The Role of rs713041 Glutathione Peroxidase 4 (GPX4) Single Nucleotide Polymorphism on Disease Susceptibility in Humans: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Search Strategy
2.3. Inclusion and Exclusion Criteria
2.4. Data Extraction
2.5. Quality Assessment
2.6. Data Analysis
3. Results and Discussion
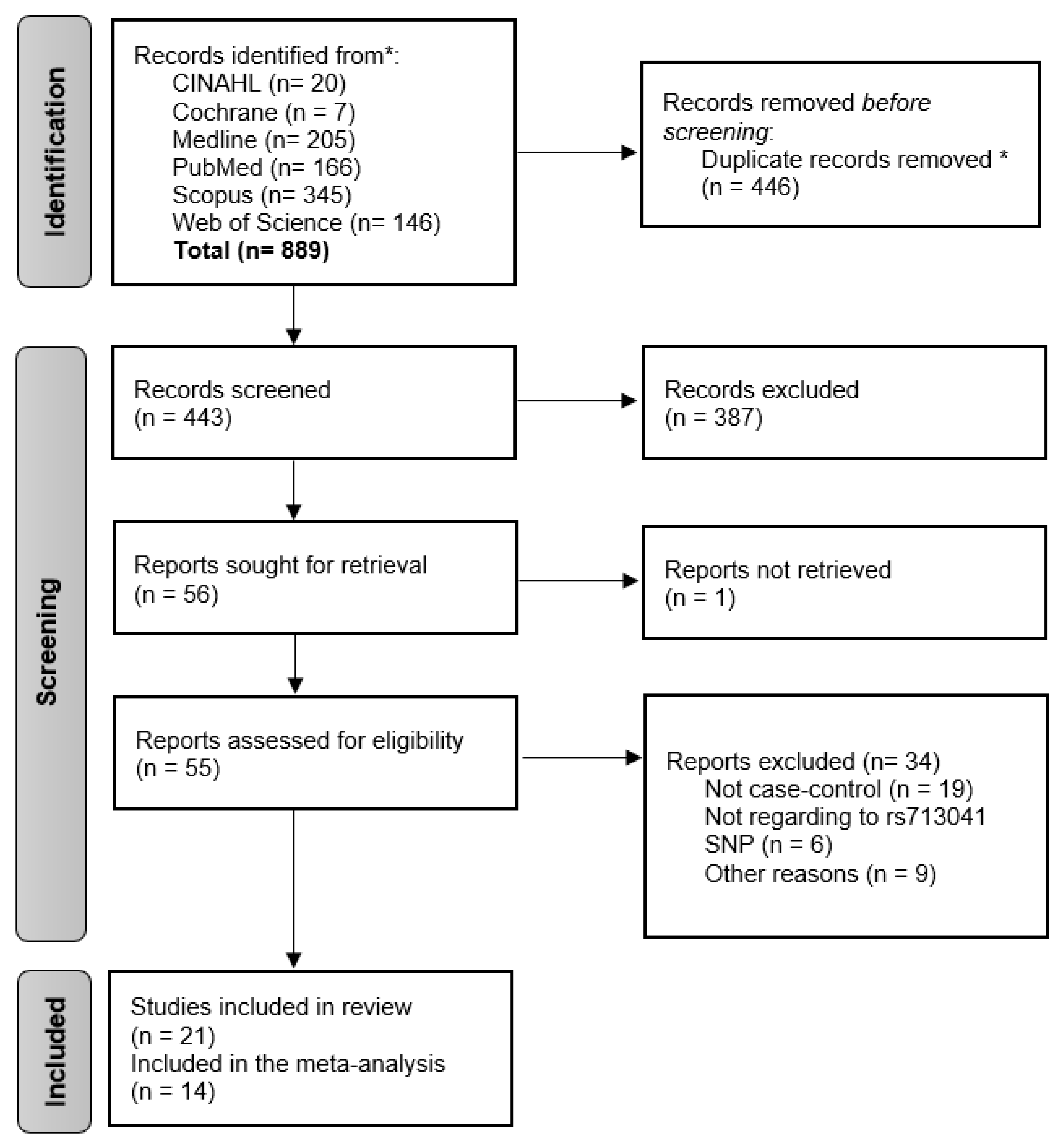
3.1. Description of Included Studies
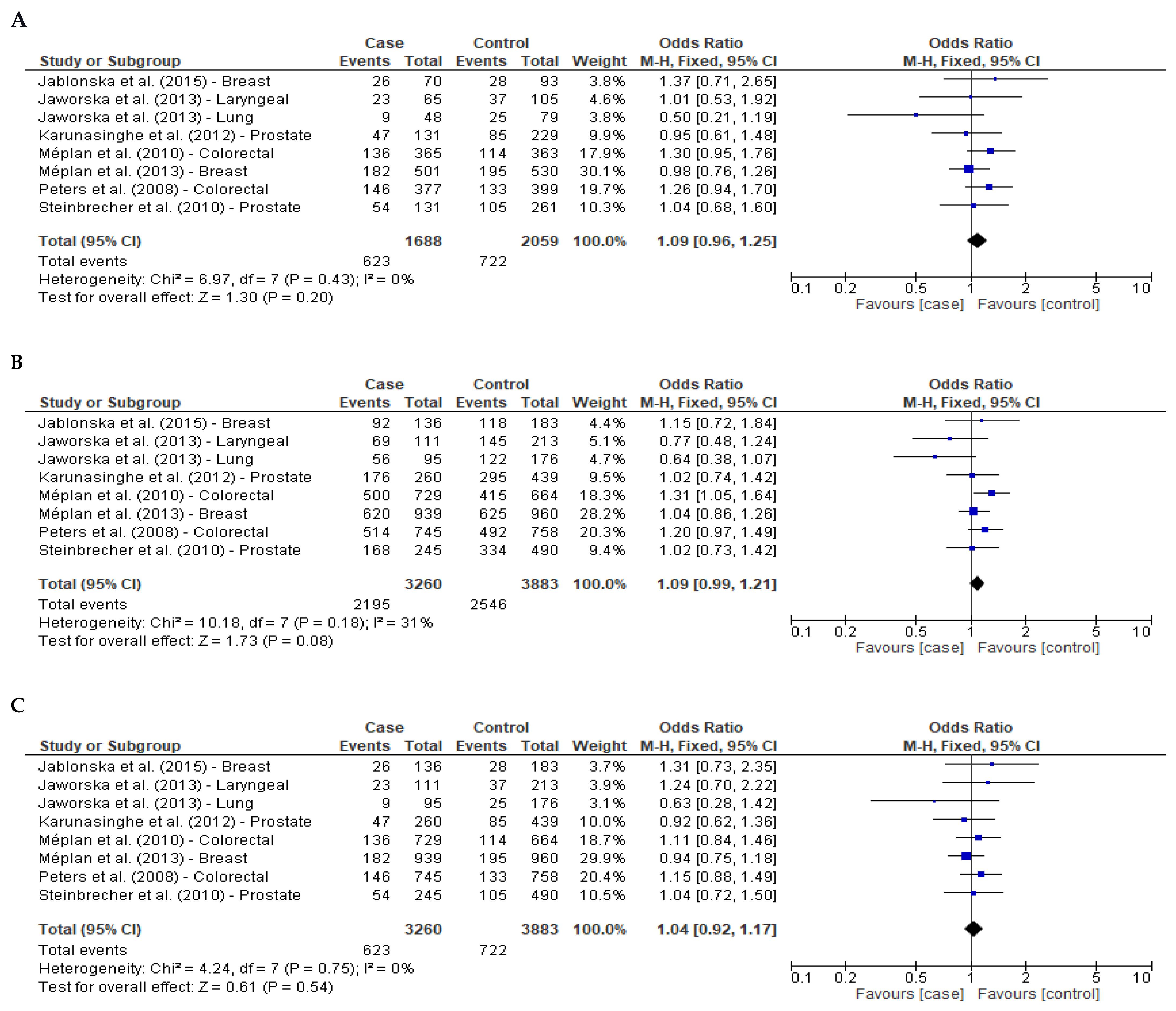
3.2. Meta-Analysis Cancer
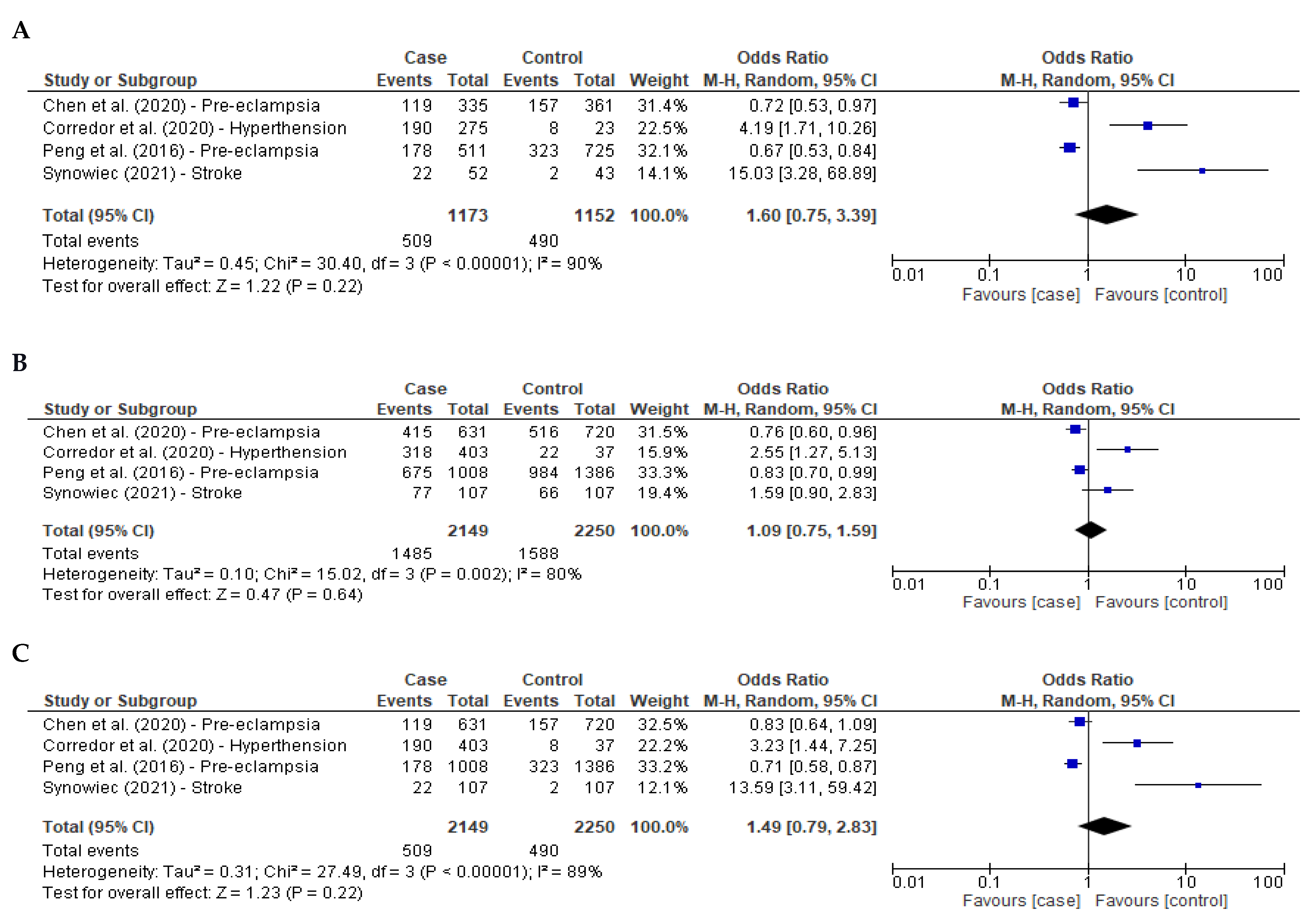
3.3. Meta-Analysis Hypertension-Related Diseases
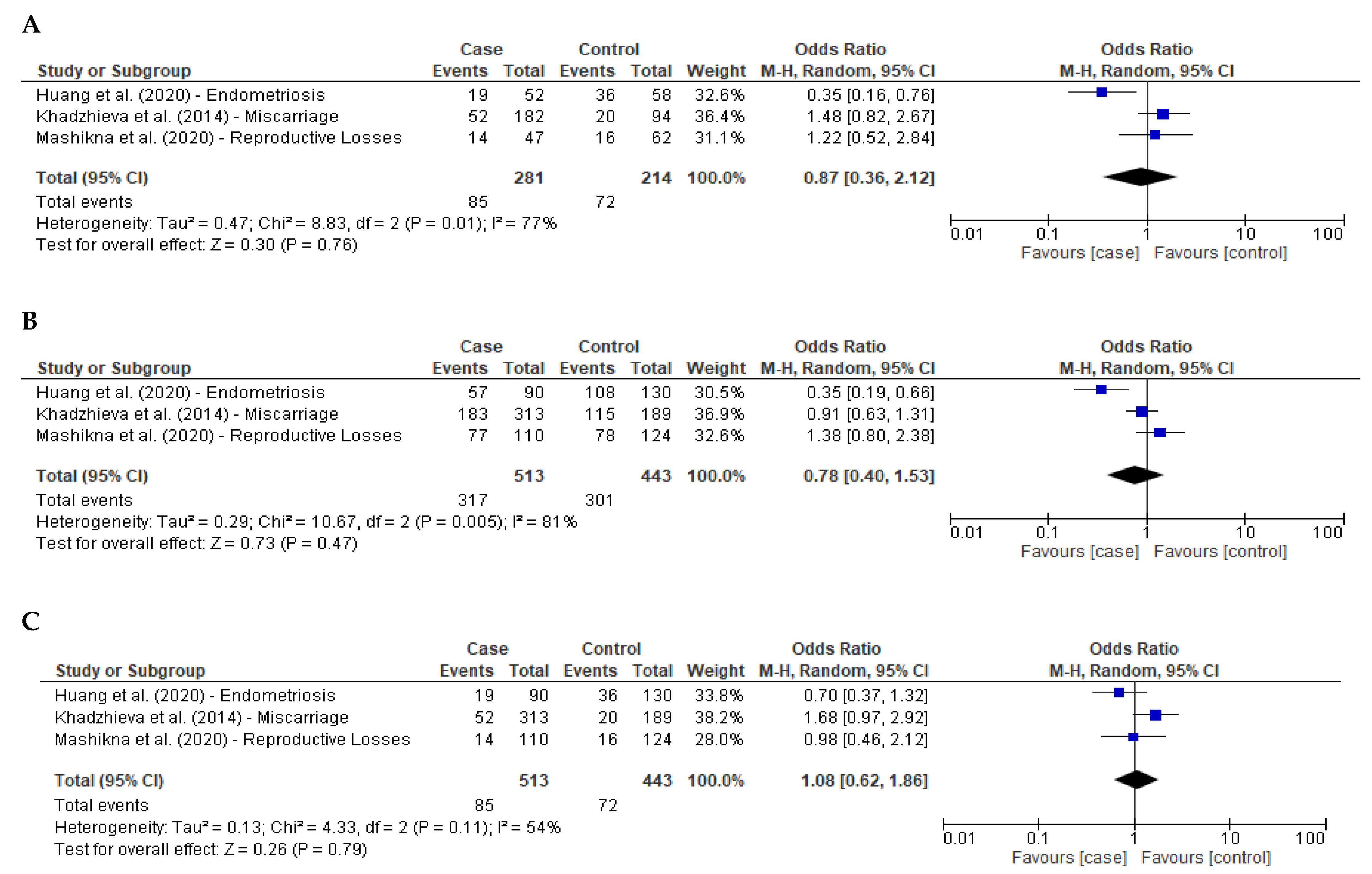
3.4. Meta-Analysis Reproduction-Related Diseases
3.5. Link of GPX4 (rs713041) Genotype to Other Diseases
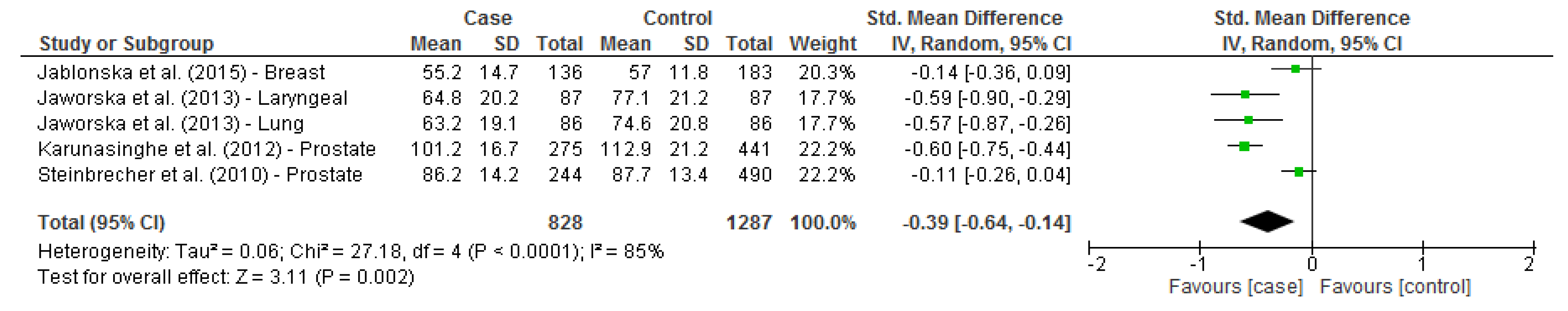
3.6. Selenium Status and Cancer Risk
3.7. Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shi, Y.; Yang, W.; Tang, X.; Yan, Q.; Cai, X.; Wu, F. Keshan Disease: A Potentially Fatal Endemic Cardiomyopathy in Remote Mountains of China. Front. Pediatr. 2021, 9, 576916. [Google Scholar] [CrossRef] [PubMed]
- Kadkol, S.; Diamond, A.M. The Interaction between Dietary Selenium Intake and Genetics in Determining Cancer Risk and Outcome. Nutrients 2020, 12, 2424. [Google Scholar] [CrossRef] [PubMed]
- Ursini, F.; Bosello Travain, V.; Cozza, G.; Miotto, G.; Roveri, A.; Toppo, S.; Maiorino, M. A white paper on Phospholipid Hydroperoxide Glutathione Peroxidase (GPx4) forty years later. Free Radic. Biol. Med. 2022, 188, 117–133. [Google Scholar] [CrossRef] [PubMed]
- Brigelius-Flohe, R.; Maiorino, M. Glutathione peroxidases. Biochim. Biophys. Acta 2013, 1830, 3289–3303. [Google Scholar] [CrossRef] [PubMed]
