map is created. The third step is to choose the vector from each group using the decision map. After that, the sparse representation method is utilized to fuse the other vector pairs. Finally, the approach then uses the decision map to create the fusion result, and the average is determined for the overlapping patches.
4.3. Decision-Level Fusion (Dictionary)
The interpreted/labeled data are used in decision-level fusion, where the input images are treated separately. Selective principles are linked to consolidate the data, enhance fundamental translation, and provide a better understanding of the seen items. The main advantage of this approach is that multi-modality fusion is strengthened and made more reliable according to the higher-level representations. There are three approaches that are usually combined at the decision level to produce fused images. These three approaches are information theory, logical reasoning, and statistical approaches. Joint measures, Bayesian fusion techniques, hybrid consensus methods, voting, and fuzzy decision rules are some illustrations of the three approaches. In order to produce the single fused image in decision-level fusion, each input image is selected using predefined criteria and then fuses, depending on the validity of each conclusion, into the global optimum. In order to generate the most information possible, a pre-established principles strategy is used. The most often utilized strategies at the decision fusion level are Bayesian techniques and dictionary learning [18]. The Bayesian strategy relies on the Bayes hypothesis, which is based on probabilities for merging data from numerous sensors. Nonparametric Bayesian, HWT Bayesian, and DWT Swarm Optimized are examples of Bayesian approaches.
5. Image Fusion Techniques
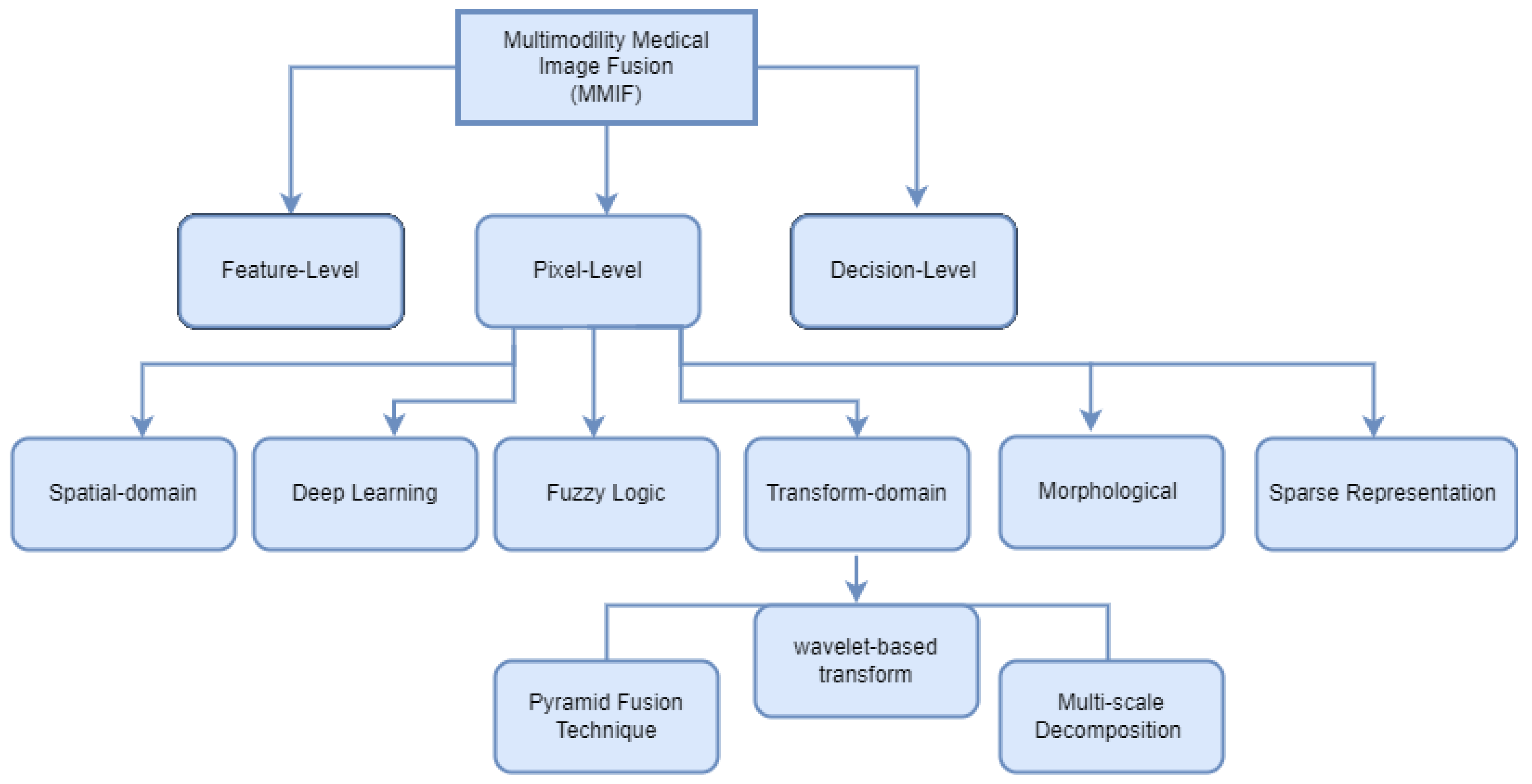
Image fusion combines images from many modalities to produce results that are more accurate, comprehensive, and may be more easily interpreted. Combining multimodal images has several benefits, such as precise geometric adjustments, completing data for improved categorization, improving characteristics for investigations, and so forth. In several fields of study, including computer vision, multimedia analysis, medicinal research, and material sciences, image fusion is used extensively, and its effectiveness has been demonstrated. Registration is viewed as an optimization problem that is utilized to take advantage of similarities while also lowering costs. Image registration is a technique for aligning the subsequent aspects of multiple images regarding a reference image. Multiple source images are utilized for registration in which the original image serves as a reference image, and the original images are aligned using the reference image. The important features of registered images are extracted in feature extraction to generate various feature maps. Although there are many image fusion methods, this study focused on six models: spatial domain, transform domain, fuzzy logic-based methods, morphological methods, sparse representation fusion, and deep learning Fusion Methods, as shown in Figure 4.
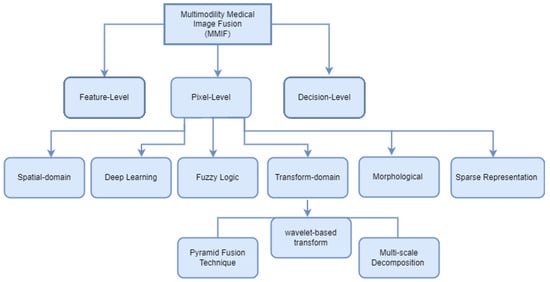
Figure 4.
Multimodal Medical Image Fusion techniques classification.
5.1. Spatial Domain Fusion Techniques
Early research has focused heavily on spatial domain-based medical image fusion techniques. In this type, the fusion rules are applied directly to pixels of source images, i.e., the actual values of pixels in the source images are combined to produce the fused image.
