Deep Learning-Based Classification of Fine Hand Movements from Low Frequency EEG
Abstract
1. Introduction
2. Related Works
3. Materials and Methods
3.1. Experimental Protocol
3.1.1. Experiment 1—Touch and Grasp
3.1.2. Experiment 2—Palmar and Lateral
3.2. Pre-Processing
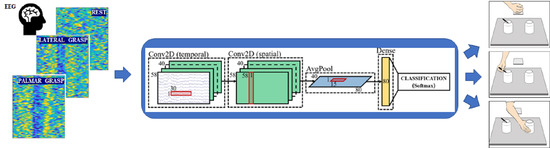
3.3. Classification with CNN
3.4. Classification with Baseline Models
3.5. Cross-Validation and Performance Evaluation
4. Results and Discussion
4.1. Pre-Processing, Feature Extraction, and MRCPs
4.2. Classification Results
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BCI | brain–computer interface |
| CAR | common average reference |
| CNN | convolutional neural network |
| DL | deep learning |
| EEG | electroencephalography |
| eLu | exponential linear unit |
| EOG | electrooculogram |
| ICA | independent component analysis |
| LDA | linear discriminant analysis |
| MRCPs | movement-related cortical potentials |
| RF | random forest |
| sLDA | shrinkage linear discriminant analysis |
Appendix A
| Subject | CNN | sLDA (0.6 s) | sLDA (0.8 s) | sLDA (1 s) | RF (0.6 s) | RF (0.8 s) | RF (1 s) |
|---|---|---|---|---|---|---|---|
| S000 | 0.62 | 0.60 | 0.64 | 0.64 | 0.58 | 0.58 | 0.58 |
| S001 | 0.66 | 0.50 | 0.52 | 0.60 | 0.54 | 0.56 | 0.69 |
| S004 | 0.74 | 0.75 | 0.73 | 0.75 | 0.69 | 0.73 | 0.69 |
| S006 | 0.83 | 0.76 | 0.72 | 0.70 | 0.86 | 0.74 | 0.83 |
| S007 | 0.68 | 0.75 | 0.79 | 0.75 | 0.77 | 0.73 | 0.80 |
| S008 | 0.84 | 0.75 | 0.79 | 0.85 | 0.88 | 0.79 | 0.93 |
| S009 | 0.61 | 0.61 | 0.67 | 0.50 | 0.61 | 0.57 | 0.61 |
| S010 | 0.58 | 0.52 | 0.54 | 0.57 | 0.52 | 0.50 | 0.46 |
| MEAN | 0.70 | 0.66 | 0.68 | 0.67 | 0.68 | 0.65 | 0.70 |
| STD | 0.10 | 0.11 | 0.10 | 0.11 | 0.14 | 0.11 | 0.15 |
| Subject | CNN | sLDA (0.6 s) | sLDA (0.8 s) | sLDA (1 s) | RF (0.6 s) | RF (0.8 s) | RF (1 s) |
|---|---|---|---|---|---|---|---|
| S000 | 0.62 | 0.59 | 0.65 | 0.64 | 0.58 | 0.58 | 0.60 |
| S001 | 0.65 | 0.49 | 0.54 | 0.62 | 0.53 | 0.55 | 0.68 |
| S004 | 0.76 | 0.78 | 0.74 | 0.76 | 0.69 | 0.73 | 0.71 |
| S006 | 0.83 | 0.77 | 0.73 | 0.70 | 0.88 | 0.73 | 0.87 |
| S007 | 0.69 | 0.75 | 0.81 | 0.78 | 0.80 | 0.73 | 0.79 |
| S008 | 0.84 | 0.77 | 0.79 | 0.85 | 0.88 | 0.82 | 0.94 |
| S009 | 0.62 | 0.62 | 0.67 | 0.51 | 0.61 | 0.56 | 0.64 |
| S010 | 0.59 | 0.51 | 0.54 | 0.57 | 0.53 | 0.55 | 0.54 |
| MEAN | 0.70 | 0.66 | 0.68 | 0.68 | 0.69 | 0.66 | 0.72 |
| STD | 0.10 | 0.12 | 0.10 | 0.12 | 0.15 | 0.11 | 0.14 |
| Subject | CNN | sLDA (0.6 s) | sLDA (0.8 s) | sLDA (1 s) | RF (0.6 s) | RF (0.8 s) | RF (1 s) |
|---|---|---|---|---|---|---|---|
| G01 | 0.79 | 0.64 | 0.65 | 0.65 | 0.60 | 0.59 | 0.66 |
| G02 | 0.43 | 0.61 | 0.55 | 0.53 | 0.49 | 0.63 | 0.47 |
| G03 | 0.59 | 0.51 | 0.59 | 0.68 | 0.59 | 0.53 | 0.53 |
