Phage Display Selection and In Silico Characterization of Peptides as Potential GroEL Modulators
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Preparation
2.2. Selection of GroEL-Specific Binding Peptides via Phage Display
2.3. In Silico Studies
2.3.1. Protein Structure Preparation
2.3.2. Docking with AutoDock CrankPep (ADCP)
2.3.3. Molecular Dynamics
3. Results
3.1. Groel Purification
3.2. Phage Display
3.3. Docking Studies
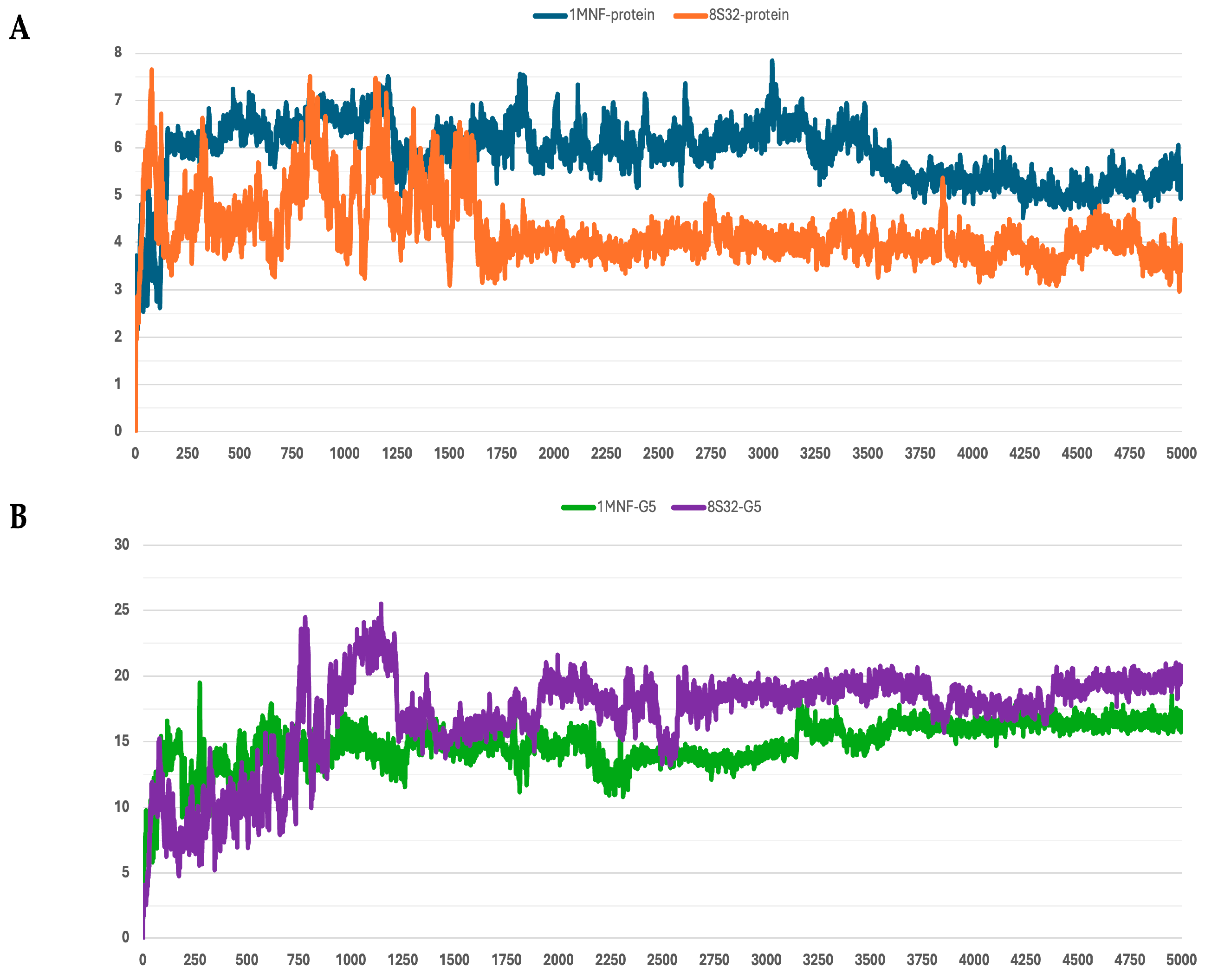
3.4. Molecular Dynamics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RMSD | Root Mean Square Deviation |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| EDTA | Ethylenediaminetetraacetic acid |
| EF-SDS | Sodium dodecyl sulfate–polyacrylamide gel electrophoresis |
| TBS | Tris-Buffered Saline |
| MMGBSA | Molecular Mechanics Generalized Born Surface Area |
References
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- World Health Organization. No Time to Wait: Securing the Future from Drug-Resistant Infections; WHO: Geneva, Switzerland, 2019; p. 28. [Google Scholar]
- Lund, P.A. Microbial molecular chaperones. Adv. Microb. Physiol. 2001, 44, 93–140. [Google Scholar] [CrossRef] [PubMed]
- Henderson, B.; Fares, M.A.; Lund, P.A. Chaperonin 60: A paradoxical, evolutionarily conserved protein family with multiple moonlighting functions. Biol. Rev. 88, 955–987. [CrossRef]
- Horwich, A.L.; Fenton, W.A. Chaperonin-mediated protein folding: Using a central cavity to kinetically assist polypeptide chain folding. Q. Rev. Biophys. 2009, 42, 83–116. [Google Scholar] [CrossRef]
- Clare, D.K.; Vasishtan, D.; Stagg, S.; Quispe, J.; Farr, G.W.; Topf, M.; Horwich, A.L.; Saibil, H.R. ATP-triggered conformational changes delineate substrate-binding and -folding mechanics of the GroEL chaperonin. Cell 2012, 149, 113–123. [Google Scholar] [CrossRef]
- Ranson, N.A.; Farr, G.W.; Roseman, A.M.; Gowen, B.; Fenton, W.A.; Horwich, A.L.; Saibil, H.R. ATP-bound states of GroEL captured by cryo-electron microscopy. Cell 2001, 107, 869–879. [Google Scholar] [CrossRef]
