Newborn Intravenous Injection of Liposomal CRISPR/Cas9 Complex Has No Incidence of Off-Targets or Tumors in Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Vectors
2.2. Preparation of Formulations and Complexes
2.3. Characterization of the Complexes
2.4. Animals
2.5. Histological Analyses
2.6. Immediate Biodistribution of Fluorescent Complexes After Newborn Injection
2.7. On and off Target Analysis
- Chr2: F-tcaactgtttgagccagctcaagg and R-ggctttgcctggctaacagattac;
- Chr5: F-acggcaaaggtagcaggcag and R-agcacgcccactacagggtt;
- Chr11: F-gtagataaggagctcaggtagcc and R-ctgccccagatgtagtctgaac;
- Chr17: F-gaagtgtatggctgccatgtgc and R-gtggagtttggatggccttcg;
- Chr6: F-gctctcccaaagtcgctctg and R-ggagcgggagaaatggatatgaag.
2.8. Ethics
2.9. Statistics
3. Results
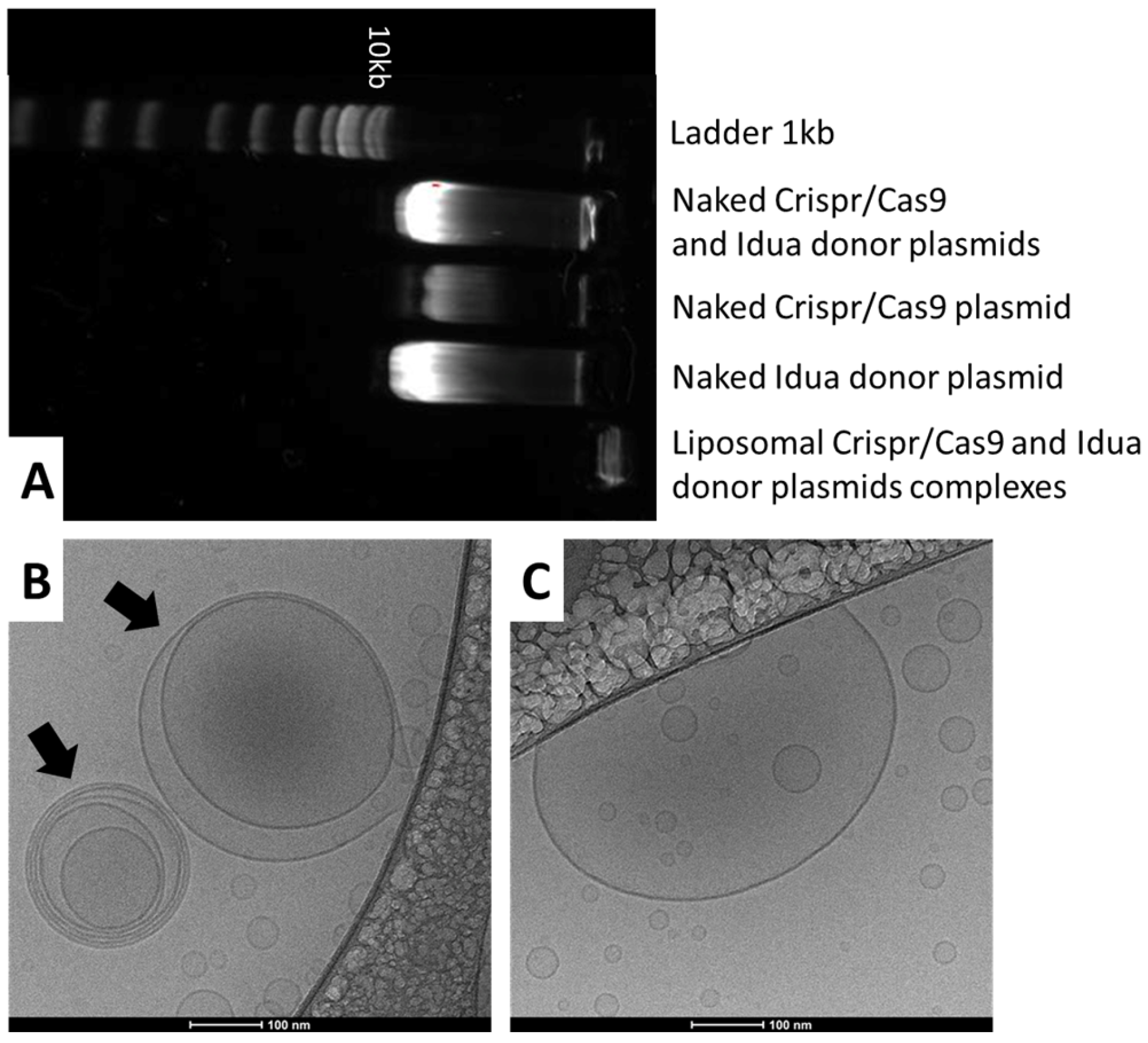
3.1. Physicochemical Characterization of Complexes
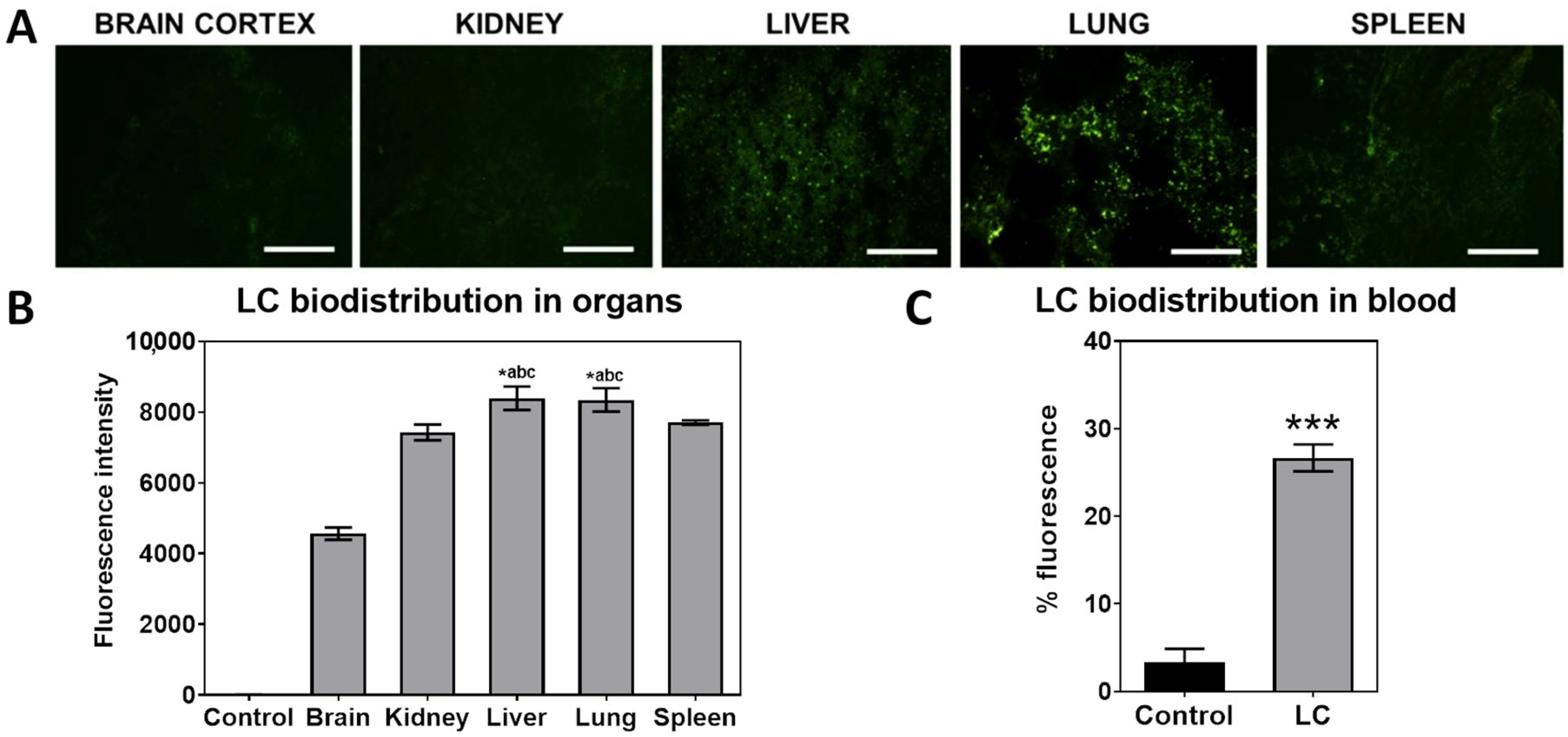
3.2. Vector Biodistribution After Intravenous Injection
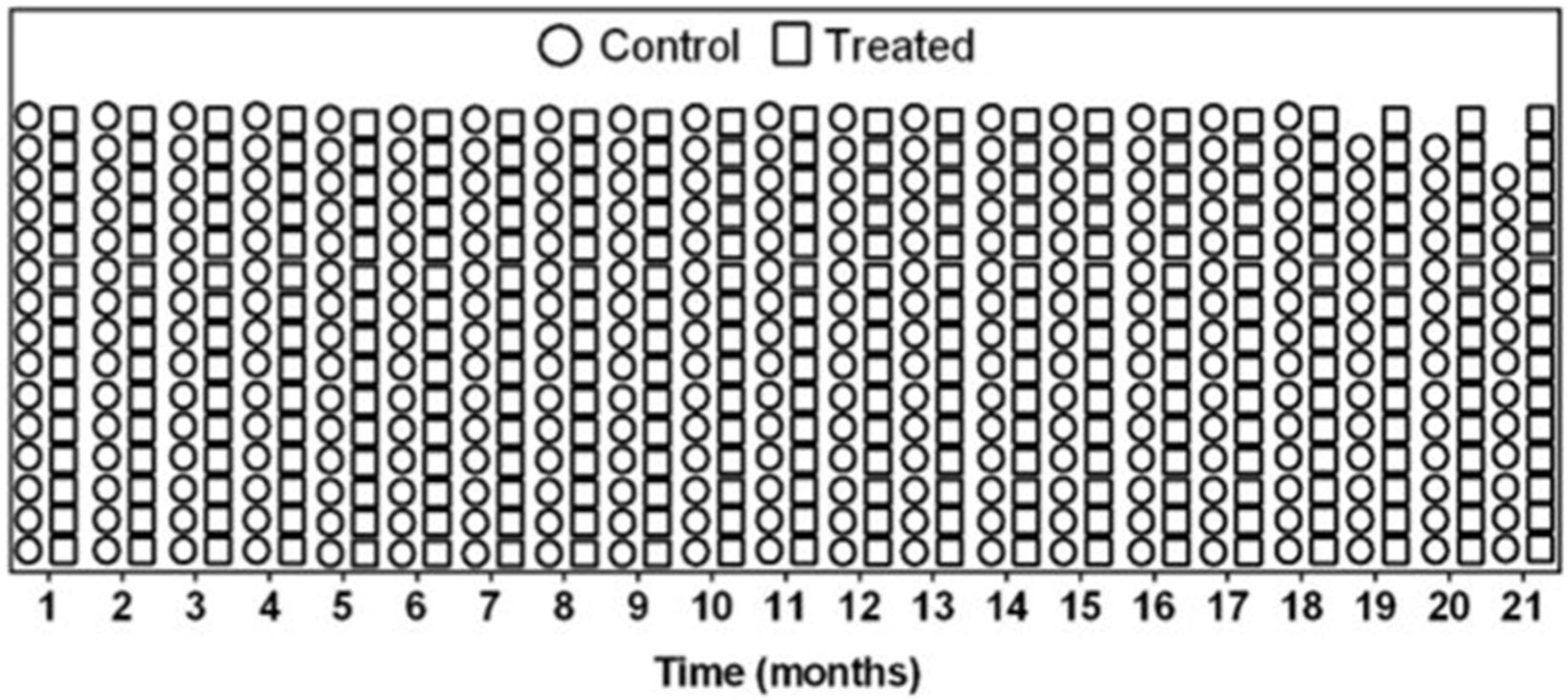
3.3. Weight and Survival Are Comparable Between Treated and Control Mice Analysis
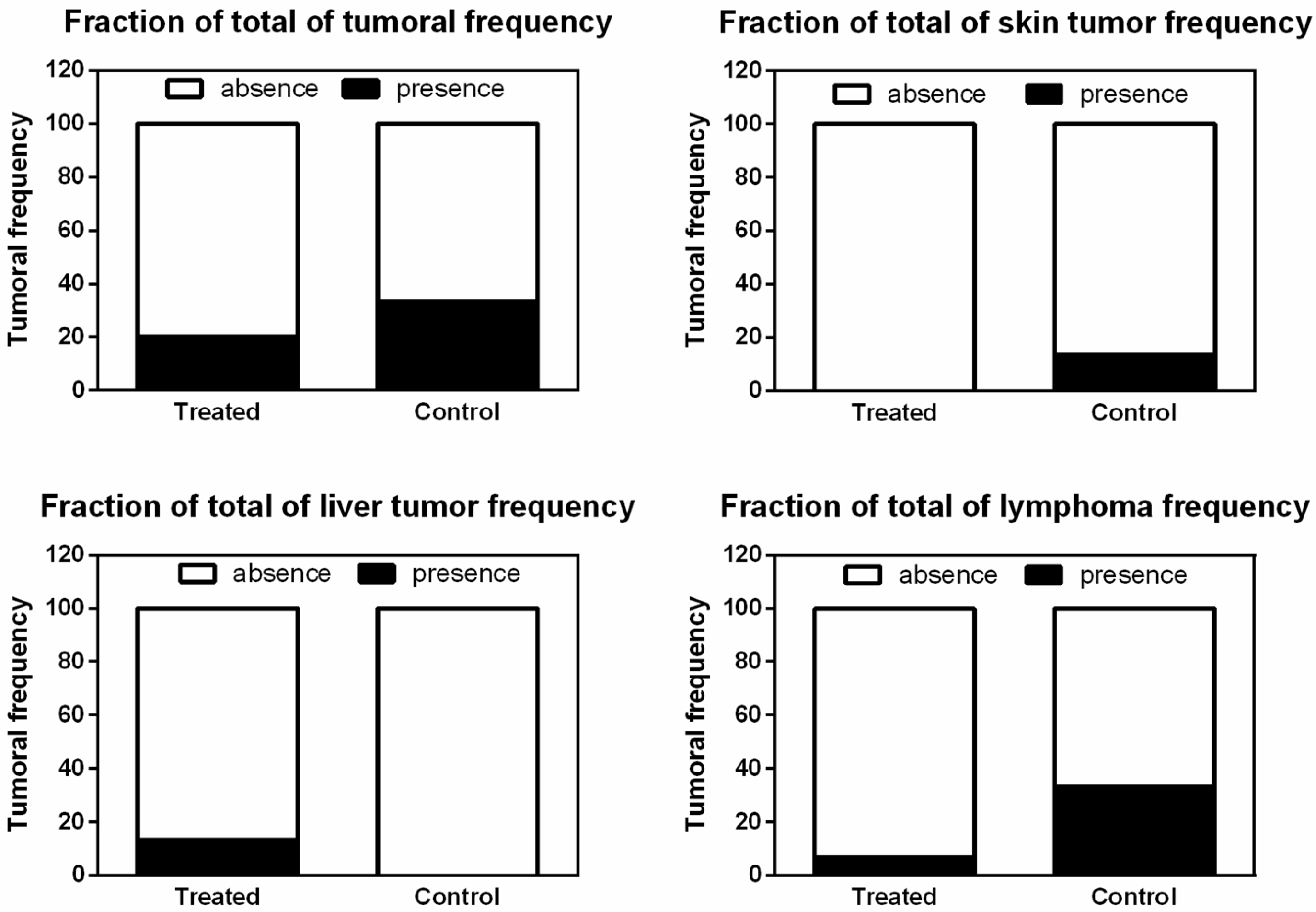
3.4. CRISPR-Cas9-Liposomal Treatment Does Not Increase Tumor Frequency