| G04 | 0.58 | 0.67 | 0.63 | 0.61 | 0.52 | 0.58 | 0.54 |
| G05 | 0.75 | 0.52 | 0.62 | 0.64 | 0.58 | 0.62 | 0.56 |
| G06 | 0.55 | 0.50 | 0.59 | 0.64 | 0.68 | 0.51 | 0.49 |
| G07 | 0.59 | 0.57 | 0.49 | 0.53 | 0.56 | 0.59 | 0.50 |
| G08 | 0.73 | 0.79 | 0.75 | 0.79 | 0.68 | 0.77 | 0.71 |
| G09 | 0.74 | 0.69 | 0.69 | 0.62 | 0.57 | 0.71 | 0.63 |
| G10 | 0.63 | 0.66 | 0.54 | 0.55 | 0.63 | 0.61 | 0.63 |
| G11 | 0.61 | 0.55 | 0.59 | 0.63 | 0.56 | 0.49 | 0.55 |
| G12 | 0.81 | 0.56 | 0.58 | 0.58 | 0.66 | 0.66 | 0.52 |
| G13 | 0.58 | 0.53 | 0.45 | 0.56 | 0.51 | 0.63 | 0.51 |
| G14 | 0.60 | 0.63 | 0.68 | 0.74 | 0.54 | 0.59 | 0.54 |
| G15 | 0.65 | 0.63 | 0.71 | 0.59 | 0.57 | 0.61 | 0.55 |
| MEAN | 0.64 | 0.60 | 0.61 | 0.62 | 0.58 | 0.61 | 0.56 * |
| STD | 0.10 | 0.08 | 0.08 | 0.07 | 0.06 | 0.07 | 0.07 |
| Subject | CNN | sLDA (0.6 s) | sLDA (0.8 s) | sLDA (1 s) | RF (0.6 s) | RF (0.8 s) | RF (1 s) |
|---|---|---|---|---|---|---|---|
| G01 | 0.80 | 0.64 | 0.64 | 0.65 | 0.60 | 0.58 | 0.67 |
| G02 | 0.46 | 0.62 | 0.54 | 0.53 | 0.49 | 0.63 | 0.47 |
| G03 | 0.58 | 0.53 | 0.60 | 0.68 | 0.58 | 0.55 | 0.54 |
| G04 | 0.59 | 0.66 | 0.63 | 0.62 | 0.52 | 0.57 | 0.52 |
| G05 | 0.74 | 0.54 | 0.62 | 0.64 | 0.57 | 0.62 | 0.56 |
| G06 | 0.55 | 0.52 | 0.57 | 0.63 | 0.68 | 0.67 | 0.51 |
| G07 | 0.60 | 0.58 | 0.45 | 0.53 | 0.57 | 0.58 | 0.49 |
| G08 | 0.73 | 0.81 | 0.75 | 0.80 | 0.67 | 0.77 | 0.73 |
| G09 | 0.75 | 0.69 | 0.69 | 0.64 | 0.59 | 0.72 | 0.64 |
| G10 | 0.64 | 0.65 | 0.44 | 0.51 | 0.62 | 0.59 | 0.63 |
| G11 | 0.61 | 0.55 | 0.62 | 0.64 | 0.56 | 0.48 | 0.53 |
| G12 | 0.80 | 0.57 | 0.57 | 0.57 | 0.66 | 0.66 | 0.54 |
| G13 | 0.59 | 0.53 | 0.46 | 0.54 | 0.51 | 0.64 | 0.50 |
| G14 | 0.60 | 0.64 | 0.67 | 0.73 | 0.54 | 0.58 | 0.54 |
| G15 | 0.67 | 0.63 | 0.72 | 0.59 | 0.57 | 0.62 | 0.55 |
| MEAN | 0.65 | 0.61 | 0.60 | 0.62 | 0.58 * | 0.62 | 0.56 * |
| STD | 0.10 | 0.08 | 0.10 | 0.08 | 0.06 | 0.07 | 0.07 |
References
- Cisotto, G.; Pupolin, S.; Silvoni, S.; Cavinato, M.; Agostini, M.; Piccione, F. Brain-computer interface in chronic stroke: An application of sensorimotor closed-loop and contingent force feedback. In Proceedings of the 2013 IEEE International Conference on Communications (ICC), Budapest, Hungary, 9–13 June 2013; pp. 4379–4383. [Google Scholar]
- Silvoni, S.; Cavinato, M.; Volpato, C.; Cisotto, G.; Genna, C.; Agostini, M.; Turolla, A.; Ramos-Murguialday, A.; Piccione, F. Kinematic and neurophysiological consequences of an assisted-force-feedback brain-machine interface training: A case study. Front. Neurol. 2013, 4, 173. [Google Scholar] [CrossRef]
- Cisotto, G.; Pupolin, S.; Cavinato, M.; Piccione, F. An EEG-based BCI platform to improve arm reaching ability of chronic stroke patients by means of an operant learning training with a contingent force feedback. Int. J. E-Health Med. Commun. (IJEHMC) 2014, 5, 114–134. [Google Scholar] [CrossRef][Green Version]