- Kumar, C.M.; Mande, S.C.; Mahajan, G. Multiple chaperonins in bacteria—Novel functions and non-canonical behaviors. Cell Stress Chaperones 2015, 20, 555–574. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Sigler, P.B. GroEL/GroES: Structure and function of a two-stroke folding machine. J. Struct. Biol. 1998, 124, 129–141. [Google Scholar] [CrossRef]
- Gardner, S.; Darrow, M.C.; Lukoyanova, N.; Thalassinos, K.; Saibil, H.R. Structural basis of substrate progression through the bacterial chaperonin cycle. Proc. Natl. Acad. Sci. USA 2023, 120, e2308933120. [Google Scholar] [CrossRef]
- Dupuy, J.F.; Collet, J.F. Entering deeper into the mysteries of the GroEL–GroES nanomachine. Curr. Opin. Microbiol. 2024, 79, 102480. [Google Scholar] [CrossRef]
- Fourie, K.R.; Wilson, H.L. Understanding GroEL and DnaK stress response proteins as antigens for bacterial diseases. Vaccines 2020, 8, 773. [Google Scholar] [CrossRef]
- Abdeen, S.; Salim, N.; Mammadova, N.; Summers, C.M.; Frankson, R.; Ambrose, A.J.; Anderson, G.G.; Schultz, P.G.; Horwich, A.L.; Chapman, E.; et al. GroEL/ES inhibitors as potential antibiotics. Bioorg. Med. Chem. Lett. 2016, 26, 3127–3134. [Google Scholar] [CrossRef]
- Stevens, M.; Howe, C.; Ray, A.M.; Washburn, A.; Chitre, S.; Sivinski, J.; Park, Y.; Hoang, Q.Q.; Chapman, E.; Johnson, S.M. Analogs of nitrofuran antibiotics are potent GroEL/ES inhibitor pro-drugs. Bioorg. Med. Chem. 2020, 28, 115710. [Google Scholar] [CrossRef] [PubMed]
- Kunkle, T.; Abdeen, S.; Salim, N.; Ray, A.M.; Stevens, M.; Ambrose, A.J.; Victorino, J.; Park, Y.; Hoang, Q.Q.; Chapman, E.; et al. Hydroxybiphenylamide GroEL/ES inhibitors are potent antibacterials against planktonic and biofilm forms of Staphylococcus aureus. J. Med. Chem. 2018, 61, 10651–10664. [Google Scholar] [CrossRef]
- Abdeen, S.; Kunkle, T.; Salim, N.; Ray, A.M.; Mammadova, N.; Summers, C.; Stevens, M.; Ambrose, A.J.; Park, Y.; Schultz, P.G.; et al. Sulfonamido-2-arylbenzoxazole GroEL/ES inhibitors as potent antibacterials against methicillin-resistant Staphylococcus aureus (MRSA). J. Med. Chem. 2018, 61, 7345–7357. [Google Scholar] [CrossRef]