- Seibt, T.M.; Proneth, B.; Conrad, M. Role of GPX4 in ferroptosis and its pharmacological implication. Free Radic. Biol. Med. 2019, 133, 144–152. [Google Scholar] [CrossRef]
- Forcina, G.C.; Dixon, S.J. GPX4 at the Crossroads of Lipid Homeostasis and Ferroptosis. Proteomics 2019, 19, e1800311. [Google Scholar] [CrossRef]
- Ingold, I.; Berndt, C.; Schmitt, S.; Doll, S.; Poschmann, G.; Buday, K.; Roveri, A.; Peng, X.; Porto Freitas, F.; Seibt, T.; et al. Selenium Utilization by GPX4 Is Required to Prevent Hydroperoxide-Induced Ferroptosis. Cell 2018, 172, 409–422.e1. [Google Scholar] [CrossRef]
- Copeland, P.R. Regulation of gene expression by stop codon recoding: Selenocysteine. Gene 2003, 312, 17–25. [Google Scholar] [CrossRef]
- Squires, J.E.; Berry, M.J. Eukaryotic selenoprotein synthesis: Mechanistic insight incorporating new factors and new functions for old factors. IUBMB Life 2008, 60, 232–235. [Google Scholar] [CrossRef]
- Kipp, A.; Banning, A.; van Schothorst, E.M.; Meplan, C.; Schomburg, L.; Evelo, C.; Coort, S.; Gaj, S.; Keijer, J.; Hesketh, J.; et al. Four selenoproteins, protein biosynthesis, and Wnt signalling are particularly sensitive to limited selenium intake in mouse colon. Mol. Nutr. Food Res. 2009, 53, 1561–1572. [Google Scholar] [CrossRef]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. Selenoproteins: Molecular pathways and physiological roles. Physiol. Rev. 2014, 94, 739–777. [Google Scholar] [CrossRef] [PubMed]
- Meplan, C.; Crosley, L.K.; Nicol, F.; Horgan, G.W.; Mathers, J.C.; Arthur, J.R.; Hesketh, J.E. Functional effects of a common single-nucleotide polymorphism (GPX4c718t) in the glutathione peroxidase 4 gene: Interaction with sex. Am. J. Clin. Nutr. 2008, 87, 1019–1027. [Google Scholar] [CrossRef] [PubMed]