- Biasiucci, A.; Leeb, R.; Iturrate, I.; Perdikis, S.; Al-Khodairy, A.; Corbet, T.; Schnider, A.; Schmidlin, T.; Zhang, H.; Bassolino, M.; et al. Brain-actuated functional electrical stimulation elicits lasting arm motor recovery after stroke. Nat. Commun. 2018, 9, 2421. [Google Scholar] [CrossRef] [PubMed]
- Ofner, P.; Schwarz, A.; Pereira, J.; Wyss, D.; Wildburger, R.; Müller-Putz, G.R. Attempted Arm and Hand Movements can be Decoded from Low-Frequency EEG from Persons with Spinal Cord Injury. Sci. Rep. 2019, 9, 7134. [Google Scholar] [CrossRef]
- Müller-Putz, G.R.; Ofner, P.; Pereira, J.; Pinegger, A.; Schwarz, A.; Zube, M.; Eck, U.; Hessing, B.; Schneiders, M.; Rupp, R. Applying intuitive EEG-controlled grasp neuroprostheses in individuals with spinal cord injury: Preliminary results from the MoreGrasp clinical feasibility study. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 5949–5955. [Google Scholar] [CrossRef]
- Furuya, S.; Hanakawa, T. The curse of motor expertise: Use-dependent focal dystonia as a manifestation of maladaptive changes in body representation. Neurosci. Res. 2016, 104, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Cisotto, G.; Kita, K.; Uehara, K.; Yoshinaga, K.; Hashimoto, Y.; Sakamoto, T.; Junichi, U.; Takashi, H. Abnormal electroencephalographic oscillations in β and low γ bands in patients with writer’s cramp. (poster presentation). In Proceedings of the Annual Meeting of Society for Neuroscience, Washington, DC, USA, 11–15 November 2017. [Google Scholar]
- Packheiser, J.; Schmitz, J.; Pan, Y.; El Basbasse, Y.; Friedrich, P.; Güntürkün, O.; Ocklenburg, S. Using mobile EEG to investigate alpha and beta asymmetries during hand and foot use. Front. Neurosci. 2020, 14, 109. [Google Scholar] [CrossRef]
- Shibasaki, H.; Hallett, M. What is the Bereitschaftspotential? Clin. Neurophysiol. 2006, 117, 2341–2356. [Google Scholar] [CrossRef]
- Sharma, N.; Pomeroy, V.M.; Baron, J.C. Motor imagery: A backdoor to the motor system after stroke? Stroke 2006, 37, 1941–1952. [Google Scholar] [CrossRef]
- Pereira, J.; Sburlea, A.I.; Müller-Putz, G.R. EEG patterns of self-paced movement imaginations towards externally-cued and internally-selected targets. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef]
- Jochumsen, M.; Niazi, I.K.; Dremstrup, K.; Kamavuako, E.N. Detecting and classifying three different hand movement types through electroencephalography recordings for neurorehabilitation. Med. Biol. Eng. Comput. 2016, 54, 1491–1501. [Google Scholar] [CrossRef]