Spatial domain approaches work on the pixel level of the image, which is a fundamental pixel-level strategy. The generated images have less spatial distortion and a lower signal-to-noise ratio (SNR) since it is applied to the original images. It works by manipulating the values of an image’s pixels to achieve the desired effects. PCA, IHS, Brovey, High Pass Filtering Techniques, ICA, Simple Maximum, Simple Average, Weighted Average, and gradient filtering are some spatial domain methods. However, spatial domain techniques produce spectral and spatial distortion in the final fused image, which is viewed as a detriment to the fusion process.
Changtao et al. [27] proposed a methodology that enhances the quality of the fused images by combining the benefits of his and PCA fusion techniques. They compare their proposed approach with other fusion methods such as PCA, Brovey, and discrete wavelet transform (DWT). Visual and quantitative analysis shows that their suggested algorithm greatly enhances the fusion quality. Bashir et al. [20] introduced a PCA and Stationary Wavelet Transform (SWT) based model that was evaluated using a range of medical images. Results show that PCA appears to give better performance when the input images have different contrast and brightness levels in multimodal fusion. SWT appears to perform better when the input images are multimodal and multi-sensor images. There are five different sets of images: X-ray, MRI, CT, satellite, and stereo images. In order to determine whether the approach performs better for the type of imagery, several evaluation performance matrices were used for this set of images. Depoian et al. [28] proposed a unique approach to obtaining better image fusion by combining PCA with a neural network. In comparison to conventional weighted fusion approaches, the employment of an auto-encoder neural network that is used to combine this information leads to better degree results in data visualization. Rehal et al. [29] suggested a new method that used a hybrid fast Fourier transform to extract features and intensity from a group of images. They used the gray wolf optimization technique in the hue saturation to obtain the ideal result. The proposed methodology shows better results than the conventional one that was employed in previous research according to testing of the method.
5.2. Transform Fusion Techniques (Multi-Scale Decomposition Methods)
Recently, due to the spectral and spatial distortion in the fused image by using the spatial domain, many researchers have turned their research focus to the study of the transform domain for a better fusion effect. The transform domain approach has transformed the input images using a Fourier transform to obtain the low-frequency coefficient and high-frequency coefficient. The transform quantities are then subjected to the fusion process, which is then followed by an inverse transformation to generate the fused image within the final form. Transform domain techniques of image fusion are highly effective in handling spatial distortion. However, it is difficult to extend its one-dimensional property to two dimensions or several dimensions. A highly helpful technique for evaluating images from remote sensing, medical imaging, etc., is multi-resolution analysis. Discrete wavelet transform is now becoming a key technique for fusion. The transform domain is based on the multi-scale-based transform. The multi-scale-based transform fusion method is classified into three steps: decomposition, fusion, and reconstruction.
The techniques used in the transform domain are divided into pyramidal fusion techniques, wavelet fusion techniques, and multi-scale decomposition techniques. Firstly, the most common methods used in pyramidal fusion techniques are the Laplacian pyramid and the morphological pyramid, Ratio Pyramids, and Gaussian Pyramids [30]. Liu et al. [31] proposed an approach that combined Laplacian Pyramid (LP) and Convolutional Sparse Representation (CSR). Each set of pre-registered computed tomography images and magnetic resonance images performs the LP transform to produce its detail layers and base layers. The base layer is then combined using a CSR-based method, and the detail layers are combined using the well-known “max absolute” criteria. By applying the inverse LP transform to the combined base layer and detail layers, the fused image is finally rebuilt. Their approach has the advantages of being able to properly extract the texture detail information from the source images and maintaining the overall contrast of the combined image. Experimental results show how superior their suggested approach is. Zhu et al. [32] proposed a novel multi-modality medical image fusion method based on phase congruency and local Laplacian energy. On pairs of medical image sources, the non-subsampled contourlet transform is used to separate the source images into high-pass and low-pass sub-bands. A phase congruency-based fusion rule that integrates the high-pass sub-bands can improve the detailed characteristics of the fused image for medical diagnosis. Local Laplacian energy-based fusion rule is suggested for low-pass sub-bands. The weighted local energy and the weighted sum of the Laplacian coefficients reflect the structured information and specific characteristics of source image pairings, respectively, and make up the local Laplacian energy. As a result, the proposed fusion rule can incorporate two crucial elements for the fusion of low-pass sub-bands at the same time. In order to create the fused image, the combined high-pass and low-pass sub-bands are inversely converted. Three kinds of multi-modality medical image pairings are employed in the comparison tests to test the efficacy of the suggested approach. The outcomes of the experiment demonstrate that the suggested approach performs competitively in terms of both image quantity and processing expenses.
Secondly, wavelet fusion techniques include the following type Discrete Wavelet Transform (DWT), Stationary Wavelet Transform (SWT), Redundant Wavelet Transform (RWT) Dual-Tree Complex Wavelet Transforms (DT CWT). Most published research on multimodal medical image fusion algorithms depends on DWT [33]. DWT creates various input frequency signals to keep stable output perfect rank in the time domain and frequency domain which leads to maintaining the specific information of the image [34]. It has a good visual and quantitative fusion effect and overcomes PCA’s drawbacks. MRI and PET image fusion is used in DWT-based fusion methods and also used in others [35,36,37]. Bhavana et al. [35] proposed a novel fusion method using MRI and PET brain images on DWT without any reducing anatomical information and with less color distortion. A novel fusion method of DWT is different from the usual DWT fusion method, where the proposed method applied wavelet decomposition with four stages for low- and high-activity regions, respectively. Cheng et al. [37] produce a good fusion image of PET/CT, which discover and locates the disease region in the pancreatic gland using wavelet transform by a weighted fusion-based medical image algorithm. Georgieva et al. [38] introduce a brief overview of the benefits and current directions of multidimensional wavelet transform-based medical image processing techniques. They also introduce how it might be combined with other approaches that treat each coefficient set as a tensor. The use of computers in different medical processing methods, including noise reduction, segmentation, classification, and medical image fusion is made simpler by the fact that wavelet tensor modifications have only one parameter to be tweaked, as opposed to the standard wavelet decompositions. Due to the uniqueness of medical imaging data, these approaches have not yet been widely applied in research and practical applications despite their advantages. The latest recommendations in this area concern the selection of suitable techniques and algorithms to enhance their advantages in accordance with the uniqueness of various medical items in the images.
Wang et al. [39] Presented a multimodal color medical image fusion algorithm based on the discrete cosine transform in geometric algebra (GA DCT). The GADCT algorithm combines the characteristics of GA, which represents the multi-vector signal as a whole, to increase the quality of the fusion image and reduce the number of complex operations associated with encoding and decoding.
Finally, multi-scale decomposition techniques include Non-Subsampled Contourlet Transform (NSCT), Non-Subsampled Shearlet Transform (NSST), and Pulse-Coupled Neural Network (PCNN) techniques. The following sections will focus on the most common techniques used in Multi-scale Decomposition techniques.
5.2.1. Non-Subsampled Contourlet Transform (NSCT)
Do et al. [40] created the contourlet transform with the intention of capturing the inherent geometrical features of an image and enhancing the isotropic wavelet property. The decomposition consists of two filtering steps because it is a directional multi-resolution transform. First, the Laplacian pyramid is applied to selected point discontinuities. Second, performing a local directional decomposition through a directional filter bank to create a linear structure linking these discontinuities. A contourlet transform is a useful tool for selecting intrinsic contours rather than curvelets or ridgelets because of the set of basis oriented along various scales and orientations.