- Gautrey, H.; Nicol, F.; Sneddon, A.A.; Hall, J.; Hesketh, J. A T/C polymorphism in the GPX4 3’UTR affects the selenoprotein expression pattern and cell viability in transfected Caco-2 cells. Biochim. Biophys. Acta 2011, 1810, 584–591. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.; Sirotkin, K. dbSNP—Database for Single Nucleotide Polymorphisms and Other Classes of Minor Genetic Variation. Genome Res. 1999, 9, 677–679. [Google Scholar] [CrossRef]
- Cebrian, A.; Pharoah, P.D.; Ahmed, S.; Smith, P.L.; Luccarini, C.; Luben, R.; Redman, K.; Munday, H.; Easton, D.F.; Dunning, A.M.; et al. Tagging single-nucleotide polymorphisms in antioxidant defense enzymes and susceptibility to breast cancer. Cancer Res. 2006, 66, 1225–1233. [Google Scholar] [CrossRef] [PubMed]
- Meplan, C.; Dragsted, L.O.; Ravn-Haren, G.; Tjonneland, A.; Vogel, U.; Hesketh, J. Association between polymorphisms in glutathione peroxidase and selenoprotein P genes, glutathione peroxidase activity, HRT use and breast cancer risk. PLoS ONE 2013, 8, e73316. [Google Scholar] [CrossRef] [PubMed]
- Jablonska, E.; Gromadzinska, J.; Peplonska, B.; Fendler, W.; Reszka, E.; Krol, M.B.; Wieczorek, E.; Bukowska, A.; Gresner, P.; Galicki, M.; et al. Lipid peroxidation and glutathione peroxidase activity relationship in breast cancer depends on functional polymorphism of GPX1. BMC Cancer 2015, 15, 657. [Google Scholar] [CrossRef] [PubMed]
- Bermano, G.; Pagmantidis, V.; Holloway, N.; Kadri, S.; Mowat, N.A.; Shiel, R.S.; Arthur, J.R.; Mathers, J.C.; Daly, A.K.; Broom, J.; et al. Evidence that a polymorphism within the 3’UTR of glutathione peroxidase 4 is functional and is associated with susceptibility to colorectal cancer. Genes Nutr. 2007, 2, 225–232. [Google Scholar] [CrossRef]