- Jochumsen, M.; Niazi, I.K.; Mrachacz-Kersting, N.; Farina, D.; Dremstrup, K. Detection and classification of movement-related cortical potentials associated with task force and speed. J. Neural Eng. 2013, 10, 056015. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Dremstrup, K.; Farina, D. Single-trial discrimination of type and speed of wrist movements from EEG recordings. Clin. Neurophysiol. 2009, 120, 1596–1600. [Google Scholar] [CrossRef]
- Schwarz, A.; Ofner, P.; Pereira, J.; Sburlea, A.I.; Mueller-Putz, G.R. Decoding natural reach-and-grasp actions from human EEG. J. Neural Eng. 2017, 15, 016005. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, A.; Pereira, J.; Kobler, R.; Müller-Putz, G.R. Unimanual and Bimanual Reach-and-Grasp Actions Can Be Decoded From Human EEG. IEEE Trans. Biomed. Eng. 2019, 67, 1684–1695. [Google Scholar] [CrossRef] [PubMed]
- Ofner, P.; Schwarz, A.; Pereira, J.; Müller-Putz, G.R. Upper limb movements can be decoded from the time-domain of low-frequency EEG. PLoS ONE 2017, 12, e0182578. [Google Scholar]
- Lotte, F.; Congedo, M.; Lécuyer, A.; Lamarche, F.; Arnaldi, B. A review of classification algorithms for EEG-based brain–computer interfaces. J. Neural Eng. 2007, 4, R1. [Google Scholar] [CrossRef]
- Jadhav, P.; Rajguru, G.; Datta, D.; Mukhopadhyay, S. Automatic sleep stage classification using time–frequency images of CWT and transfer learning using convolution neural network. Biocybern. Biomed. Eng. 2020, 40, 494–504. [Google Scholar] [CrossRef]
- Cisotto, G.; Zanga, A.; Chlebus, J.; Zoppis, I.; Manzoni, S.; Markowska-Kaczmar, U. Comparison of Attention-based Deep Learning Models for EEG Classification. arXiv 2020, arXiv:2012.01074. [Google Scholar]
- Zhang, G.; Davoodnia, V.; Sepas-Moghaddam, A.; Zhang, Y.; Etemad, A. Classification of hand movements from EEG using a deep attention-based LSTM network. IEEE Sens. J. 2019, 20, 3113–3122. [Google Scholar] [CrossRef]
- Dose, H.; Møller, J.S.; Iversen, H.K.; Puthusserypady, S. An end-to-end deep learning approach to MI-EEG signal classification for BCIs. Expert Syst. Appl. 2018, 114, 532–542. [Google Scholar] [CrossRef]
- Lee, B.H.; Jeong, J.H.; Shim, K.H.; Kim, D.J. Motor Imagery Classification of Single-Arm Tasks Using Convolutional Neural Network based on Feature Refining. In 2020 8th International Winter Conference on Brain-Computer Interface (BCI); IEEE: Piscataway, NJ, USA, 2020; pp. 1–5. [Google Scholar]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A compact convolutional neural network for EEG-based brain—Computer interfaces. J. Neural Eng. 2018, 15, 056013. [Google Scholar] [CrossRef]
- Robinson, N.; Vinod, A.P.; Ang, K.K.; Tee, K.P.; Guan, C.T. EEG-based classification of fast and slow hand movements using wavelet-CSP algorithm. IEEE Trans. Biomed. Eng. 2013, 60, 2123–2132. [Google Scholar] [CrossRef]