- Li, Y.; Zheng, Z.; Ramsey, A.; Chen, L. Analysis of peptides and proteins in their binding to GroEL. J. Pept. Sci. 2010, 16, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, H.; Lu, X.; Chi, H.; Li, S.; Huang, F. Probing the interaction mechanisms between transmembrane peptides and the chaperonin GroEL with fluorescence anisotropy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 194, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Otvos, L., Jr.; O, I.; Rogers, M.E.; Consolvo, P.J.; Condie, B.A.; Lovas, S.; Bulet, P.; Blaszczyk-Thurin, M. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry 2000, 39, 14150–14159. [Google Scholar] [CrossRef]
- Volke, D.; Krizsan, A.; Berthold, N.; Knappe, D.; Hoffmann, R. Identification of Api88 binding partners in Escherichia coli using a photoaffinity-cross-link strategy and label-free quantification. J. Proteome Res. 2015, 14, 3274–3283. [Google Scholar] [CrossRef]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef]
- Paschke, M.; Höhne, W. Phage display systems and their applications. Appl. Microbiol. Biotechnol. 2005, 70, 2–11. [Google Scholar] [CrossRef]
- Burston, S.G.; Weissman, J.S.; Farr, G.W.; Fenton, W.A.; Horwich, A.L. Release of both native and non-native proteins from a cis-only GroEL ternary complex. Nature 1996, 383, 96–99. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Wang, J.; Chen, L. Domain Motions in GroEL upon Binding of an Oligopeptide. J. Mol. Biol. 2003, 334, 489–499. [Google Scholar] [CrossRef]
- Bartolucci, C.; Lamba, D.; Grazulis, S.; Manakova, E.; Heumann, H. Crystal Structure of Wild-type Chaperonin GroEL. J. Mol. Biol. 2005, 354, 940–951. [Google Scholar] [CrossRef]
- Izert-Nowakowska, M.A.; Klimecka, M.M.; Antosiewicz, A.; Wróblewski, K.; Kowalski, J.J.; Bandyra, K.J.; Góral, T.; Kmiecik, S.; Serwa, R.A.; Górna, M.W. Targeted protein degradation in Escherichia coli using CLIPPERs. EMBO Rep. 2025, 26, 3994–4016. [Google Scholar] [CrossRef]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Søndergaard, C.R.; Olsson, M.H.; Rostkowski, M.; Jensen, J.H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 2011, 7, 2284–2295. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sanner, M.F. AutoDock CrankPep: Combining folding and docking to predict protein–peptide complexes. Bioinformatics 2019, 35, 5121–5127. [Google Scholar] [CrossRef] [PubMed]
- Desmond Molecular Dynamics System, version 2024-3; D.E. Shaw Research; Schrödinger, LLC: New York, NY, USA, 2024.
