Distributed Big Data Storage Infrastructure for Biomedical Research Featuring High-Performance and Rich-Features
Abstract
1. Introduction
2. Methods
2.1. Optimization for Biomedical Big Data
- The majority of IO operations are read—especially sequential read—not write. Therefore, when designing the F3BFS overall architecture and underlying data structure, we prioritize improving read performance while sacrificing some write performance. As a result, the read operations of the cluster are greatly accelerated, and the architecture of F3BFS is simplified as it does not require advanced data write features such as copy-on-write or write-ahead logging;
- A biomedical data file typically contains more metadata, which is needed to be queried frequently, such as, e.g., the species it belongs to, the sequence technology, etc. As such, we automate the extraction of metadata from data files and save it to a centralized database when the data is uploaded to F3BFS;
- As there are only a few types of common biomedical datasets, more targeted optimizations can be made. For example, we intentionally design an additional layer of a file system to integrate functional utilities, which we call the peripheral layer.
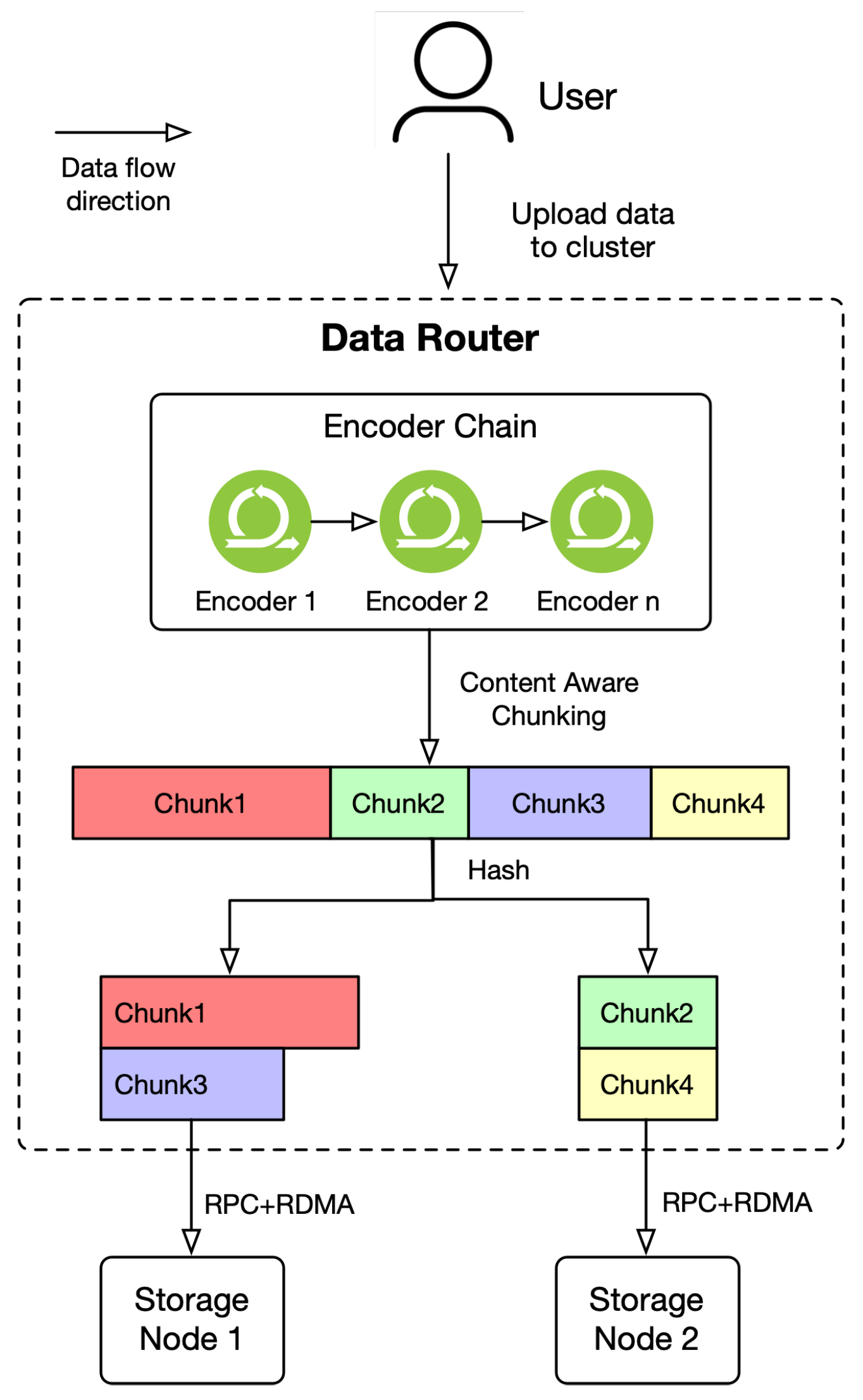