- Lotte, F.; Bougrain, L.; Cichocki, A.; Clerc, M.; Congedo, M.; Rakotomamonjy, A.; Yger, F. A review of classification algorithms for EEG-based brain–computer interfaces: A 10 year update. J. Neural Eng. 2018, 15, 031005. [Google Scholar] [CrossRef]
- Kumar, P.; Saini, R.; Roy, P.P.; Sahu, P.K.; Dogra, D.P. Envisioned speech recognition using EEG sensors. Pers. Ubiquitous Comput. 2018, 22, 185–199. [Google Scholar] [CrossRef]
- Steingrüber, H.J.; Lienert, G.A. Hand-Dominanz-Test; HDT: Verlag für Psychologie, Hogrefe, 1971. [Google Scholar]
- Oostenveld, R.; Praamstra, P. The five percent electrode system for high-resolution EEG and ERP measurements. Clin. Neurophysiol. 2001, 112, 713–719. [Google Scholar] [CrossRef]
- The Mathworks, Inc. MATLAB Version 9.8.0.1359463 (R2020a); The Mathworks, Inc.: Natick, MA, USA, 2020. [Google Scholar]
- Makeig, S.; Bell, A.J.; Jung, T.P.; Sejnowski, T.J. Independent Component Analysis of Electroencephalographic Data. Available online: https://papers.nips.cc/paper/1995/file/754dda4b1ba34c6fa89716b85d68532b-Paper.pdf (accessed on 15 April 2021).
- Choi, H.; Li, X.; Lau, S.T.; Hu, C.; Zhou, Q.; Shung, K.K. Development of integrated preamplifier for high-frequency ultrasonic transducers and low-power handheld receiver. IEEE Trans. Ultrason. Ferroelectr. Freq. Control. 2011, 58, 2646–2658. [Google Scholar] [CrossRef]
- Ludwig, K.A.; Miriani, R.M.; Langhals, N.B.; Joseph, M.D.; Anderson, D.J.; Kipke, D.R. Using a common average reference to improve cortical neuron recordings from microelectrode arrays. J. Neurophysiol. 2009, 101, 1679–1689. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, A.; Scherer, R.; Steyrl, D.; Faller, J.; Müller-Putz, G.R. A co-adaptive sensory motor rhythms brain-computer interface based on common spatial patterns and random forest. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; pp. 1049–1052. [Google Scholar]
- Statthaler, K.; Schwarz, A.; Steyrl, D.; Kobler, R.; Höller, M.K.; Brandstetter, J.; Hehenberger, L.; Bigga, M.; Müller-Putz, G. Cybathlon experiences of the Graz BCI racing team Mirage91 in the brain-computer interface discipline. J. Neuroeng. Rehabil. 2017, 14, 1–16. [Google Scholar] [CrossRef]
- Faller, J.; Vidaurre, C.; Solis-Escalante, T.; Neuper, C.; Scherer, R. Autocalibration and recurrent adaptation: Towards a plug and play online ERD-BCI. IEEE Trans. Neural Syst. Rehabil. Eng. 2012, 20, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Bengio, Y.; Courville, A.; Bengio, Y. Deep Learning; MIT Press Cambridge: Cambridge, MA, USA, 2016; Volume 1. [Google Scholar]
- Bartz, D.; Müller, K.R. Covariance Shrinkage for Autocorrelated Data. Available online: https://papers.nips.cc/paper/2014/file/fa83a11a198d5a7f0bf77a1987bcd006-Paper.pdf (accessed on 15 April 2021).