Li et al. [41] created a unique fusion technique for multi-modality medical images by combining it with NSCT. They use an improved innovative sum-modified Laplacian (INSML) feature in which the complementary information of multi-modality images is retrieved and utilized in the fusion rules for the low-transform NSCT coefficients. Additionally, the WLE-INSML features are used to extract the high-transform NSCT coefficients and build the fusion rules for these coefficients. They assess their suggested fusion approach by using an open dataset generated from twelve pairs of multi-modality medical images.
Li et al. [42] proposed an MMIF method that combines the advantages of the NSCT. They proposed fuzzy entropy to improve the quality of target recognition and the accuracy of fused images and provide the basis for clinical diagnosis. The image is divided into high- and low-frequency subbands through NSCT. According to the different features of the high- and low-transform components, the fusion rules are adopted. It’s necessary to calculate the membership degree of low-frequency coefficients. In order to guide the fusion of coefficients that preserve image details, fuzzy entropy is calculated and subsequently used. Maximizing regional energy is used to fuse high-frequency components. The transformation is inversed to obtain the final fused image. Based on the subjective and objective assessment criteria, experimental results show that their approach provides a satisfactory fusion effect. Additionally, this technique may successfully preserve the features of the fused image while obtaining excellent average gradient, standard deviation, and edge preservation. The outcomes of the suggested methodology can serve as an efficient guide for clinicians to evaluate patient conditions.
Alseelawi et al. [43] proposed an appeached hybrid medical image fusion using wavelet and curvelet transform with multi-resolution processing. They proposed an algorithm for enhancing the fused image quality by combining wavelet and curvelet transform techniques after the decomposition stage. They use a sub-band coding algorithm instead of curvelet fusion and the wavelet transform fusion algorithm, which leads to this technique being more efficient.
Xia et al. [44] proposed a medical image fusion technique by combining a pulse-coupling neural network and sparse representation. First, the NSCT transform is used to break down the source image into low and high-frequency sub-band coefficients. Second, the low-frequency sub-band coefficients are trained using the K Singular Value Decomposition (K-SVD) method to obtain the over-complete dictionary D. It is sparsed using the Orthogonal Matching Pursuit (OMP) algorithm to complete the fusion of the low-frequency sub-band sparse coefficients. The spatial transform of the high transform sub-band coefficients is then used to excite the Pulse Coupling Neural Network (PCNN), and the fusion coefficients of the high transform sub-band coefficients are chosen based on the number of ignition times. Finally, the NSCT inverter reconstructs the fusion medical image. The experimental results and analysis shows that the performance of the fusion result is better than the existing algorithms, and the algorithm of gray and color image fusion is higher than the contrast algorithm in the edge information transfer factor QAB/F index.
5.2.2. Pulse-Coupled Neural Network (PCNN)
The purpose of image fusion is not only to keep the characteristic information of the source image but also to ensure less distortion and good visual effects; therefore, the high-frequency coefficient is decomposed using PCNN. PCNN is a single-layer two-dimensional connected neuron array horizontally, which is used widely in the image processing field [45]. It’s a biologically inspired feedback neural network in which the neurons consist of a connected modulation field, receiving field, and a pulse generator. It has a more significant advantage in the biological background which can be used to obtain useful information from source images without a training process. It has many defects, such as difficulty in setting parameters.
Ouerghi et al. [46] proposed a new fusion method based on a simplified pulse-coupled neural network (S-PCNN) and NSST. First, PET images are converted into YIQ components. The NSST transform is only applied for Y components of PET images and MRI images. The standard deviation of the weight region and the local energy is used to fuse the low-frequency sub-band. The high-frequency coefficients are fused using S-PCNN. This algorithm achieves good quality in the fused image.
Duan et al. [47] proposed an MMIF framework based on PCNN and low-rank representation of image blocks. The NSCT is used to decompose the image. The low-frequency sub-band adopts the low-rank fusion strategy based on the K-SVD dictionary learning algorithm, which leads to strengthening the extraction of local features. The high-frequency sub-band adopts the PCNN fusion. Finally, the guided filter to deepen the edge details is used to fuse the image after NSCT inverse transform with MRI gray image. The proposed fusion method achieves great results over other existing algorithms in objective metrics and visual effects.
5.2.3. Non-Subsampled Shearlet Transform (NSST)
NSCT complication in computation shearlet is a new tool that can obtain mathematical properties and geometric, for example, scales, elongated shapes, oscillations, and directionality from images. The shearlet is optimally sparse in displaying images with edges because they form a tight frame in a variety of scales and directions. The decomposition of an image shearlet transforms such as that of a contourlet, but there is no restriction in directions for shearing. In the inverse ST, to improve the computational efficiency, the shearing filters need only to be aggregated instead of inverting a directional filter bank in the contourlet. Qiu et al. [48] proposed an image fusion method that transformed both CT and MR images into the NSST domain to obtain low and high-frequency components. They use the absolute-maximum rule to merge high-frequency components and use a sparse representation-based approach to merge the low-frequency components. To improve the performance of the sparse representation-based approach, they propose a dynamic group sparsity recovery algorithm. Finally, they performed the inverse NSST on the merged component to obtain the fused image. Their approach provides better fusion results in terms of objective and subjective quality evaluation.
Yin et al. [49] proposed an innovative multimodal medical image fusion method in NSST. They perform NSST decomposition on the source images to obtain their multidirectional and multi-scale representations. They use a Parameter-Adaptive Pulse-Coupled Neural Network (PA-PCNN) model to fuse high-frequency bands in which all parameters of PCNN are estimated by the input band. They use a new strategy, namely, energy preservation and detail extraction, that addresses two crucial issues in medical image fusion simultaneously. Finally, inverse NSST is performed on the fused low-frequency and high-frequency bands to reconstruct the fused image. In order to test the viability of the suggested approach, extensive tests were carried out employing 83 pairings of source photos across four categories of medical image fusion difficulties. The experimental results show that the suggested method can achieve state-of-the-art performance in terms of both visual perception and objective assessment.
5.3. Fuzzy-Logic-Based Methods
In 1965, Zadeh [50] was the first to establish the Fuzzy Logic (FL) theory, and since then, MMIF algorithms have made heavy use of it. Medical images have indistinct regions as a result of inadequate lighting. In light of this, the Fuzzy Sets Theory (FST) is used in medical image processing. The idea of FST has made tremendous progress in overcoming uncertainty. FL model consists of a fuzzifier, an inference engine, a de-fuzzifier, fuzzy sets, and fuzzy rules [51]. FL is used for image fusion as both a feature transform operator and a decision operator [52].
Kumar et al. [53] fused the medical images by implementing intuitionistic fuzzy logic-based image fusion. They repress the noise and enhance the input images, and integrate them into the IHS domain efficiently. Fuzzy sets are integrated to overcome the uncertainties caused by them due to the vagueness and ambiguity of the intuitionistic.
Tirupal et al. [54] proposed a new method, namely, Sugeno’s Intuitionistic Fuzzy Set, to fuse medical images. First, the medical images are converted into SIFI images. Second, the SIFIs split the images into blocks to calculate the count of whiteness and blackness of the blocks. Finally, the fused image is rebuilt from the recombination of SIFI image blocks.