2.2. Content Aware Chunking
2.3. Transparent Compression
2.4. Metadata as Extended Attributes
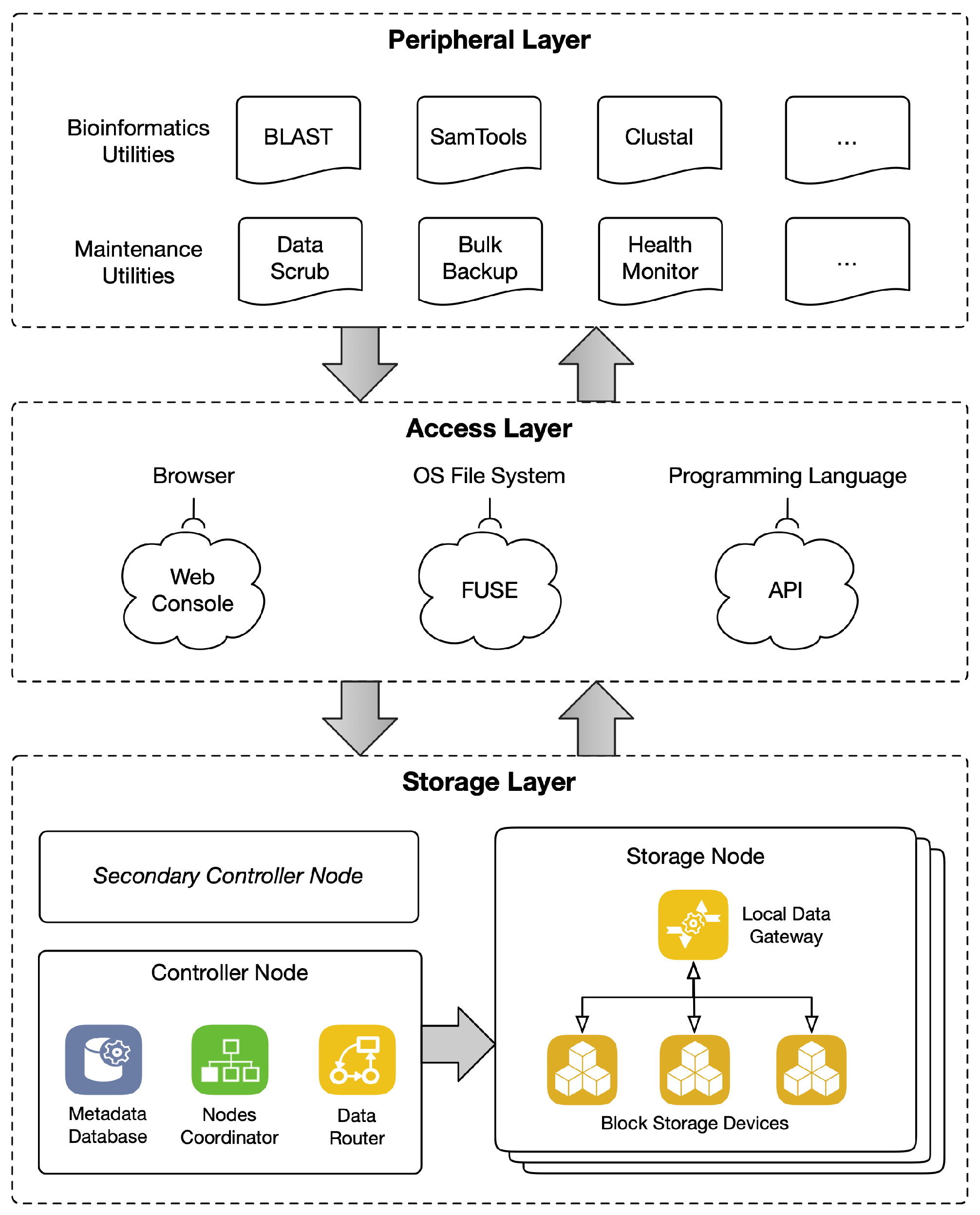
3. Implementation
3.1. Storage Layer
3.1.1. Metadata Database
3.1.2. Nodes Coordinator
3.1.3. Data Router
3.1.4. Local Data Gateway
3.2. Access Layer
3.2.1. Web Console
3.2.2. FUSE Mount Point
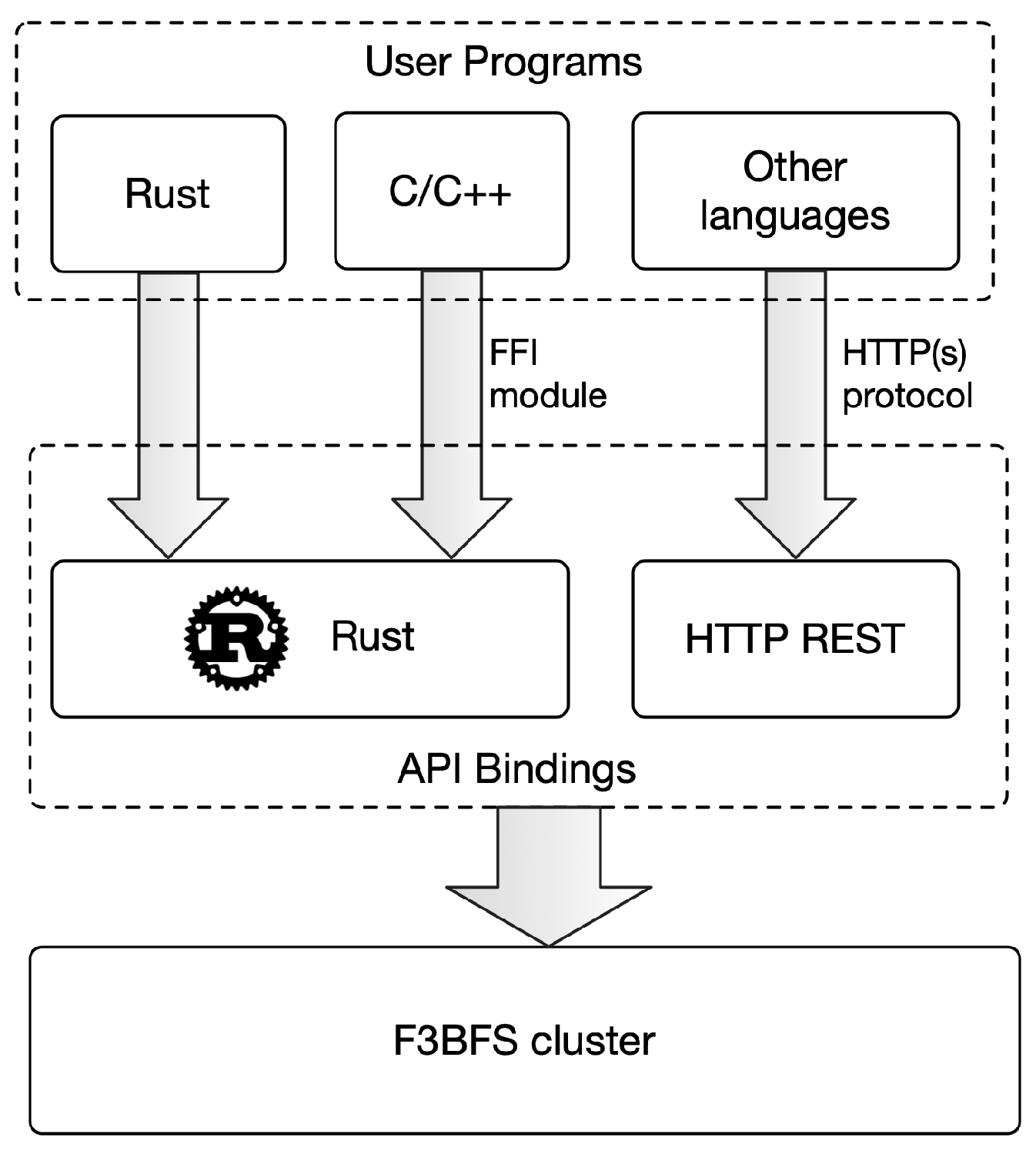
3.2.3. API
3.3. Peripheral Layer
4. Evaluation
4.1. Experimental Setup
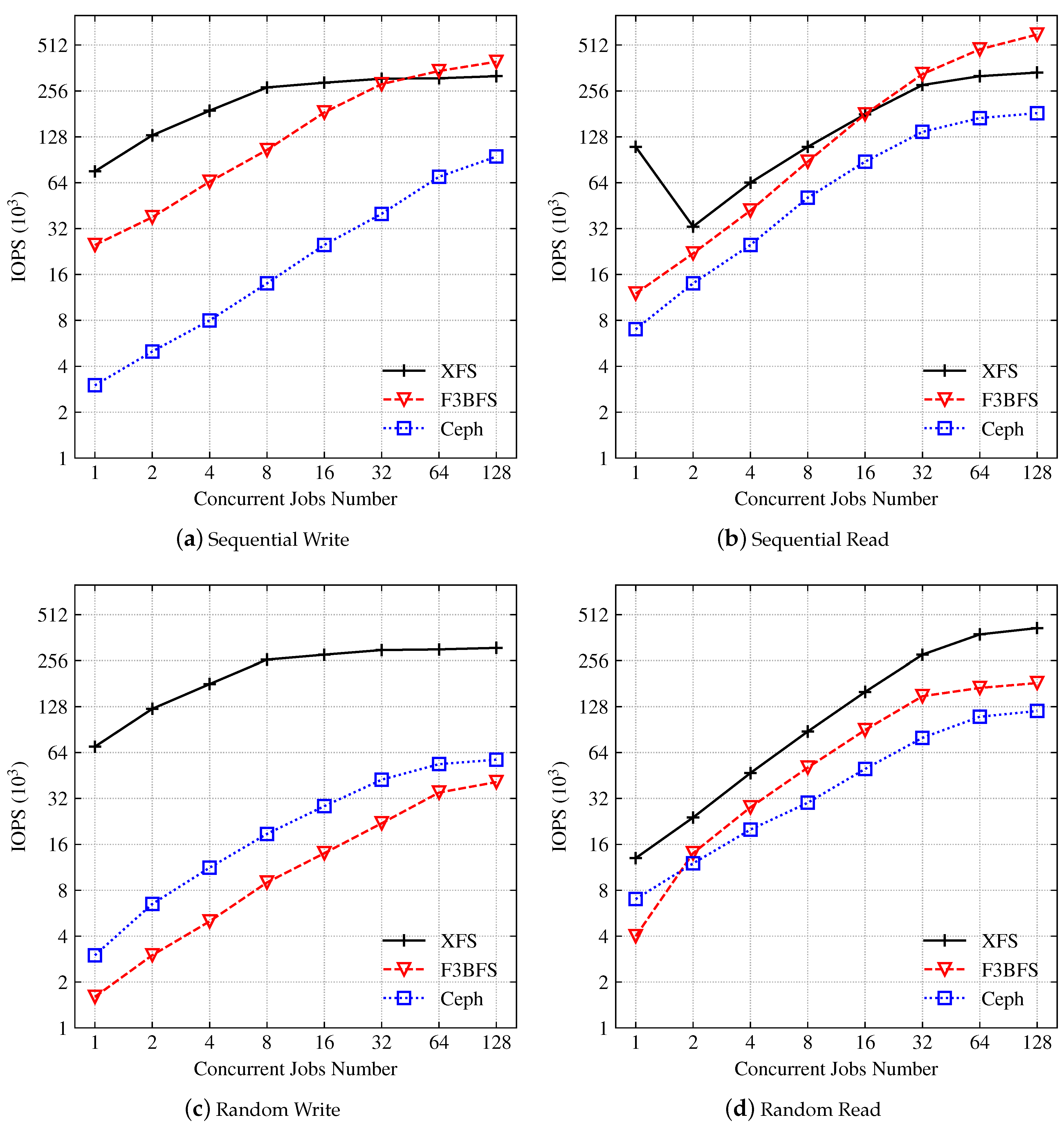
4.2. Throughput
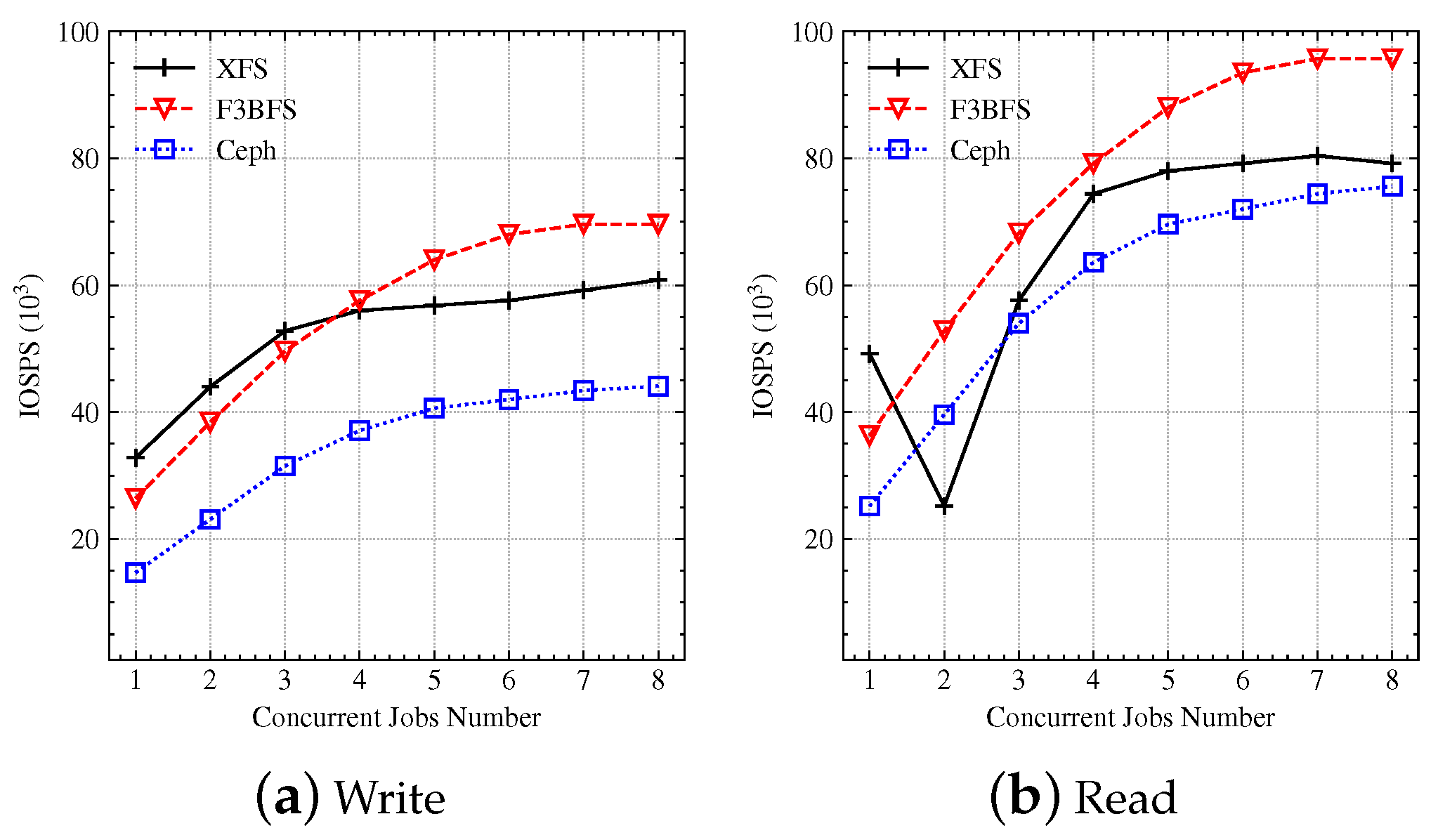
4.2.1. IOPS
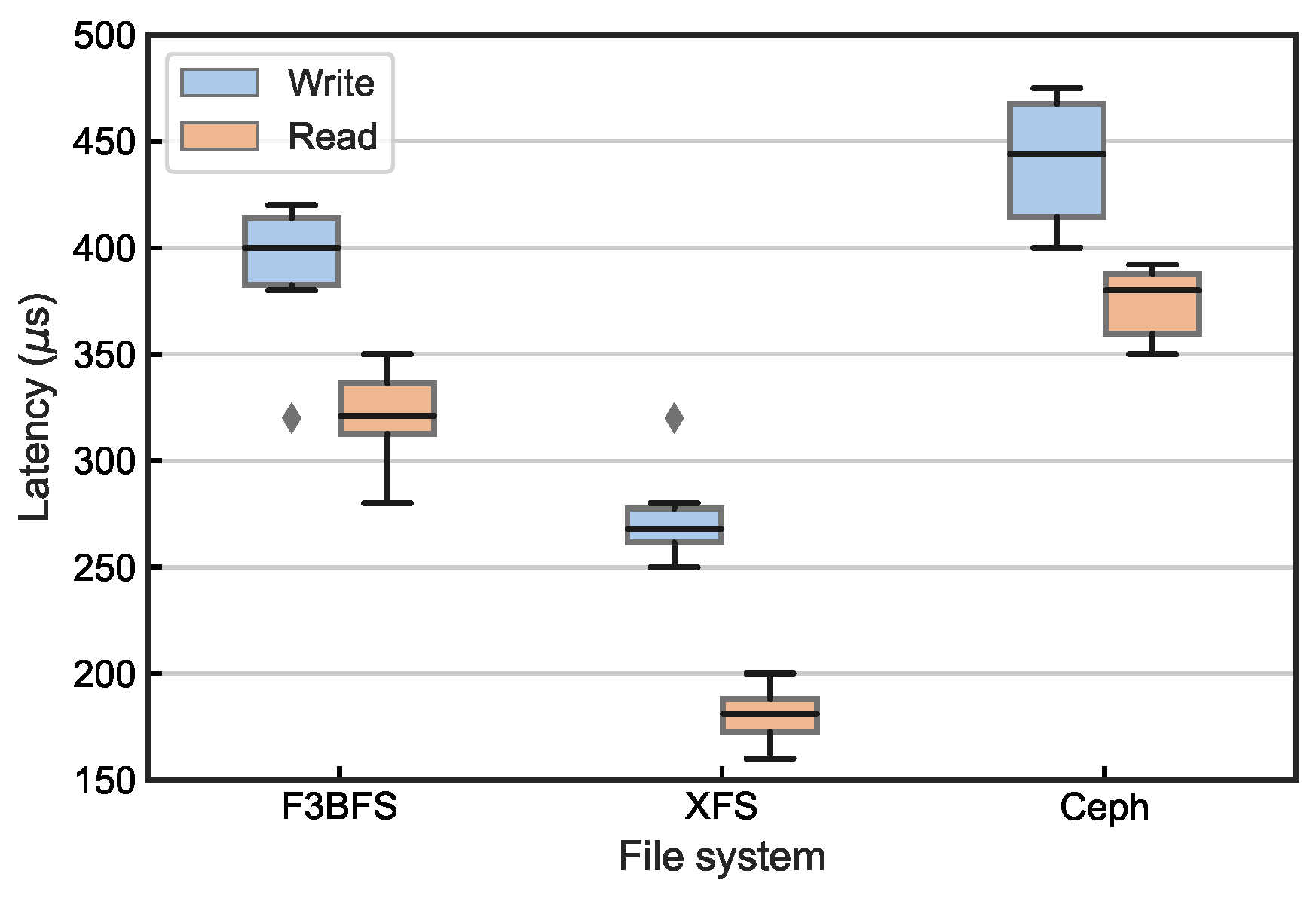
4.2.2. IOPS Latency
4.2.3. Sequence Throughput
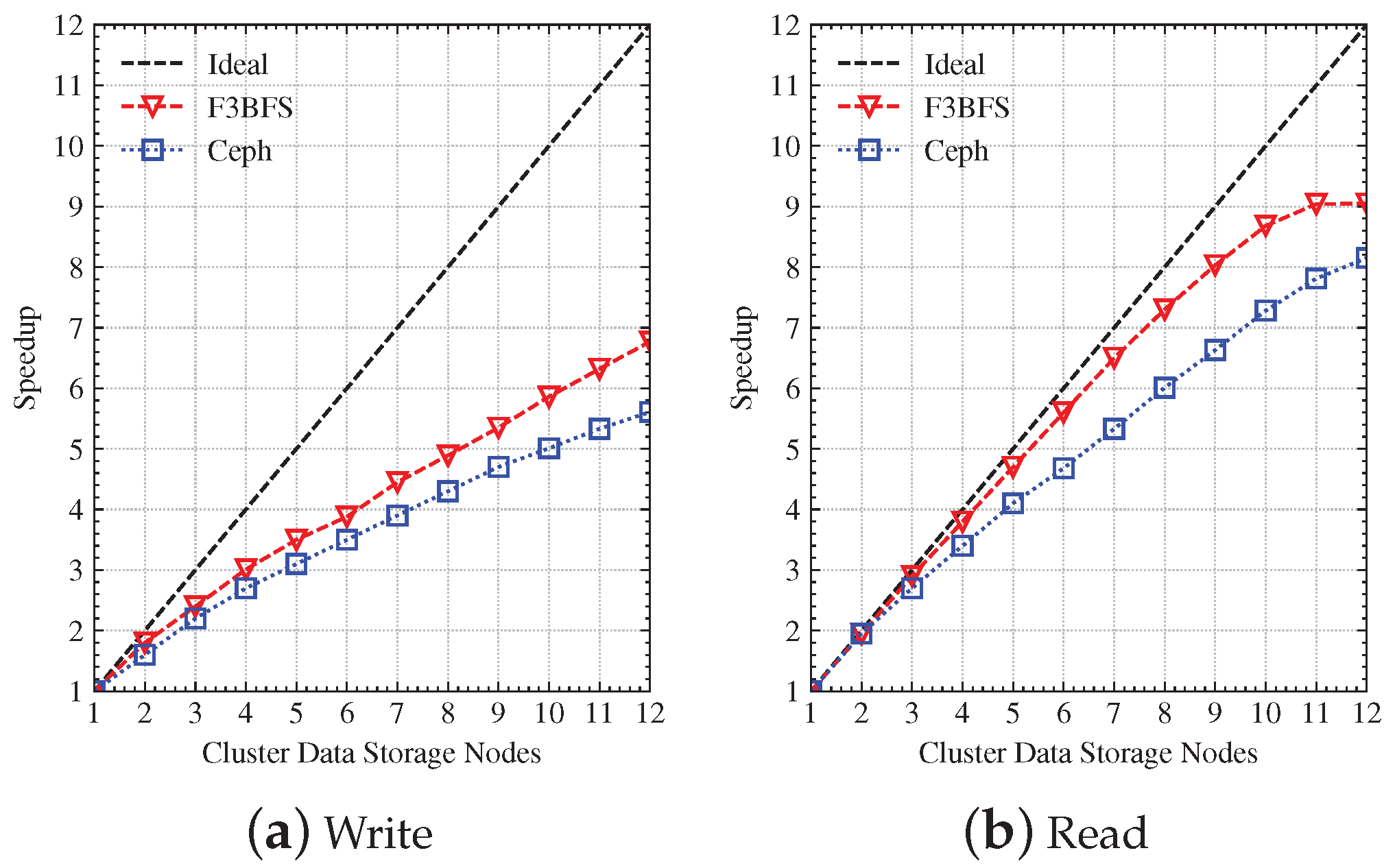
4.3. Scalability
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| IOPS | Input/Output Operations Per Second |
| IOSPS | Input/Output Sequences Per Second |
References
- Stephens, Z.D.; Lee, S.Y.; Faghri, F.; Campbell, R.H.; Zhai, C.; Efron, M.J.; Iyer, R.; Schatz, M.C.; Sinha, S.; Robinson, G.E. Big data: Astronomical or genomical? PLoS Biol. 2015, 13, e1002195. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.; Uddin, M.I.; AlSaeed, D.H.; Khan, A.; Adnan, M. Early detection of seasonal outbreaks from twitter data using machine learning approaches. Complexity 2021, 2021, 5520366. [Google Scholar] [CrossRef]