3.5. On and Off-Target Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilbie, D.; Walther, J.; Mastrobattista, E. Delivery Aspects of CRISPR/Cas for In Vivo Genome Editing. Acc. Chem. Res. 2019, 52, 1555–1564. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Krupovic, M.; Forterre, P. History of CRISPR-Cas from encounter with a mysterious repeated sequence to genome editing technology. J. Bacteriol. 2018, 200, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Lundstrom, K. Viral Vectors in Gene Therapy: Where Do We Stand in 2023? Viruses 2023, 15, 698. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, H.F.; Bruxel, F.; Fraga, M.; Schuh, R.S.; Zorzi, G.K.; Matte, U.; Fattal, E. Cationic nanoemulsions as nucleic acids delivery systems. Int. J. Pharm. 2017, 534, 356–367. [Google Scholar] [CrossRef]
- Jiang, C.; Mei, M.; Li, B.; Zhu, X.; Zu, W.; Tian, Y.; Wang, Q.; Guo, Y.; Dong, Y.; Tan, X. A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 2017, 27, 440–443. [Google Scholar] [CrossRef]
- Glass, Z.; Lee, M.; Li, Y.; Xu, Q. Engineering the Delivery System for CRISPR-based Genome Editing. Trends Biotechnol. 2018, 36, 173. [Google Scholar] [CrossRef]