- Shalev-Shwartz, S.; Ben-David, S. Understanding Machine Learning: From Theory to Algorithms; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Müller-Putz, G.; Scherer, R.; Brunner, C.; Leeb, R.; Pfurtscheller, G. Better than random: A closer look on BCI results. Int. J. Bioelectromagn. 2008, 10, 52–55. [Google Scholar]
- Rice, N.J.; Tunik, E.; Cross, E.S.; Grafton, S.T. On-line grasp control is mediated by the contralateral hemisphere. Brain Res. 2007, 1175, 76–84. [Google Scholar] [CrossRef] [PubMed]
Short Biography of Authors
 | Giulia Bressan currently works as data science and data warehouse engineer at Reply, Italy. She received her B.Sc. in Information Engineering in 2018 and her M.Sc. in ICT for Internet and Multimedia in 2020 from the University of Padova. She focused her studies on the healthcare applications of ICT, in particular regarding telemedicine and e-health. In 2020 she was a visiting student at the Institute of Neural Engineering (BCI-Lab), Graz University of Technology (TUG) where, in collaboration with the Dept. Information Engineering of the University of Padova, she developed her M.Sc. Thesis project involving the comparison of classification techniques for EEG signals. |
 | Giulia Cisotto received her M.Sc. in Telecommunication Engineering in 2010 and PhD in Information Engineering in 2014 from University of Padova (Italy). From 2014 to 2015, she was Research Associate at Keio University. Since 2019, she is non-tenured Assistant Professor at University of Padova and member of SIGNET Lab. She is also Visiting Scientist at NCNP of Tokyo (Japan). In her ten-year research, she gained experience on EEG analysis and BCI for rehabilitation, working with clinical Institutes and healthcare companies (IRCCS San Camillo, IRCCS Santa Lucia, BrainTrends srl, Italy). She has published several Journal papers, conference articles and 2 book chapters (no.citations = 299, h-index = 7). In 2018, she was awarded an IEEE Outstanding Paper Award at IEEE Healthcom. She is reviewer and TPC member for several MDPI, IEEE, Elsevier Journals and international conferences. She is Guest Editor for Frontiers in Human Neuroscience: Brain-Computer Interface. In 2021, she joined the IEEE ComSoc e-Health Technical Committee. |
 | Gernot R. Müller-Putz is Head of the Institute of Neural Engineering and PI of the BCI-Lab at the Graz University of Technology (TUG). He received his MSc in electrical and biomedical engineering in 2000, PhD in electrical engineering in 2004, and habilitation in 2008 from TUG. Since 2014 he is Full Professor for semantic data analysis. He has gained extensive experience in biosignal analysis, BCI and EEG-based neuroprosthesis control. He has authored more than 175 peer-reviewed publications and more than 180 contributions to conferences (no.citations = 18,159, h-index = 68). He serves as Editor for Frontiers in Neuroscience, IEEE T-BME and BCI Journal. In 2018, he joined the Board of Directors of the International BCI Society. Since 2019, he is Speciality Editor-in-Chief of Frontiers in Human Neuroscience: Brain-Computer Interfaces. In 2015, he was awarded with an ERC Consolidator Grant “Feel your Reach”. He is founding member and Co-Director of the NeuroIS Society. |