- Thakur, N.; Han, C.Y. An Exploratory Study of Tweets about the SARS-CoV-2 Omicron Variant: Insights from Sentiment Analysis, Language Interpretation, Source Tracking, Type Classification, and Embedded URL Detection. COVID 2022, 2, 1026–1049. [Google Scholar] [CrossRef]
- Tang, B.; Pan, Z.; Yin, K.; Khateeb, A. Recent advances of deep learning in bioinformatics and computational biology. Front. Genet. 2019, 10, 214. [Google Scholar] [CrossRef]
- Allan, C.; Burel, J.M.; Moore, J.; Blackburn, C.; Linkert, M.; Loynton, S.; MacDonald, D.; Moore, W.J.; Neves, C.; Patterson, A.; et al. OMERO: Flexible, model-driven data management for experimental biology. Nat. Methods 2012, 9, 245–253. [Google Scholar] [CrossRef]
- Bauch, A.; Adamczyk, I.; Buczek, P.; Elmer, F.J.; Enimanev, K.; Glyzewski, P.; Kohler, M.; Pylak, T.; Quandt, A.; Ramakrishnan, C.; et al. openBIS: A flexible framework for managing and analyzing complex data in biology research. BMC Bioinform. 2011, 12, 468. [Google Scholar] [CrossRef]
- Vashist, S.; Gupta, A. A Review on Distributed File System and Its Applications. Int. J. Adv. Res. Comput. Sci. 2014, 5, 235–237. [Google Scholar]