Tirupal et al. [55] introduced a technique based on an interval-valued intuitionistic fuzzy set (IVIFS) for effectively fusing multimodal medical images, with a median filter used to eliminate noise from the final fused image. Several sets of multimodal medical images are simulated and compared to the available fusion techniques, such as an intuitionistic fuzzy set and fuzzy transform.
5.4. Morphological Methods
In the early eighties, most multimodal medical image fusion algorithms used mathematical morphology broadly, which determined its objects as a set of points and operations between two sets [56]. The structuring and the objects element are observed when using filters created with morphological operators. Extracting features from a subset of spatially localized pixels has been consistently successful [8]. The image opening and image closing filters used in the morphological pyramid decomposition were found to be ineffective for edge detection. The mathematical morphology algorithm has retained important image regions and details with increased calculation time [57].
Yang et al. [58] present a recent algorithm for medical image fusion by CT and MRI images with a shift-invariant multi-scale decomposition scheme. By eliminating the downsampling operators from a morphological wavelet, the decomposition scheme is produced. An experiment using an actual medical image demonstrates how much the suggested strategy enhances the quality of the fused image. The proposed method outperforms competing approaches in terms of maintaining both “pixel” and “edge” information.
5.5. Sparse Representation Methods
In recent years, Sparse Representation (SR) methods applied in image fusion applications have become a prevalent and important research point among the research community. It attracted significant attention and performed successfully, so the transform domain image fusion algorithms that combine sparse representation techniques have been used [59]. Wang et al. [60] proposed an image fusion framework that combines images by integrating NSCT with SR, which resulted in enhanced fusion than the fusion algorithms of single transformation. However, the processing time of the proposed framework was longer than the multi-scale transform-based methods. Li et al. [61] proposed an image fusion framework that integrates SR with NSCT, which achieves an improved fusion with respect to detail preservation and structural similarity for the visible-infrared images. Maqsood et al. [62] proposed a multimodal image fusion scheme based on two-scale medical image decomposition merged with SR which the edge details in CT-MRI image fusion are improved, but it cannot use with color images. Chen et al. [63] proposed a target-enhanced multi-scale decomposition fusion technique for infrared and visible image fusion in which the texture details in visible images are preserved, and the thermal target in infrared images is enhanced.
Shabanzade et al. [64] present an image fusion framework for image modalities (i.e., PET and MRI) based on sparse representation in the NSCT domain. Source images were obtained from their low-pass and high-pass sub-bands by performing NSCT. Then, low-pass sub-bands are fused by sparse representation based by using clustering-based dictionary learning. While high-pass sub-bands are merged by application of the salience match measure rule.
Kim et al. [65] proposed an efficient dictionary learning for the multimodal image fusion method based on joint patch clustering. They build an over-complete dictionary to represent a fused image with a sufficient number of useful atoms. Different sensor modalities transfer the image information and structural similarities, and all patches from different source images are clustered together. The joint patch clusters are collected and integrated to build the over-complete dictionary to structure an informative dictionary. Finally, sparse coefficients are evaluated in multimodal images with the common dictionary learned.
Polinati et al. [66] presented a unique approach, convolutional sparse image decomposition (CSID), that combines CT and MR images. To locate edges in source images, CSID employs contrast stretching and the spatial gradient approach, as well as cartoon-texture decomposition, which produces an over-complete dictionary. In addition, they introduced a modified convolutional sparse coding approach and used enhanced decision maps and the fusion rule to create the final fused image.
5.6. Deep Learning Fusion Methods
In recent years, deep learning has been a new research field in medical image fusion. Compared with medical image fusion, it’s widely used in medical image registration [67,68,69] and medical image segmentation [70,71,72]. It uses a number of layers, and each layer takes its information from the previous layer. It helps to structure the complicated framework architecturally layered and has the capability to handle enormous amounts of data [73]. Convolutional Neural networks (CNN), Convolution Sparse Representation (CSR), and Deep Convolution Neural Networks (DCCNs) are examples of deep learning fusion approaches. The CNN model is most often used in deep learning approaches. Each layer in CNN is composed of a number of feature maps that contain neurons as coefficients. The feature maps are connected to each stage in the numerous stages using various methods such as spatial pooling, convolution, and non-linear activation [74]. Convolutional Sparse Coding (CSC) is another popular deep-learning fusion technique.
Liu et al. [75] proposed a convolutional neural network-based technique for fusing medical images. They used a Siamese convolutional network to create a weight map that integrates the pixel activity information from source images. They perform a fusion process that is more consistent with human visual perception by conducting it in a multi-scale manner via image pyramids. Some well-known image fusion techniques, such as multi-scale processing and adaptive fusion mode selection, are appropriately used to provide perceptually pleasing outcomes. The method can produce high-quality outcomes in terms of visual quality and objective metrics according to experimental results.
Rajalingam et al. [76] introduced an effective multimodal medical image fusion method based on deep learning convolutional neural networks. They use CT, MRI, and PET as the input multi-modality medical images for the experimental work. They use a Siamese convolutional network to produce a weight map that incorporates the pixel movement information from two or more multi-modality medical images. To make the medical image fusion process more accurate with human visual insight, they carried out the procedure of the medical image fusion in a multi-scale manner via medical image pyramids. To correct the fusion mode for the decomposed coefficients, they apply a local comparison-based strategy. An experimental result shows that the suggested method outperforms alternative methods currently used in terms of processing performance and outcomes for both subjective and objective evaluation criteria.
Xia et al. [77] proposed a novel muti-modal medical image fusion schema that uses both the features of deep convolutional neural network-based and multi-scale transformation. Firstly, the Gauss–Laplace and Gaussian filters divide the source images into several sub-images in the first layer of the network. Then, the convolution kernel of the rest layers is initialized, and the basic unit is constructed using the HeK-based method. The basic unit is trained using a backpropagation algorithm. To create a deep stacked neural network, train a number of fundamental units that are sacked with the idea of SAE. The suggested network is used to decompose the input images to obtain their own low-frequency and high-frequency images, merge the fusion rule to fuse low-frequency and high-frequency images, and then return those merged images to the network’s final layer to produce the final fusion images. By doing numerous experiments on various medical image datasets, the effectiveness of their suggested fusion method is assessed. Experimental results show that, in comparison to other approaches now in use, their proposed method not only successfully fuses the numerous images to provide superior results but also ensures an improvement in the many quantitative parameters. Furthermore, the revised CNN approach runs significantly more quickly than similar algorithms with high fusion quality.
Wang et al. [78] introduced medical image fusion algorithms that can combine medical images from many morphologies to improve the accuracy and reliability of a clinical diagnosis, which plays a greatly important role in many clinical applications. This research suggests a CNN-based medical image fusion algorithm to produce a fused image with good visual quality and distinct structural information. The suggested approach combines the pixel activity data from the source images with the trained Siamese convolutional network to produce the weight map. Meanwhile, the source image is decomposed using a contrast pyramid. Source images are combined using various spatial transform bands and a weighted fusion operator. Comparative experiments’ results demonstrated that the suggested fusion method might successfully retain the source images’ intricate structural details while producing pleasing aesthetic effects for humans.