- Jaworska, K.; Gupta, S.; Durda, K.; Muszynska, M.; Sukiennicki, G.; Jaworowska, E.; Grodzki, T.; Sulikowski, M.; Waloszczyk, P.; Wojcik, J.; et al. A low selenium level is associated with lung and laryngeal cancers. PLoS ONE 2013, 8, e59051. [Google Scholar] [CrossRef]
- Karunasinghe, N.; Han, D.Y.; Goudie, M.; Zhu, S.; Bishop, K.; Wang, A.; Duan, H.; Lange, K.; Ko, S.; Medhora, R.; et al. Prostate disease risk factors among a New Zealand cohort. J. Nutr. Nutr. 2012, 5, 339–351. [Google Scholar] [CrossRef]
- Costa-Urrutia, P.; Flores-Buendia, A.M.; Ascencio-Montiel, I.; Solares-Tlapechco, J.; Medina-Campos, O.N.; Pedraza-Chaverri, J.; Granados, J.; Jimenez-Osorio, A.S.; Rodriguez-Arellano, M.E. Antioxidant Enzymes Haplotypes and Polymorphisms Associated with Obesity in Mexican Children. Antioxidants 2020, 9, 684. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.Y.; Wu, C.H.; Liu, C.H.; Yang, S.F.; Wang, P.H.; Lin, L.Y.; Lee, T.H.; Lee, M.S. Association between the Genetic Variants of Glutathione Peroxidase 4 and Severity of Endometriosis. Int. J. Environ. Res. Public Health 2020, 17, 5089. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Yuan, J.; Yao, Q.; Yan, N.; Song, R.; Jiang, W.; Li, D.; Shi, L.; Zhang, J.A. A case-control study of selenoprotein genes polymorphisms and autoimmune thyroid diseases in a Chinese population. BMC Med. Genet. 2017, 18, 54. [Google Scholar] [CrossRef]
- da Rocha, T.J.; Silva Alves, M.; Guisso, C.C.; de Andrade, F.M.; Camozzato, A.; de Oliveira, A.A.; Fiegenbaum, M. Association of GPX1 and GPX4 polymorphisms with episodic memory and Alzheimer’s disease. Neurosci. Lett. 2018, 666, 32–37. [Google Scholar] [CrossRef]