- Pillai, T.S.; Chidambaram, V.; Alagappan, R.; Al-Kiswany, S.; Arpaci-Dusseau, A.C.; Arpaci-Dusseau, R.H. All File Systems Are Not Created Equal: On the Complexity of Crafting {Crash-Consistent} Applications. In Proceedings of the 11th USENIX Symposium on Operating Systems Design and Implementation (OSDI 14), Broomfield, CO, USA, 6–8 October 2014; pp. 433–448. [Google Scholar]
- Navarro, G.; Sepúlveda, V.; Marín, M.; González, S. Compressed filesystem for managing large genome collections. Bioinformatics 2019, 35, 4120–4128. [Google Scholar] [CrossRef]
- Hoogstrate, Y.; Jenster, G.W.; van de Werken, H.J. FASTAFS: File system virtualisation of random access compressed FASTA files. BMC Bioinform. 2021, 22, 535. [Google Scholar] [CrossRef]
- Ison, J.; Ménager, H.; Brancotte, B.; Jaaniso, E.; Salumets, A.; Raček, T.; Lamprecht, A.L.; Palmblad, M.; Kalaš, M.; Chmura, P.; et al. Community curation of bioinformatics software and data resources. Brief. Bioinform. 2020, 21, 1697–1705. [Google Scholar] [CrossRef]
- Liu, B.; Madduri, R.K.; Sotomayor, B.; Chard, K.; Lacinski, L.; Dave, U.J.; Li, J.; Liu, C.; Foster, I.T. Cloud-based bioinformatics workflow platform for large-scale next-generation sequencing analyses. J. Biomed. Inform. 