- Schuh, R.S.; Poletto, É.; Pasqualim, G.; Tavares, A.M.V.; Meyer, F.S.; Gonzalez, E.A.; Giugliani, R.; Matte, U.; Teixeira, H.F.; Baldo, G. In vivo genome editing of mucopolysaccharidosis I mice using the CRISPR/Cas9 system. J. Control. Release Off. J. Control. Release Soc. 2018, 288, 23–33. [Google Scholar] [CrossRef]
- Schuh, R.S.; Gonzalez, E.A.; Tavares, A.M.V.; Seolin, B.G.; Elias, L.S.; Vera, L.N.P.; Kubaski, F.; Poletto, E.; Giugliani, R.; Teixeira, H.F.; et al. Neonatal nonviral gene editing with the CRISPR/Cas9 system improves some cardiovascular, respiratory, and bone disease features of the mucopolysaccharidosis I phenotype in mice. Gene Ther. 2020, 27, 74–84. [Google Scholar] [CrossRef]
- Guo, C.; Ma, X.; Gao, F.; Guo, Y. Off-target effects in CRISPR/Cas9 gene editing. Front. Bioeng. Biotechnol. 2023, 11, 1143157. [Google Scholar] [CrossRef]
- Fujii, W.; Kawasaki, K.; Sugiura, K.; Naito, K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 2013, 41, e187. [Google Scholar] [CrossRef]
- Cradick, T.J.; Qiu, P.; Lee, C.M.; Fine, E.J.; Bao, G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas off-target Sites. Mol. Ther. Nucleic Acids 2014, 3, e214. [Google Scholar] [CrossRef] [PubMed]