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Lu, C.; Wu, C.; Ghoreishi, D.; Chen, W.; Wang, L.; Damm, W.; Ross, G.A.; Dahlgren, M.K.; Russell, E.; Von Bargen, C.D.; et al. OPLS4: Improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput. 2021, 17, 4291–4300. [Google Scholar] [CrossRef] [PubMed]
- Grønbech-Jensen, N.; Farago, O. Constant pressure and temperature discrete-time Langevin molecular dynamics. J. Chem. Phys. 2014, 141, 194108. [Google Scholar] [CrossRef] [PubMed]
- Frey, B.J.; Dueck, D. Clustering by passing messages between data points. Science 2007, 315, 972–976. [Google Scholar] [CrossRef]
- Kragol, G.; Lovas, S.; Varadi, G.; Condie, B.A.; Hoffmann, R.; Otvos, L. The antibacterial peptide pyrrhocoricin inhibits the ATPase actions of DnaK and prevents chaperone-assisted protein folding. Biochemistry 2001, 40, 3016–3026. [Google Scholar] [CrossRef] [PubMed]






| Peptide | Net Charge | GRAVY | WW Whole-Residue Hydrophobicity | Amphipaticity Index | H-Moment | Predicted Membrane Penetration * |
|---|---|---|---|---|---|---|
| G1 | +0.75 | −2.125 | 1.48 | 0.45 | 0.85 | low |
| G2 | +2 | 0.275 | 1.01 | 0.27 | 1.24 | low |
| G3 | +0.25 | −1.03 | 5.75 | 0.45 | 0.76 | low |
| G4 | +0.5 | −0.38 | 1.59 | 0.36 | 0.89 | low |
| G5 | 0 | −0.833 | 0.97 | 0.36 | 0.94 | low |
| Peptide | Results Docking 8S32 Kcal/mol | Results Docking 1MNF Kcal/mol | Results Docking 1XCK Kcal/mol |
|---|---|---|---|
| G1 | −22.1 | −22.2 | −21.6 |
| G2 | −20.8 | −19.5 | −20.4 |
| G3 | −19.9 | −17.1 | −17.8 |
| G4 | −20.5 | −19.8 | −20.7 |
| G5 | −19.6 | −19.5 | −18.7 |
| Peptide | RMSD 8S32 | RMSD 1MNF | RMSD 1XCK |
|---|---|---|---|
| G1 peptide | 15.58 ± 2.33 | 29.92 ± 14.74 | |
| protein | 3.09 ± 0.32 | 3.64 ± 0.49 | |
| G2 peptide | 29.72 ± 5.99 | 14.26 ± 3.85 | |
| protein | 4.47 ± 0.98 | 7.35 ± 2.12 | |
| G3 peptide | 20.91 ± 3.49 | 19.19 ± 4.37 | |
| protein | 3.98 ± 0.79 | 4.44 ± 0.84 | |
| G4 peptide | 6.33 ± 1.0 | 12.04 ± 2.04 | |
| protein | 3.26 ± 0.66 | 4.08 ± 0.97 | |
| G5 peptide | 17.09 ± 3.57 | 14.82 ± 1.72 | |
| protein | 4.27 ± 0.71 | 5.88 ± 0.70 |
| Peptide | MMGBSA 8S32 Kcal/mol | MMGBSA 1MNF Kcal/mol | MMGBSA 1XCK Kcal/mol |
|---|---|---|---|
| G1 | −45.08 ± 10.85 | −48.10 ± 14.78 | |
| G2 | −59.42 ± 18.35 | −41.65 ± 11.13 | |
| G3 | −37.88 ± 10.62 | −53.29 ± 11.71 | |
| G4 | −116.68 ± 15.85 | −51.32 ± 14.48 | |
| G5 | −58.07 ± 24.04 | −57.48 ± 19.19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Olla, S.; Colombarolli, S.G.; Siguri, C.; Murrau, D.; Vitali, A. Phage Display Selection and In Silico Characterization of Peptides as Potential GroEL Modulators. Pharmaceutics 2026, 18, 46. https://doi.org/10.3390/pharmaceutics18010046
Olla S, Colombarolli SG, Siguri C, Murrau D, Vitali A. Phage Display Selection and In Silico Characterization of Peptides as Potential GroEL Modulators. Pharmaceutics. 2026; 18(1):46. https://doi.org/10.3390/pharmaceutics18010046
Chicago/Turabian StyleOlla, Stefania, Stella Garcia Colombarolli, Chiara Siguri, Davide Murrau, and Alberto Vitali. 2026. "Phage Display Selection and In Silico Characterization of Peptides as Potential GroEL Modulators" Pharmaceutics 18, no. 1: 46. https://doi.org/10.3390/pharmaceutics18010046
APA StyleOlla, S., Colombarolli, S. G., Siguri, C., Murrau, D., & Vitali, A. (2026). Phage Display Selection and In Silico Characterization of Peptides as Potential GroEL Modulators. Pharmaceutics, 18(1), 46. https://doi.org/10.3390/pharmaceutics18010046