Wang et al. [79] introduced a new MMIF algorithm based on CNN and NSCT. To obtain better fusion results, they exploit the advantages of both NSCT and CNN. In the proposed algorithm, the source images are divided into high and low-frequency sub-bands. They use a new fusion rule, namely, Perceptual High-Frequency CNN (PHF-CNN), which produces high-frequency sub-bands. In the case of the low-frequency sub-band, the decision map is generated by adopting two result maps. Finally, they inverse NSCT to integrate fused frequency sub-bands. According to experimental results, the suggested approach is superior to existing algorithms in terms of assessment, and it increases the quality of fused images.
Li et al. [80] examine the most recent developments in DL image fusion and suggest some directions for future research in the area. Deep learning models can automatically extract the most useful characteristics from data to get around the challenge of manual design and integration of multimodal medical images. This approach can successfully complete image fusion in batches, fulfill diagnostic demands, and significantly improve the effectiveness of medical image fusion. It makes sense in terms of enhancing the precision of medical diagnosis. All these techniques mentioned above are illustrated and summarized in Table 2 and Table 3 to highlight the advantages and disadvantages of these techniques.

Table 2.
MMIF technique classification and major contributions in research papers.

Table 3.
Advantages and disadvantages of MMIF techniques.
6. Evaluation Metrics
Each fusion method has advantages, and the effectiveness of image fusion algorithms is assessed by considering several parameter measures documented in the literature [81,82]. There are two fusion quality evaluation metrics that can be categorized as subjective/qualitative and objective/quantitative evaluation methods.
The subjective quality assessment compares the original input images with the final fused image based on visual examination. There are various parameters, such as color, spatial details, image size, etc., must be considered in the examination of the fused image. Nevertheless, the absence of ground truth images that are completely fused causes these quality assessment methods to be costly, inconvenient, and consume time.
The objective method is categorized based on two methods. The first method is used when the reference image is available. The second method is used when the reference image is not available. The validation of the fusion algorithm used the ground truth image as the reference image. The ground truth medical image is only available in very rare circumstances, or it can be constructed manually. In the case the ground truth image is not available, the resultant fused image and the source medical images are used to calculate the quality metric. Objective quality assessment parameters of the reference image include Peak Signal to Noise Ratio (PSNR), Root Mean Square Error (RMSE), Structural Similarity (SSIM), Mutual Information (MI), Universal Quality Index (UQI), and Correlation Coefficient (CC). Objective quality assessment parameters without a reference image include Standard Deviation (SD), Entropy (EN), Spatial Frequency (SF), and Gradient-Based Index (QAB/F). The objective quality evaluation results are arranged in Table 3. The objective methods are categorized based on whether a reference image is available or not. The following subsections show the calculations involved for each quantitative metric. quantitative metric. Table 4 presents a detailed comparison between different MMIF techniques using quantitative metrics.

Table 4.
Comparison of performance evaluation metric.
6.1. Metrics Requiring a Reference Image
6.1.1. Root Mean Square Error Ratio (RMSE)
By comparing the actual or ideal fused medical image to the ground truth image, RMSE determines the final medical image’s quality. Its value ought to be close to zero for optimal merged image results. i and j stand for horizontal and vertical pixels, respectively, where R stands for input and F stands for fused images. The RMSE is calculated as follows:
6.1.2. Mutual Information (MI)
The similarities between the two images are determined by their mutual information. It should have a high value for better fusion. Where F is the combined image and A and B are the two input images. MI can be easily calculated using Equation (2).
6.1.3. Structural Similarity Index Measure (SSIM)
The structural similarity between a fused image and a source image is calculated using the SSIM metric. Its value ranges from 0 to 1, where 0 signifying a complete lack of similarity to the source image and 1 signifying an exact match. The fusion effect is better when the SSIM value is higher since it indicates how similar the fused image is to the source image. SSIM can be calculated using Equations (3) and (4).
where the average values of source images are represented by and the average value of the fused image is represented by . The variances of the source image and the fused image are represented by the symbols and . And reflect the combined variance of the two source images, respectively.
6.1.4. Correlation Coefficient (CC)
The correlation between the reference image and the fused image is shown by the correlation coefficient. It reflects spectrum information, and values should be near +1. The correlation coefficient between the source and the combined image is abbreviated as C_rf. When the reference and the fused images are perfectly identical, the ideal value is one; as the similarity decreases, it becomes less than one. The formula for calculating CC is as follows:
6.1.5. Universal Quality Index (UQI)
The Universal Quality Index is utilized to create the fused image, which is used to calculate how much salient information is present in the reference image. The reference and fused image must be identical for this measure to achieve its optimum value of 1, which ranges from −1 to 1. Information transformation from two images is denoted by x and y, where μ stands for average and σ for variation. The formula for calculating UQI is as follows:
6.1.6. Peak Signal to Noise Ratio (PSNR)
Peak Signal to Noise Ratio is frequently employed as a measure of the effectiveness of reconstruction in image fusion. It is the ratio between the highest value of an image and the magnitude of background noise. When the reference and fused images are similar, the PSNR value is high. Better fusion is suggested by a higher value. The letters I and max stand for the original image and maximum pixel grey level, respectively. MSE stands for mean square error. I and J are separate photos that have been combined. The formula for calculating PSNR is as follows:
6.2. Metrics Requiring a Reference Image
6.2.1. Standard Deviation (SD)
Standard Deviation refers to a statistic used to assess the contrast in the fused image. The fused image exhibits significant contrast when the standard deviation value is high. The formula for calculating SD is as follows:
6.2.2. Entropy (EN)
The fused image with average information content is measured using entropy. The fused image has a high level of information richness, as shown by the high entropy value. Bits per pixel are used to quantify entropy. Where is the probability corresponding to grey level i. The formula for calculating EN is as follows:
6.2.3. Spatial Frequency (SF)
The metric that reflects distinct contrasts and surface changes and measures the overall amount of activity in a fused image is known as spatial frequency. The fused image is better when the value of SF is high. M and N are the image width and height. The formula for calculating SF is as follows:
6.2.4. Gradient-Based Index (QAB/F)
QAB/F measures the amount of edge data that are conveyed from the input source images to the fused image. The optimum value of 1 is achieved for this measure which has a range of 0 to 1, and then all the edges of the source images are transmitted to the fused image. The loss of all edge information is represented by a value of 0. Where the edge information storage is value; is the weighting map. The formula for calculating Q^(AB/F) is as follows:
7. Conclusions
MMIF technique is widely used to enhance the visual properties of the output image for more effective therapy and accurate diagnosis. There is a huge number of research papers that propose different techniques for image fusion. This paper presents a comprehensive study on MMIF, which includes: (1) a classification of medical modalities used in MMIF; (2) an illustration of procedures included in MMIF; (3) an explanation of the different levels of MMIF: pixel level fusion, feature level fusion, and decision level fusion; (4) comparison between different domains for MMIF techniques: spatial fusion, transform domain such as pyramidal fusion techniques, wavelet fusion techniques, and multi-scale decomposition techniques (NSCT, NSST, PNCC), fuzzy logic method, morphological methods, sparse representation methods, and deep learning-based methods; (5) illustration of the evaluation metrics including subjective quality assessment and objective quality assessment which is further divided into objective quality assessment parameters with a reference image, and objective quality assessment parameters without a reference image; (6) a comparison between image quality assessment metrics of different existing techniques.