- Schuh, R.S.; Poletto, É.; Fachel, F.N.S.; Matte, U.; Baldo, G.; Teixeira, H.F. Physicochemical properties of cationic nanoemulsions and liposomes obtained by microfluidization complexed with a single plasmid or along with an oligonucleotide: Implications for CRISPR/Cas technology. J. Colloid Interface Sci. 2018, 530, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Bonamassa, B.; Hai, L.; Liu, D. Hydrodynamic gene delivery and its applications in pharmaceutical research. Pharm. Res. 2011, 28, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Ishiwata, H.; Suzuki, N.; Ando, S.; Kikuchi, H.; Kitagawa, T. Characteristics and biodistribution of cationic liposomes and their DNA complexes. J. Control. Release Off. J. Control. Release Soc. 2000, 69, 139–148. [Google Scholar] [CrossRef]
- Zhang, J.S.; Liu, F.; Conwell, C.C.; Tan, Y.; Huang, L. Mechanistic studies of sequential injection of cationic liposome and plasmid DNA. Mol. Ther. J. Am. Soc. Gene Ther. 2006, 13, 429–437. [Google Scholar] [CrossRef]
- Allen, T.M.; Cullis, P.R. Liposomal drug delivery systems: From concept to clinical applications. Adv. Drug Deliv. Rev. 2013, 65, 36–48. [Google Scholar] [CrossRef]
- Cataldi, M.; Vigliotti, C.; Mosca, T.; Cammarota, M.R.; Capone, D. Emerging role of the spleen in the pharmacokinetics of monoclonal antibodies, nanoparticles and exosomes. Int. J. Mol. Sci. 2017, 18, 1249. [Google Scholar] [CrossRef]