2014, 49, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Zou, X.; Jiang, H.; Zhou, Y.; Liu, C.; Feng, D.; Hua, Y.; Hu, Y.; Zhang, Y. The design of fast content-defined chunking for data deduplication based storage systems. IEEE Trans. Parallel Distrib. Syst. 2020, 31, 2017–2031. [Google Scholar] [CrossRef]
- Kuruppu, S.; Puglisi, S.J.; Zobel, J. Optimized relative Lempel-Ziv compression of genomes. In Proceedings of the Thirty-Fourth Australasian Computer Science Conference, Perth, Australia, 17–20 January 2011; Volume 113, pp. 91–98. [Google Scholar]
- Manber, U.; Myers, G. Suffix arrays: A new method for on-line string searches. SIAM J. Comput. 1993, 22, 935–948. [Google Scholar] [CrossRef]
- Ye, J.; McGinnis, S.; Madden, T.L. BLAST: Improvements for better sequence analysis. Nucleic Acids Res. 2006, 34, W6–W9. [Google Scholar] [CrossRef]
- Buels, R.; Yao, E.; Diesh, C.M.; Hayes, R.D.; Munoz-Torres, M.; Helt, G.; Goodstein, D.M.; Elsik, C.G.; Lewis, S.E.; Stein, L.; et al. JBrowse: A dynamic web platform for genome visualization and analysis. Genome Biol. 2016, 17, 66. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal omega. Curr. Protoc. Bioinform. 2014, 48, 3–13. [Google Scholar] [CrossRef]