Also, a detailed comparison between recently proposed MMIF techniques is presented, along with numeric values obtained for different metrics. Several objective metrics are reported to assess the quality of the fused image. It is challenging to judge completely if one technique is better in all aspects. Each technique has its own advantages and disadvantages. However, our findings; based on the detailed performed experiments; showed that spatial image fusion techniques are computationally fast and simple, but they are not efficient in terms of quality. The fused image is not satisfied and contains many spectral deteriorations. However, frequency domain techniques reduce spectral distortion and produce a higher signal-to-noise ratio, which makes them better than the spatial fusion techniques in terms of the obtained quality. For better performance, the researchers always use a hybrid between spatial domain techniques and frequency domain techniques. Other well-known image fusion techniques are sparse representation-based techniques, in which the coefficients are the most essential parameters that improve the fusion results by enhancing the image’s contrast while keeping the visual information and preserving the structure of source images. However, the sparse representation technique has some drawbacks, such as misregistration and minimal detail preservation capacity. Recently, deep learning-based techniques have been widely used; to greatly enhance the quality of the fused image by using CNNs. Deep learning approaches work better than other fusion techniques when there is a large amount of input data with several dimensions and variety. These methods rely on a dynamic process with numerous parameters to train a fusion model. However, these methods require more time for training and special GPUs to work correctly, which makes them computationally expensive compared with other techniques. Another drawback of deep learning techniques is their inability to deliver accurate outcomes for smaller image datasets.
Author Contributions
Conceptualization, M.A.S., K.A. and A.A.A.; methodology, M.A.S., K.A. and A.A.A.; validation, M.A.S. and K.A.; formal analysis, A.M.S.; investigation, A.M.S.; resources, K.A. and A.A.A.; writing—original draft preparation, K.A and A.M.S.; writing—review and editing, M.A.S. and A.A.A.; visualization, K.A. and A.M.S.; supervision, A.A.A., M.A.S. and K.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Blum, R.S.; Xue, Z.; Zhang, Z. An Overview of lmage Fusion. In Multi-Sensor Image Fusion and Its Applications; CRC Press: Boca Raton, FL, USA, 2018; pp. 1–36. [Google Scholar]
- Vajgl, M.; Perfilieva, I.; Hod’áková, P. Advanced f-transform-based image fusion. Adv. Fuzzy Syst. 2012, 2012, 4. [Google Scholar] [CrossRef]
- Tawfik, N.; Elnemr, H.A.; Fakhr, M.; Dessouky, M.I.; El-Samie, A.; Fathi, E. Survey study of multimodality medical image fusion methods. Multimed. Tools Appl. 2021, 80, 6369–6396. [Google Scholar] [CrossRef]
- Ganasala, P.; Kumar, V. Feature-motivated simplified adaptive PCNN-based medical image fusion algorithm in NSST domain. J. Digit. Imaging 2016, 29, 73–85. [Google Scholar] [CrossRef]
- PubMed. Available online: https://www.ncbi.nlm.nih.gov/pubmed/ (accessed on 22 December 2022).
- Venkatrao, P.H.; Damodar, S.S. HWFusion: Holoentropy and SP-Whale optimisation-based fusion model for magnetic resonance imaging multimodal image fusion. IET Image Process. 2018, 12, 572–581. [Google Scholar] [CrossRef]
- Kaur, M.; Singh, D. Multi-modality medical image fusion technique using multi-objective differential evolution based deep neural networks. J. Ambient Intell. Humaniz. Comput. 2021, 12, 2483–2493. [Google Scholar] [CrossRef]
- James, A.P.; Dasarathy, B.V. Medical image fusion: A survey of the state of the art. Inf. Fusion 2014, 19, 4–19. [Google Scholar] [CrossRef]
- Bhat, S.; Koundal, D. Multi-focus image fusion techniques: A survey. Artif. Intell. Rev. 2021, 54, 5735–5787. [Google Scholar] [CrossRef]
- Zhou, Y.; Yu, L.; Zhi, C.; Huang, C.; Wang, S.; Zhu, M.; Ke, Z.; Gao, Z.; Zhang, Y.; Fu, S. A Survey of Multi-Focus Image Fusion Methods. Appl. Sci. 2022, 12, 6281. [Google Scholar] [CrossRef]
- Li, B.; Xian, Y.; Zhang, D.; Su, J.; Hu, X.; Guo, W. Multi-sensor image fusion: A survey of the state of the art. J. Comput. Commun. 2021, 9, 73–108. [Google Scholar] [CrossRef]
- Bai, L.; Xu, C.; Wang, C. A review of fusion methods of multi-spectral image. Optik 2015, 126, 4804–4807. [Google Scholar] [CrossRef]
- Madkour, M.; Benhaddou, D.; Tao, C. Temporal data representation, normalization, extraction, and reasoning: A review from clinical domain. Comput. Methods Programs Biomed. 2016, 128, 52–68. [Google Scholar] [CrossRef] [PubMed]
- Ghassemian, H. A review of remote sensing image fusion methods. Inf. Fusion 2016, 32, 75–89. [Google Scholar] [CrossRef]
- Dinh, P.-H. Multi-modal medical image fusion based on equilibrium optimizer algorithm and local energy functions. Appl. Intell. 2021, 51, 8416–8431. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, H.; Tian, X.; Jiang, J.; Ma, J. Image fusion meets deep learning: A survey and perspective. Inf. Fusion 2021, 76, 323–336. [Google Scholar] [CrossRef]
- Jose, J.; Gautam, N.; Tiwari, M.; Tiwari, T.; Suresh, A.; Sundararaj, V.; Rejeesh, M. An image quality enhancement scheme employing adolescent identity search algorithm in the NSST domain for multimodal medical image fusion. Biomed. Signal Process. Control 2021, 66, 102480. [Google Scholar] [CrossRef]
- Meher, B.; Agrawal, S.; Panda, R.; Abraham, A. A survey on region based image fusion methods. Inf. Fusion 2019, 48, 119–132. [Google Scholar] [CrossRef]
- MITA. Available online: https://www.medicalimaging.org/about-mita/modalities (accessed on 22 December 2022).