- Herweijer, H.; Wolff, J.A. Gene therapy progress and prospects: Hydrodynamic gene delivery. Gene Ther. 2007, 14, 99–107. [Google Scholar] [CrossRef]
- Al-Dosari, M.S.; Knapp, J.E.; Liu, D. Hydrodynamic Delivery. Adv. Genet. 2005, 54, 65–82. [Google Scholar] [CrossRef]
- He, Y.; Pimenov, A.A.; Nayak Jv Plowey, J.; Falo, L.D.; Huang, L. Intravenous Injection of Naked DNA Encoding Secreted flt3 Ligand Dramatically Increases the Number of Dendritic Cells and Natural Killer Cells In Vivo. Hum. Gene Ther. 2000, 11, 547–554. [Google Scholar] [CrossRef]
- Schile, A.; Dion, S.; Imai-Leonard, D.; Doty, R. Spontaneous Tumors in Aging Colonies of C57BL/6J Mice. Cancer Res. 2018, 78, 5113. Available online: https://aacrjournals.org/cancerres/article-abstract/78/13_Supplement/5113/629668 (accessed on 10 June 2024). [CrossRef]
- Some Tumor Data for This Paper were Retrieved from the Mouse Models of Human Cancer Database (MMHCdb), Mouse Genome Informatics, The Jackson Laboratory, Bar Harbor, Maine. World Wide Web. Available online: https://tumor.informatics.jax.org/mtbwi/dynamicGrid.do (accessed on 29 September 2024).