 | Selina C.Wriessnegger is Assistant Professor and Deputy Head at the Institute of Neural Engineering (BCI-Lab) of the Graz University of Technology (TUG). She received her PhD from the Ludwig- Maximilians University in 2005 for Human Cognitive and Brain Sciences. In 2004, she was research assistant the IRCCS Santa Lucia Foundation of Rome (Italy). From 2005 to 2008 she was University Assistant at the Karl-Franzens-University Graz. She was visiting professor at SISSA of Trieste (2017) and guest professor at the University of Padova (2019). She has authored more than 90 peer reviewed publications (no.citations = 1732, h-index = 20). Since 2019, she is Associate Editor of Frontiers in Human Neuroscience: Brain-Computer Interfaces. In addition, she was in the organizing committee of several international BCI conferences. Her research interests are neural correlates of motor imagery, subliminal visual information processing, novel applications of BCIs for healthy users, VR in cognitive neuroscience and affective computing. |



| Subject | CNN | sLDA (0.6 s) | sLDA (0.8 s) | sLDA (1 s) | RF (0.6 s) | RF (0.8 s) | RF (1 s) |
|---|---|---|---|---|---|---|---|
| S000 | 0.62 | 0.60 | 0.64 | 0.64 | 0.58 | 0.58 | 0.58 |
| S001 | 0.66 | 0.50 | 0.52 | 0.60 | 0.54 | 0.56 | 0.69 |
| S004 | 0.74 | 0.75 | 0.73 | 0.75 | 0.69 | 0.73 | 0.69 |
| S006 | 0.84 | 0.76 | 0.72 | 0.70 | 0.86 | 0.74 | 0.82 |
| S007 | 0.68 | 0.76 | 0.80 | 0.76 | 0.78 | 0.73 | 0.80 |
| S008 | 0.85 | 0.78 | 0.78 | 0.83 | 0.88 | 0.80 | 0.93 |
| S009 | 0.61 | 0.62 | 0.67 | 0.50 | 0.62 | 0.58 | 0.62 |
| S010 | 0.59 | 0.52 | 0.54 | 0.57 | 0.52 | 0.50 | 0.46 |
| MEAN | 0.70 | 0.66 | 0.68 | 0.67 | 0.68 | 0.65 | 0.70 |
| STD | 0.10 | 0.11 | 0.10 | 0.11 | 0.14 | 0.11 | 0.15 |
| Subject | CNN | sLDA (0.6 s) | sLDA (0.8 s) | sLDA (1 s) | RF (0.6 s) | RF (0.8 s) | RF (1 s) |
|---|---|---|---|---|---|---|---|
| G01 | 0.79 | 0.65 | 0.65 | 0.65 | 0.61 | 0.59 | 0.67 |
| G02 | 0.43 | 0.61 | 0.55 | 0.53 | 0.49 | 0.63 | 0.47 |
| G03 | 0.58 | 0.51 | 0.59 | 0.67 | 0.59 | 0.53 | 0.53 |
| G04 | 0.58 | 0.67 | 0.63 | 0.61 | 0.53 | 0.59 | 0.55 |
| G05 | 0.75 | 0.52 | 0.63 | 0.65 | 0.58 | 0.63 | 0.56 |
| G06 | 0.55 | 0.51 | 0.60 | 0.64 | 0.68 | 0.53 | 0.49 |
| G07 | 0.60 | 0.58 | 0.50 | 0.54 | 0.58 | 0.60 | 0.50 |
| G08 | 0.72 | 0.78 | 0.75 | 0.78 | 0.67 | 0.76 | 0.73 |
| G09 | 0.73 | 0.69 | 0.69 | 0.62 | 0.58 | 0.71 | 0.63 |
| G10 | 0.65 | 0.65 | 0.54 | 0.54 | 0.63 | 0.61 | 0.63 |
| G11 | 0.61 | 0.56 | 0.60 | 0.63 | 0.58 | 0.50 | 0.56 |
| G12 | 0.80 | 0.56 | 0.58 | 0.58 | 0.66 | 0.66 | 0.52 |
| G13 | 0.57 | 0.53 | 0.45 | 0.55 | 0.51 | 0.63 | 0.51 |
| G14 | 0.60 | 0.64 | 0.68 | 0.75 | 0.55 | 0.59 | 0.55 |
| G15 | 0.65 | 0.63 | 0.71 | 0.59 | 0.57 | 0.61 | 0.55 |
| MEAN | 0.64 | 0.61 | 0.61 | 0.62 | 0.59 | 0.61 | 0.56 * |
| STD | 0.10 | 0.08 | 0.08 | 0.07 | 0.06 | 0.07 | 0.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bressan, G.; Cisotto, G.; Müller-Putz, G.R.; Wriessnegger, S.C. Deep Learning-Based Classification of Fine Hand Movements from Low Frequency EEG. Future Internet 2021, 13, 103. https://doi.org/10.3390/fi13050103
Bressan G, Cisotto G, Müller-Putz GR, Wriessnegger SC. Deep Learning-Based Classification of Fine Hand Movements from Low Frequency EEG. Future Internet. 2021; 13(5):103. https://doi.org/10.3390/fi13050103
Chicago/Turabian StyleBressan, Giulia, Giulia Cisotto, Gernot R. Müller-Putz, and Selina Christin Wriessnegger. 2021. "Deep Learning-Based Classification of Fine Hand Movements from Low Frequency EEG" Future Internet 13, no. 5: 103. https://doi.org/10.3390/fi13050103
APA StyleBressan, G., Cisotto, G., Müller-Putz, G. R., & Wriessnegger, S. C. (2021). Deep Learning-Based Classification of Fine Hand Movements from Low Frequency EEG. Future Internet, 13(5), 103. https://doi.org/10.3390/fi13050103