- Wigner, P.; Czarny, P.; Synowiec, E.; Bijak, M.; Bialek, K.; Talarowska, M.; Galecki, P.; Szemraj, J.; Sliwinski, T. Variation of genes involved in oxidative and nitrosative stresses in depression. Eur. Psychiatry 2018, 48, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Wigner, P.; Dziedzic, A.; Synowiec, E.; Miller, E.; Bijak, M.; Saluk-Bijak, J. Variation of genes encoding nitric oxide synthases and antioxidant enzymes as potential risks of multiple sclerosis development: A preliminary study. Sci. Rep. 2022, 12, 10603. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Group, P. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Green, S. Cochrane Handbook for Systematic Reviews of Interventions, Version 5.1.0. 2011. Available online: www.handbook.cochrane.org (accessed on 12 December 2022).
- Stang, A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur. J. Epidemiol. 2010, 25, 603–605. [Google Scholar] [CrossRef] [PubMed]
- Egger, M.; Davey Smith, G.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef]
- Duval, S.; Tweedie, R. A Nonparametric “Trim and Fill” Method of Accounting for Publication Bias in Meta-Analysis. J. Am. Stat. Assoc. 2000, 95, 89–98. [Google Scholar] [CrossRef]
- Andrade, C. Mean Difference, Standardized Mean Difference (SMD), and Their Use in Meta-Analysis: As Simple as It Gets. J. Clin. Psychiatry 2020, 81, 20f13681. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.; Cochrane Consumers and Communication Review Group. Cochrane Consumers and Communication Group: Meta-Analysis. Available online: http://cccrg.cochrane.org (accessed on 18 July 2022).