- Zhang, X.H.; Tee, L.Y.; Wang, X.G.; Huang, Q.S.; Yang, S.H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol. Ther. Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Cradick, T.J.; Brown, M.T.; Deshmukh, H.; Ranjan, P.; Sarode, N.; Wile, B.M.; Vertino, P.M.; Stewart, F.J.; Bao, G. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014, 42, 7473–7485. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Lin, C.Y.; Gootenberg, J.S.; Konermann, S.; Trevino, A.E.; Scott, D.A.; Inoue, A.; Matoba, S.; Zhang, Y.; et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 2013, 154, 1380–1389. [Google Scholar] [CrossRef]
- Zischewski, J.; Fischer, R.; Bortesi, L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol. Adv. 2017, 35, 95–104. [Google Scholar] [CrossRef]
- Bao, X.R.; Pan, Y.; Lee, C.M.; Davis, T.H.; Bao, G. Tools for experimental and computational analyses of off-target editing by programmable nucleases. Nat. Protoc. 2021, 16, 10–26. [Google Scholar] [CrossRef]
- Liang, D.; Gutierrez, N.M.; Chen, T.; Lee, Y.; Park, S.-W.; Ma, H.; Koski, A.; Ahmed, R.; Darby, H.; Li, Y.; et al. Frequent Gene Conversion in Human Embryos Induced by Double Strand Breaks; CHS: São Paulo, Brazil, 2020. [Google Scholar] [CrossRef]
- Chen, M.; Mao, A.; Xu, M.; Weng, Q.; Mao, J.; Ji, J. CRISPR-Cas9 for cancer therapy: Opportunities and challenges. Cancer Lett. 2019, 447, 48–55. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, M.; Gu, L.; Wang, Y. CRISPR/Cas9-mediated knock-in strategy at the Rosa26 locus in cattle fetal fibroblasts. PLoS ONE 2022, 17, e0276811. [Google Scholar] [CrossRef]
- Han, H.A.; Pang, J.K.S.; Soh, B.S. Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing. J. Mol. Med. 2020, 98, 615–632. [Google Scholar] [CrossRef]
- Naeem, M.; Alkhnbashi, O.S. Current Bioinformatics Tools to Optimize CRISPR/Cas9 Experiments to Reduce off-Target Effects. Int. J. Mol. Sci. 2023, 24, 6261. [Google Scholar] [CrossRef]
- Naeem, M.; Majeed, S.; Hoque, M.Z.; Ahmad, I. Latest Developed Strategies to Minimize the off-Target Effects in CRISPR-Cas-Mediated Genome Editing. Cells 2020, 9, 1608. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Sánchez-Hernández, S.; Gutiérrez-Guerrero, A.; Pinedo-Gomez, J.; Benabdellah, K. Biased and unbiased methods for the detection of off-target cleavage by CRISPR/Cas9: An overview. Int. J. Mol. Sci. 2016, 17, 1507. [Google Scholar] [CrossRef] [PubMed]
- Atkins, A.; Chung, C.H.; Allen, A.G.; Dampier, W.; Gurrola, T.E.; Sariyer, I.K.; Nonnemacher, M.R.; Wigdahl, B. Off-Target Analysis in Gene Editing and Applications for Clinical Translation of CRISPR/Cas9 in HIV-1 Therapy. Front. Genome Ed. 2021, 3, 673022. [Google Scholar] [CrossRef] [PubMed]
- Yaish, O.; Asif, M.; Orenstein, Y. A systematic evaluation of data processing and problem formulation of CRISPR off-target site prediction. Brief. Bioinform. 2022, 23, bbac157. [Google Scholar] [CrossRef]