- Bashir, R.; Junejo, R.; Qadri, N.N.; Fleury, M.; Qadri, M.Y. SWT and PCA image fusion methods for multi-modal imagery. Multimed. Tools Appl. 2019, 78, 1235–1263. [Google Scholar] [CrossRef]
- Hermessi, H.; Mourali, O.; Zagrouba, E. Multimodal medical image fusion review: Theoretical background and recent advances. Signal Process. 2021, 183, 108036. [Google Scholar] [CrossRef]
- Dinh, P.-H. A novel approach based on three-scale image decomposition and marine predators algorithm for multi-modal medical image fusion. Biomed. Signal Process. Control 2021, 67, 102536. [Google Scholar] [CrossRef]
- Chang, L.; Ma, W.; Jin, Y.; Xu, L. An image decomposition fusion method for medical images. Math. Probl. Eng. 2020, 2020, 4513183. [Google Scholar] [CrossRef]
- Daniel, E. Optimum wavelet-based homomorphic medical image fusion using hybrid genetic–grey wolf optimization algorithm. IEEE Sens. J. 2018, 18, 6804–6811. [Google Scholar] [CrossRef]
- Li, S.; Kang, X.; Fang, L.; Hu, J.; Yin, H. Pixel-level image fusion: A survey of the state of the art. Inf. Fusion 2017, 33, 100–112. [Google Scholar] [CrossRef]
- Fei, Y.; Wei, G.; Zongxi, S. Medical image fusion based on feature extraction and sparse representation. Int. J. Biomed. Imaging 2017, 2017, 3020461. [Google Scholar] [CrossRef]
- He, C.; Liu, Q.; Li, H.; Wang, H. Multimodal medical image fusion based on IHS and PCA. Procedia Eng. 2010, 7, 280–285. [Google Scholar] [CrossRef]
- Depoian, A.C.; Jaques, L.E.; Xie, D.; Bailey, C.P.; Guturu, P. Neural network image fusion with PCA preprocessing. In Proceedings of the Big Data III: Learning, Analytics, and Applications, Online Event, 12–16 April 2021; pp. 132–147. [Google Scholar]
- Rehal, M.; Goyal, A. Multimodal Image Fusion based on Hybrid of Hilbert Transform and Intensity Hue Saturation using Fuzzy System. Int. J. Comput. Appl. 2021, 975, 8887. [Google Scholar] [CrossRef]
- Azam, M.A.; Khan, K.B.; Ahmad, M.; Mazzara, M. Multimodal medical image registration and fusion for quality Enhancement. CMC-Comput. Mater. Contin 2021, 68, 821–840. [Google Scholar] [CrossRef]
- Liu, F.; Chen, L.; Lu, L.; Ahmad, A.; Jeon, G.; Yang, X. Medical image fusion method by using Laplacian pyramid and convolutional sparse representation. Concurr. Comput. Pract. Exp. 2020, 32, e5632. [Google Scholar] [CrossRef]
- Zhu, Z.; Zheng, M.; Qi, G.; Wang, D.; Xiang, Y. A phase congruency and local Laplacian energy based multi-modality medical image fusion method in NSCT domain. IEEE Access 2019, 7, 20811–20824. [Google Scholar] [CrossRef]
- Kavitha, C.; Chellamuthu, C.; Rajesh, R. Medical image fusion using combined discrete wavelet and ripplet transforms. Procedia Eng. 2012, 38, 813–820. [Google Scholar] [CrossRef]
- Osadchiy, A.; Kamenev, A.; Saharov, V.; Chernyi, S. Signal processing algorithm based on discrete wavelet transform. Designs 2021, 5, 41. [Google Scholar] [CrossRef]
- Bhavana, V.; Krishnappa, H. Multi-modality medical image fusion using discrete wavelet transform. Procedia Comput. Sci. 2015, 70, 625–631. [Google Scholar] [CrossRef]
- Jaffery, Z.A.; Zaheeruddin; Singh, L. Computerised segmentation of suspicious lesions in the digital mammograms. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2017, 5, 77–86. [Google Scholar] [CrossRef]
- Cheng, S.; He, J.; Lv, Z. Medical image of PET/CT weighted fusion based on wavelet transform. In Proceedings of the 2008 2nd International Conference on Bioinformatics and Biomedical Engineering, Shanghai, China, 16–18 May 2008; pp. 2523–2525. [Google Scholar]
- Georgieva, V.; Petrov, P.; Zlatareva, D. Medical image processing based on multidimensional wavelet transforms-Advantages and trends. In Proceedings of the AIP Conference Proceedings, Sofia, Bulgaria, 7–3 June 2022; p. 020001. [Google Scholar]
- Wang, R.; Fang, N.; He, Y.; Li, Y.; Cao, W.; Wang, H. Multi-modal Medical Image Fusion Based on Geometric Algebra Discrete Cosine Transform. Adv. Appl. Clifford Algebr. 2022, 32, 1–23. [Google Scholar] [CrossRef]
- Do, M.N.; Vetterli, M. The contourlet transform: An efficient directional multiresolution image representation. IEEE Trans. Image Process. 2005, 14, 2091–2106. [Google Scholar] [CrossRef]
- Li, B.; Peng, H.; Wang, J. A novel fusion method based on dynamic threshold neural P systems and nonsubsampled contourlet transform for multi-modality medical images. Signal Process. 2021, 178, 107793. [Google Scholar] [CrossRef]
- Li, W.; Lin, Q.; Wang, K.; Cai, K. Improving medical image fusion method using fuzzy entropy and nonsubsampling contourlet transform. Int. J. Imaging Syst. Technol. 2021, 31, 204–214. [Google Scholar] [CrossRef]
- Alseelawi, N.; Hazim, H.T.; Salim ALRikabi, H.T. A Novel Method of Multimodal Medical Image Fusion Based on Hybrid Approach of NSCT and DTCWT. Int. J. Online Biomed. Eng. 2022, 18, 28011. [Google Scholar] [CrossRef]
- Xia, J.; Chen, Y.; Chen, A.; Chen, Y. Medical image fusion based on sparse representation and PCNN in NSCT domain. Comput. Math. Methods Med. 2018, 2018, 2806047. [Google Scholar] [CrossRef]
- Xiong, Y.; Wu, Y.; Wang, Y.; Wang, Y. A medical image fusion method based on SIST and adaptive PCNN. In Proceedings of the 2017 29th Chinese Control and Decision Conference (CCDC), Chongqing, China, 28–30 May 2017; pp. 5189–5194. [Google Scholar]
- Ouerghi, H.; Mourali, O.; Zagrouba, E. Non-subsampled shearlet transform based MRI and PET brain image fusion using simplified pulse coupled neural network and weight local features in YIQ colour space. IET Image Process. 2018, 12, 1873–1880. [Google Scholar] [CrossRef]
- Duan, Y.; He, K.; Xu, D. Medical Image Fusion Technology Based on Low-Rank Representation of Image Blocks and Pulse Coupled Neural Network. In Proceedings of the 2022 7th International Conference on Image, Vision and Computing (ICIVC), Xi’an, China, 26–28 June 2022; pp. 473–479. [Google Scholar]
- Qiu, C.; Wang, Y.; Zhang, H.; Xia, S. Image fusion of CT and MR with sparse representation in NSST domain. Comput. Math. Methods Med. 2017, 2017, 9308745. [Google Scholar] [CrossRef]
- Yin, M.; Liu, X.; Liu, Y.; Chen, X. Medical image fusion with parameter-adaptive pulse coupled neural network in nonsubsampled shearlet transform domain. IEEE Trans. Instrum. Meas. 2018, 68, 49–64. [Google Scholar] [CrossRef]
- Zadeh, L.A. Fuzzy sets. Inf. Control 1965, 8, 338–353. [Google Scholar] [CrossRef]