- Vanhecke, T.E. Zotero. J. Med. Libr. Assoc. 2008, 96, 275–276. [Google Scholar] [CrossRef]
- Meplan, C.; Hughes, D.J.; Pardini, B.; Naccarati, A.; Soucek, P.; Vodickova, L.; Hlavata, I.; Vrana, D.; Vodicka, P.; Hesketh, J.E. Genetic variants in selenoprotein genes increase risk of colorectal cancer. Carcinogenesis 2010, 31, 1074–1079. [Google Scholar] [CrossRef] [PubMed]
- Peters, U.; Chatterjee, N.; Hayes, R.B.; Schoen, R.E.; Wang, Y.; Chanock, S.J.; Foster, C.B. Variation in the selenoenzyme genes and risk of advanced distal colorectal adenoma. Cancer Epidemiol. Biomark. Prev. 2008, 17, 1144–1154. [Google Scholar] [CrossRef]
- Steinbrecher, A.; Meplan, C.; Hesketh, J.; Schomburg, L.; Endermann, T.; Jansen, E.; Akesson, B.; Rohrmann, S.; Linseisen, J. Effects of selenium status and polymorphisms in selenoprotein genes on prostate cancer risk in a prospective study of European men. Cancer Epidemiol. Biomark. Prev. 2010, 19, 2958–2968. [Google Scholar] [CrossRef]
- Chen, A.; Zhao, H.; Wang, J.; Zhang, R.; Liu, J.; Zhao, X.; Li, C.; Jia, X.; Li, X.; Lin, Y.; et al. Haplotype Analysis of Candidate Genes Involved in Inflammation and Oxidative Stress and the Susceptibility to Preeclampsia. J. Immunol. Res. 2020, 2020, 4683798. [Google Scholar] [CrossRef]
- Peng, X.; Lin, Y.; Li, J.; Liu, M.; Wang, J.; Li, X.; Liu, J.; Jia, X.; Jing, Z.; Huang, Z.; et al. Evaluation of Glutathione Peroxidase 4 role in Preeclampsia. Sci. Rep. 2016, 6, 33300. [Google Scholar] [CrossRef]
- Corredor, Z.; Filho, M.; Rodriguez-Ribera, L.; Velazquez, A.; Hernandez, A.; Catalano, C.; Hemminki, K.; Coll, E.; Silva, I.; Diaz, J.M.; et al. Genetic Variants Associated with Chronic Kidney Disease in a Spanish Population. Sci. Rep. 2020, 10, 144. [Google Scholar] [CrossRef]
- Synowiec, E.; Wigner, P.; Cichon, N.; Watala, C.; Czarny, P.; Saluk-Bijak, J.; Miller, E.; Sliwinski, T.; Zielińska-Nowak, E.; Bijak, M. Single-nucleotide polymorphisms in oxidative stress-related genes and the risk of a stroke in a polish population-a preliminary study. Brain Sci. 2021, 11, 391. [Google Scholar] [CrossRef]
- Khadzhieva, M.B.; Lutcenko, N.N.; Volodin, I.V.; Morozova, K.V.; Salnikova, L.E. Association of oxidative stress-related genes with idiopathic recurrent miscarriage. Free Radic. Res. 2014, 48, 534–541. [Google Scholar] [CrossRef]
- Mashkina, E.V.; Kovalenko, K.A.; Miktadova, A.V.; Shkurat, M.A. Association of Gene Polymorphisms of Antioxidants with Reproductive Losses. Russ. J. Genet. 2020, 56, 354–362. [Google Scholar] [CrossRef]
- Ściskalska, M.; Milnerowicz, H. Association of genetic variants in the GPX1 and GPX4 genes with the activities of glutathione-dependent enzymes, their interaction with smoking and the risk of acute pancreatitis. Biomed. Pharmacother. 2022, 146, 112591. [Google Scholar] [CrossRef] [PubMed]
- Gusti, A.M.T.; Qusti, S.Y.; Bahijri, S.M.; Toraih, E.A.; Bokhari, S.; Attallah, S.M.; Alzahrani, A.; Alshehri, W.M.A.; Alotaibi, H.; Fawzy, M.S. Glutathione s-transferase (GSTT1 rs17856199) and nitric oxide synthase (nos2 rs2297518) genotype combination as potential oxidative stress-related molecular markers for type 2 diabetes mellitus. Diabetes Metab. Syndr. Obes. Targets Ther. 2021, 14, 1385–1403. [Google Scholar] [CrossRef] [PubMed]
- Du, X.H.; Dai, X.X.; Xia Song, R.; Zou, X.Z.; Yan Sun, W.; Mo, X.Y.; Lu Bai, G.; Xiong, Y.M. SNP and mRNA expression for glutathione peroxidase 4 in Kashin-Beck disease. Br. J. Nutr. 2012, 107, 164–169. [Google Scholar] [CrossRef]
- Paglia, D.E.; Valentine, W.N. Studies on the quantitative and qualitative characterization of erythrocyte glutathione peroxidase. J. Lab. Clin. Med. 1967, 70, 158–169. [Google Scholar] [PubMed]
- Rayman, M.P. Selenium in cancer prevention: A review of the evidence and mechanism of action. Proc. Nutr. Soc. 2005, 64, 527–542. [Google Scholar] [CrossRef]
- Jiang, X.; Stockwell, B.R.; Conrad, M. Ferroptosis: Mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 266–282. [Google Scholar] [CrossRef]
- Villette, S.; Kyle, J.A.; Brown, K.M.; Pickard, K.; Milne, J.S.; Nicol, F.; Arthur, J.R.; Hesketh, J.E. A novel single nucleotide polymorphism in the 3’ untranslated region of human glutathione peroxidase 4 influences lipoxygenase metabolism. Blood Cells Mol. Dis. 2002, 29, 174–178. [Google Scholar] [CrossRef]