- Vangoor, B.K.R.; Tarasov, V.; Zadok, E. To FUSE or Not to FUSE: Performance of User-Space File Systems. In Proceedings of the 15th USENIX Conference on File and Storage Technologies (FAST 17), Santa Clara, CA, USA, 27 February–2 March 2017; pp. 59–72. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Quinlan, A.R. BEDTools: The Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinform. 2014, 47, 11–12. [Google Scholar] [CrossRef]
- Carver, T.; Böhme, U.; Otto, T.D.; Parkhill, J.; Berriman, M. BamView: Viewing mapped read alignment data in the context of the reference sequence. Bioinformatics 2010, 26, 676–677. [Google Scholar] [CrossRef]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef]
- Schmieder, R.; Lim, Y.W.; Rohwer, F.; Edwards, R. TagCleaner: Identification and removal of tag sequences from genomic and metagenomic datasets. BMC Bioinform. 2010, 11, 341. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Luo, X.; Qian, J.; Pang, X.; Song, J.; Qian, G.; Chen, J.; Chen, S. FastUniq: A fast de novo duplicates removal tool for paired short reads. PLoS ONE 2012, 7, e52249. [Google Scholar] [CrossRef] [PubMed]
- de Ruijter, A.; Guldenmund, F. The bowtie method: A review. Saf. Sci. 2016, 88, 211–218. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Wei, Z.; Wang, W.; Hu, P.; Lyon, G.J.; Hakonarson, H. SNVer: A statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic Acids Res. 2011, 39, e132. [Google Scholar] [CrossRef]
- Cao, W.; Liu, Z.; Wang, P.; Chen, S.; Zhu, C.; Zheng, S.; Wang, Y.; Ma, G. PolarFS: An ultra-low latency and failure resilient distributed file system for shared storage cloud database. Proc. VLDB Endow. 2018, 11, 1849–1862. [Google Scholar] [CrossRef]
- CNCB-NGDC Members and Partners. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2021, 50, D27–D38. [Google Scholar]
- Eager, D.L.; Zahorjan, J.; Lazowska, E.D. Speedup versus efficiency in parallel systems. IEEE Trans. Comput. 1989, 38, 408–423. [Google Scholar] [CrossRef]
| Encoder/Decoder Name | Function |
|---|---|
| compression [9] | Transparent compression |
| seqmeta [10] | Sequence metadata extraction |
| blastdb [16] | BLAST database construction |
| jbrowse [17] | JBrowse database construction |
| clustal [18] | Clustal database construction |
| Utility | Function |
|---|---|
| BLAST [16] | Basic local alignment search tool |
| Clustal Omega [18] | Multiple sequence alignment program |
| SamTools [20] | Tools for high-throughput sequencing data |
| Bedtool [21] | Genomics analysis toolkit |