- Biswas, B.; Sen, B.K. Medical image fusion technique based on type-2 near fuzzy set. In Proceedings of the 2015 IEEE International Conference on Research in Computational Intelligence and Communication Networks (ICRCICN), Kolkata, India, 20–22 November 2015; pp. 102–107. [Google Scholar]
- Das, A.; Bhattacharya, M. Evolutionary algorithm based automated medical image fusion technique: Comparative study with fuzzy fusion approach. In Proceedings of the 2009 World Congress on Nature & Biologically Inspired Computing (NaBIC), Coimbatore, India, 9–11 December 2009; pp. 269–274. [Google Scholar]
- Kumar, M.; Kaur, A.; Amita. Improved image fusion of colored and grayscale medical images based on intuitionistic fuzzy sets. Fuzzy Inf. Eng. 2018, 10, 295–306. [Google Scholar] [CrossRef]
- Tirupal, T.; Mohan, B.C.; Kumar, S.S. Multimodal medical image fusion based on Sugeno’s intuitionistic fuzzy sets. ETRI J. 2017, 39, 173–180. [Google Scholar] [CrossRef]
- Tirupal, T.; Chandra Mohan, B.; Srinivas Kumar, S. Multimodal medical image fusion based on interval-valued intuitionistic fuzzy sets. In Machines, Mechanism and Robotics; Springer: Berlin/Heidelberg, Germany, 2022; pp. 965–971. [Google Scholar]
- Soille, P. Morphological Image Analysis: Principles and Applications; Springer: Berlin/Heidelberg, Germany, 1999; Volume 2. [Google Scholar]
- Bai, X. Morphological image fusion using the extracted image regions and details based on multi-scale top-hat transform and toggle contrast operator. Digit. Signal Process. 2013, 23, 542–554. [Google Scholar] [CrossRef]
- Yang, B.; Jing, Z. Medical image fusion with a shift-invariant morphological wavelet. In Proceedings of the 2008 IEEE Conference on Cybernetics and Intelligent Systems, Chengdu, China, 21–24 September 2008; pp. 175–178. [Google Scholar]
- Zhu, Z.; Chai, Y.; Yin, H.; Li, Y.; Liu, Z. A novel dictionary learning approach for multi-modality medical image fusion. Neurocomputing 2016, 214, 471–482. [Google Scholar] [CrossRef]
- Wang, J.; Peng, J.; Feng, X.; He, G.; Wu, J.; Yan, K. Image fusion with nonsubsampled contourlet transform and sparse representation. J. Electron. Imaging 2013, 22, 043019. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Y.; Huang, X.; Qi, G.; Zheng, M.; Zhu, Z. An image fusion method based on sparse representation and sum modified-Laplacian in NSCT domain. Entropy 2018, 20, 522. [Google Scholar] [CrossRef]
- Maqsood, S.; Javed, U. Multi-modal medical image fusion based on two-scale image decomposition and sparse representation. Biomed. Signal Process. Control 2020, 57, 101810. [Google Scholar] [CrossRef]
- Chen, J.; Li, X.; Luo, L.; Mei, X.; Ma, J. Infrared and visible image fusion based on target-enhanced multiscale transform decomposition. Inf. Sci. 2020, 508, 64–78. [Google Scholar] [CrossRef]
- Shabanzade, F.; Ghassemian, H. Multimodal image fusion via sparse representation and clustering-based dictionary learning algorithm in nonsubsampled contourlet domain. In Proceedings of the 2016 8th International Symposium on Telecommunications (IST), Tehran, Iran, 27–28 September 2016; pp. 472–477. [Google Scholar]
- Kim, M.; Han, D.K.; Ko, H. Joint patch clustering-based dictionary learning for multimodal image fusion. Inf. Fusion 2016, 27, 198–214. [Google Scholar] [CrossRef]
- Polinati, S.; Bavirisetti, D.P.; Rajesh, K.N.; Naik, G.R.; Dhuli, R. The Fusion of MRI and CT Medical Images Using Variational Mode Decomposition. Appl. Sci. 2021, 11, 10975. [Google Scholar] [CrossRef]
- Balakrishnan, G.; Zhao, A.; Sabuncu, M.R.; Guttag, J.; Dalca, A.V. VoxelMorph: A learning framework for deformable medical image registration. IEEE Trans. Med. Imaging 2019, 38, 1788–1800. [Google Scholar] [CrossRef]
- Hu, Y.; Modat, M.; Gibson, E.; Li, W.; Ghavami, N.; Bonmati, E.; Wang, G.; Bandula, S.; Moore, C.M.; Emberton, M. Weakly-supervised convolutional neural networks for multimodal image registration. Med. Image Anal. 2018, 49, 1–13. [Google Scholar] [CrossRef]
- Yang, X.; Kwitt, R.; Styner, M.; Niethammer, M. Quicksilver: Fast predictive image registration–a deep learning approach. NeuroImage 2017, 158, 378–396. [Google Scholar] [CrossRef] [PubMed]
- Çiçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning dense volumetric segmentation from sparse annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Ronneberger, O. Invited talk: U-net convolutional networks for biomedical image segmentation. In Bildverarbeitung für die Medizin 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 234–241. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Zhou, T.; Ruan, S.; Canu, S. A review: Deep learning for medical image segmentation using multi-modality fusion. Array 2019, 3, 100004. [Google Scholar] [CrossRef]
- Nguyen, K.P.; Fatt, C.C.; Treacher, A.; Mellema, C.; Trivedi, M.H.; Montillo, A. Anatomically informed data augmentation for functional MRI with applications to deep learning. In Proceedings of the Medical Imaging 2020: Image Processing, Houston, TX, USA, 15–20 February 2020; pp. 172–177. [Google Scholar]
- Liu, Y.; Chen, X.; Cheng, J.; Peng, H. A medical image fusion method based on convolutional neural networks. In Proceedings of the 2017 20th International Conference on Information Fusion (Fusion), Xi’an, China, 10–13 July 2017; pp. 1–7. [Google Scholar]
- Rajalingam, B.; Priya, R. Multimodal medical image fusion based on deep learning neural network for clinical treatment analysis. Int. J. ChemTech Res. 2018, 11, 160–176. [Google Scholar]
- Xia, K.-j.; Yin, H.-s.; Wang, J.-q. A novel improved deep convolutional neural network model for medical image fusion. Clust. Comput. 2019, 22, 1515–1527. [Google Scholar] [CrossRef]
- Wang, K.; Zheng, M.; Wei, H.; Qi, G.; Li, Y. Multi-modality medical image fusion using convolutional neural network and contrast pyramid. Sensors 2020, 20, 2169. [Google Scholar] [CrossRef]
- Wang, Z.; Li, X.; Duan, H.; Su, Y.; Zhang, X.; Guan, X. Medical image fusion based on convolutional neural networks and non-subsampled contourlet transform. Expert Syst. Appl. 2021, 171, 114574. [Google Scholar] [CrossRef]
- Li, Y.; Zhao, J.; Lv, Z.; Li, J. Medical image fusion method by deep learning. Int. J. Cogn. Comput. Eng. 2021, 2, 21–29. [Google Scholar] [CrossRef]
- Eskicioglu, A.M.; Fisher, P.S. Image quality measures and their performance. IEEE Trans. Commun. 1995, 43, 2959–2965. [Google Scholar] [CrossRef]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).