- Crosley, L.K.; Bashir, S.; Nicol, F.; Arthur, J.R.; Hesketh, J.E.; Sneddon, A.A. The single-nucleotide polymorphism (GPX4c718t) in the glutathione peroxidase 4 gene influences endothelial cell function: Interaction with selenium and fatty acids. Mol. Nutr. Food Res. 2013, 57, 2185–2194. [Google Scholar] [CrossRef]
- Setu, T.; Basak, T. An Introduction to Basic Statistical Models in Genetics. Open J. Stat. 2021, 11, 1017–1025. [Google Scholar] [CrossRef]
- Hughes, D.J.; Fedirko, V.; Jenab, M.; Schomburg, L.; Meplan, C.; Freisling, H.; Bueno-de-Mesquita, H.B.; Hybsier, S.; Becker, N.P.; Czuban, M.; et al. Selenium status is associated with colorectal cancer risk in the European prospective investigation of cancer and nutrition cohort. Int. J. Cancer 2015, 136, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Fedirko, V.; Jenab, M.; Meplan, C.; Jones, J.S.; Zhu, W.; Schomburg, L.; Siddiq, A.; Hybsier, S.; Overvad, K.; Tjonneland, A.; et al. Association of Selenoprotein and Selenium Pathway Genotypes with Risk of Colorectal Cancer and Interaction with Selenium Status. Nutrients 2019, 11, 935. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Aranguren, L.C.; Prada, C.E.; Riano-Medina, C.E.; Lopez, M. Endothelial dysfunction and preeclampsia: Role of oxidative stress. Front. Physiol. 2014, 5, 372. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Li, Z.; Ju, W.; Yang, X.; Fu, X.; Gao, X. Cross-sectional Study: Relationship between Serum Selenium and Hypertension in the Shandong Province of China. Biol. Trace Elem. Res. 2018, 185, 295–301. [Google Scholar] [CrossRef]
- Pagmantidis, V.; Bermano, G.; Villette, S.; Broom, I.; Arthur, J.; Hesketh, J. Effects of Se-depletion on glutathione peroxidase and selenoprotein W gene expression in the colon. FEBS Lett. 2005, 579, 792–796. [Google Scholar] [CrossRef]

| Author (Year) | Country | Ethnicity | Disease | Genotyping Methods | Sample Size (Case/Control) | CC | CT | TT | Newcastle-Ottawa Scale | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | Case | Control | |||||||
| Peters (2008) [36] | USA | >93% Caucasian American | Advanced distal colorectal adenoma | SNPlex or TaqMan | 745/758 | 231 | 266 | 368 | 359 | 146 | 133 | High |
| Méplan (2010) [35] | Czech Republic | Czech origin adults >29 years | Colorectal cancer | KASPar | 729/664 | 229 | 249 | 364 | 301 | 136 | 114 | Moderate |
| Steinbrecher (2010) [37] | Germany | European men | Prostate cancer | MassARRAY system | 245/490 | 77 | 156 | 114 | 229 | 54 | 105 | High |
| Du (2012) [46] | China | Chinese Han population | Kashin-Beck disease | PCR-RFLP | 216/194 | 68 | 67 | 124 | 102 | 24 | 25 | High |
| Karunasinghe (2012) [20] | New Zealand | European men | Prostate cancer | TaqMan | 260/439 | 84 | 144 | 129 | 210 | 47 | 85 | High |
| Jaworska (2013) [19] | Poland | Polish adults | Laryngeal cancer | Real-time PCR | 111/213 | 42 | 68 | 46 | 108 | 23 | 37 | High |
| Lung cancer | 95/176 | 39 | 54 | 47 | 97 | 9 | 25 | |||||
| Méplan (2013) [16] | Denmark | Danish women | Breast cancer | TaqMan | 939/960 | 319 | 335 | 438 | 430 | 182 | 195 | High |
| Khadzhieva (2014) [42] | Russia | Russian women | Recurrent miscarriage | PCR-RFLP | 313/189 | 130 | 74 | 131 | 95 | 52 | 20 | Moderate |
| Jablonska (2015) [17] | Poland | Polish women | Breast cancer | TaqMan | 136/183 | 44 | 65 | 66 # | 90 # | 26 | 28 | High |
| Peng (2016) [39] | China | Chinese Han women | Pre-eclampsia (PE) | TaqMan | 1008/1386 | 333 | 402 | 497 | 661 | 178 | 323 | Moderate |
| Mild PE | 158/1386 | 64 | 402 | 70 | 661 | 24 | 323 | |||||
| Severe PE | 850/1386 | 269 | 402 | 427 | 661 | 154 | 323 | |||||
| Early onset PE | 523/1386 | 181 | 402 | 253 | 661 | 89 | 323 | |||||
| Late onset PE | 485/1386 | 152 | 402 | 244 | 661 | 89 | 323 | |||||
| Xiao (2017) [23] | China | Han Chinese | Thyroid diseases | MassARRAY system | 1022/898 | 318 | 300 | 704 * | 598 * | Moderate | ||
| Graves’ disease | 675/898 | 213 | 300 | 462 * | 598 * | |||||||
| Hashimoto’s thyroiditis | 347/898 | 105 | 300 | 242 * | 598 * | |||||||