| BamView [22] | Display alignments in BAM files |
| FastQC [23] | Quality control tool |
| TagCleaner [24] | Detect and remove tag sequences |
| FastUniq [25] | de novo duplicates removal tool |
| Bowtie2 [26] | Sequencing reads alignment |
| HISAT2 [27] | Sequencing reads mapping tool |
| SNVer [28] | SNP calling |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Sun, L.; Meng, F. Distributed Big Data Storage Infrastructure for Biomedical Research Featuring High-Performance and Rich-Features. Future Internet 2022, 14, 273. https://doi.org/10.3390/fi14100273
Xu X, Sun L, Meng F. Distributed Big Data Storage Infrastructure for Biomedical Research Featuring High-Performance and Rich-Features. Future Internet. 2022; 14(10):273. https://doi.org/10.3390/fi14100273
Chicago/Turabian StyleXu, Xingjian, Lijun Sun, and Fanjun Meng. 2022. "Distributed Big Data Storage Infrastructure for Biomedical Research Featuring High-Performance and Rich-Features" Future Internet 14, no. 10: 273. https://doi.org/10.3390/fi14100273
APA StyleXu, X., Sun, L., & Meng, F. (2022). Distributed Big Data Storage Infrastructure for Biomedical Research Featuring High-Performance and Rich-Features. Future Internet, 14(10), 273. https://doi.org/10.3390/fi14100273