| da Rocha (2018) [24] | Brazil | South Brazilian adults | Alzheimer’s disease | TaqMan | 103/108 | 28 | 34 | 49 | 42 | 25 | 29 | Moderate |
| Wigner (2018) [25] | Poland | Adults | Depression | TaqMan | 281/229 | 87 | 83 | 141 | 138 | 53 | 8 | Moderate |
| Chen (2020) [38] | China | Chinese Han women | Pre-eclampsia (PE) | TaqMan | 631/720 | 216 | 204 | 296 | 359 | 119 | 157 | Moderate |
| Mild PE | 141/720 | 47 | 204 | 66 | 359 | 28 | 157 | |||||
| Severe PE | 490/720 | 169 | 204 | 230 | 359 | 91 | 157 | |||||
| Early onset PE | 249/720 | 94 | 204 | 112 | 359 | 43 | 157 | |||||
| Late onset PE | 382/720 | 122 | 204 | 184 | 359 | 76 | 157 | |||||
| Corredor (2020) [40] | Spain | Spanish | Hypertension | TaqMan | 403/37 | 85 | 15 | 128 | 14 | 190 | 8 | Moderate |
| Huang (2020) [22] | China | Han Chinese women | Endometriosis | TaqMan | 90/130 | 33 | 22 | 38 | 72 | 19 | 36 | Moderate |
| Mashkina (2020) [43] | Russia | Russian women | Pregnancy loss | PCR-RFLP | 110/124 | 33 | 46 | 63 | 62 | 14 | 16 | Moderate |
| Gusti (2021) [45] | Saudi Arabia | Saudi adult population | Type 2 diabetes mellitus | TaqMan | 109/163 | 27 | 42 | 62 | 87 | 20 | 34 | High |
| Synowiec (2021) [41] | Poland | Caucasian | Ischemic stroke | TaqMan | 107/107 | 30 | 41 | 55 | 64 | 22 | 2 | Moderate |
| Ściskalska (2022) [44] | Poland | Adults | Acute pancreatitis | PCR-RFLP | 39/51 | 9 | 16 | 19 | 18 | 11 | 17 | High |
| Wigner (2022) [26] | Poland | Native Polish adults | Multiple sclerosis | Real-time PCR | 142/140 | 39 | 57 | 72 | 65 | 31 | 18 | High |
| Author (Year) | Ethnicity | Disease | Sample Size (Case/Control) | Selenium Status (Mean ± SD—µg/L) | Sample Size (Case/Control) | GPX3 Activity (Mean ± SD—Units/L) | ||
|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | |||||
| Steinbrecher (2010) [37] | European men | Prostate cancer | 244/490 | 86.2 ± 14.2 | 87.7 ± 13.4 | 248/492 | 647.6 ± 112.9 | 656.7± 110.2 |
| Karunasinghe (2012) [20] | European men | Prostate cancer | 275/441 | 101.2 ± 16.7 | 112.9 ± 21.2 | - | - | - |
| Jaworska (2013) [19] | Polish adults | Laryngeal cancer | 87/87 | 64.8 ± 20.2 | 77.1 ± 21.2 | - | - | - |
| Lung cancer | 86/86 | 63.2 ± 19.1 | 74.6 ± 20.8 | - | - | - | ||
| Jablonska (2015) [17] | Polish women | Breast cancer | 136/183 | 55.2 ± 14.7 | 57.0 ± 11.8 | 136/183 | 189 ± 37 | 191 ± 32 |
| Cancer Type | Cases/Controls | Additive Model (TT vs. CC) | Dominant Model (CT+TT vs. CC) | Recessive Model (TT vs. CC+CT) | |||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | OR (95% CI) | p | ||
| Breast | 1075/1143 | 1.02 (0.81–1.30) | 0.85 | 1.06 (0.89–1.26) | 0.54 | 0.98 (0.80–1.21) | 0.88 |
| Prostate | 505/929 | 1.00 (0.73–1.36) | 0.98 | 1.02 (0.81–1.29) | 0.86 | 0.98 (0.75–1.28) | 0.88 |
| Colorectal | 1474/1422 | 1.28 (1.04–1.58) | 0.02 | 1.25 (1.07–1.46) | 0.004 | 1.13 (0.93–1.36) | 0.22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbosa, P.; Abo El-Magd, N.F.; Hesketh, J.; Bermano, G. The Role of rs713041 Glutathione Peroxidase 4 (GPX4) Single Nucleotide Polymorphism on Disease Susceptibility in Humans: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2022, 23, 15762. https://doi.org/10.3390/ijms232415762
Barbosa P, Abo El-Magd NF, Hesketh J, Bermano G. The Role of rs713041 Glutathione Peroxidase 4 (GPX4) Single Nucleotide Polymorphism on Disease Susceptibility in Humans: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences. 2022; 23(24):15762. https://doi.org/10.3390/ijms232415762
Chicago/Turabian StyleBarbosa, Priscila, Nada F. Abo El-Magd, John Hesketh, and Giovanna Bermano. 2022. "The Role of rs713041 Glutathione Peroxidase 4 (GPX4) Single Nucleotide Polymorphism on Disease Susceptibility in Humans: A Systematic Review and Meta-Analysis" International Journal of Molecular Sciences 23, no. 24: 15762. https://doi.org/10.3390/ijms232415762
APA StyleBarbosa, P., Abo El-Magd, N. F., Hesketh, J., & Bermano, G. (2022). The Role of rs713041 Glutathione Peroxidase 4 (GPX4) Single Nucleotide Polymorphism on Disease Susceptibility in Humans: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences, 23(24), 15762. https://doi.org/10.3390/ijms232415762