- Qi, Q.; Liu, X.; Xiong, W.; Zhang, K.; Shen, W.; Zhang, Y.; Xu, X.; Zhong, C.; Zhang, Y.; Tian, T.; et al. Reducing CRISPR-Cas9 off-target effects by optically controlled chemical modifications of guide RNA. Cell Chem. Biol. 2024, 31, 1839–1851. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Zheng, Z.; Nguyen, N.T.; Liebers, M.; Topkar Vv Thapar, V.; Wyvekens, N.; Khayter, C.; Iafrate, A.J.; Le, L.P.; Aryee, M.J.; et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015, 33, 187–198. [Google Scholar] [CrossRef]
- Kang, S.H.; Lee Wjae An, J.H.; Lee, J.H.; Kim, Y.H.; Kim, H.; Oh, Y.; Park, Y.H.; Jin, Y.B.; Jun, B.H.; Hur, J.K.; et al. Prediction-based highly sensitive CRISPR off-target validation using target-specific DNA enrichment. Nat. Commun. 2020, 11, 3596. [Google Scholar] [CrossRef]
- Tao, J.; Bauer, D.E.; Chiarle, R. Assessing and advancing the safety of CRISPR-Cas tools: From DNA to RNA editing. Nat. Commun. 2023, 14, 212. [Google Scholar] [CrossRef]

| Formulation | Days | Mean Diameter (nm) | PDI | ζ-Potential (mV) |
|---|---|---|---|---|
| LC | 0 | 133.0 ± 3.29 | 0.101 ± 0.01 | +41.50 ± 0.95 |
| 15 | 139.0 ± 3.05 | 0.132 ± 0.02 * | +43.80 ± 1.41 | |
| 30 | 138.9 ± 1.01 | 0.143 ± 0.01 * | +46.07 ± 0.55 |
| Result | Query Type | Mismatch | Ends with RG | Chr Position | Strand | Cut Site | Score |
|---|---|---|---|---|---|---|---|
| GGATTCTCCCAGGCCCAGGGCGG—hit GGATTCTCCCAGGCCCAGGGNGG—query | No indel | 0 | Yes | Chr6:113,076,075–113,076,097 | + | 113,076,091 | 0 |
| TGATTCTCCCAGGCCCAGGGAAG—hit GGATTCTCCCAGGCCCAGGGNGG—query | No indel | 2 | Yes | Chr5:113,445,565–113,445,587 | - | 113,445,571 | 20.12 |
| GGAATCTCCCAGGCTCAGGGAGG—hit GGATTCTCCCAGGCCCAGGGNGG—query | No indel | 2 | Yes | Chr2:69,238,534–69,238,556 | - | 69,238,540 | 2.07 |
| GGAGTCTCCCAGGCCTAGGGTGG—hit GGATTCTCCCAGGCCCAGGGNGG—query | No indel | 2 | Yes | Chr11:73,171,299–73,171,321 | + | 73,171,315 | 2.47 |
| GATTCTCCCAGGCCCAGGGCGG—hit GATTCTCCCAGGCCCAGGGNGG—query | Del 19, or Del 20 | 0 | Yes | Chr6:113,076,076–113,076,097 | + | 113,076,091 | 0.63 |
| GATTCTCCCAGGCCCAGGGAAG—hit GATTCTCCCAGGCCCAGGGNGG—query | Del 19, or Del 20 | 1 | Yes | Chr5:113,445,565–113,445,586 | - | 113,445,571 | 20.63 |
| GATTCTCCCAGGCCCAGGGCGG—hit GGTTCTCCCAGGCCCAGGGNGG—query | Del 18 | 1 | Yes | Chr6:113,076,076–113,076,097 | + | 113,076,091 | 0.79 |
| GGATTTCCCAGGCCCAGAGTGG—hit GGATTTCCCAGGCCCAGGGNGG—query | Del 15 | 1 | Yes | Chr17:8,602,794–8,602,815 | + | 8,602,809 | 5.72 |
| GGATTCTCCCAGGCCCAGGGCG—hit GGATTCTCCCAGGCCCAGGNGG—query | Del 1, or Del 2, or Del 3 | 1 | No | Chr6:113,076,075–113,076,096 | + | 113,076,090 | 24.51 |
| GGATTCTCCCAGGCCCAGGGCG—hit GGATTCTCCCAGGCCCAGGGGG—query | Del PAM 3 | 1 | No | Chr6:113,076,075–113,076,096 | + | 113,076,090 | 40.51 |
| GGATTCTCCCAGGCCCAGGGCG—hit GGATTCTCCCAGGCCCAGGGNG—query | Del PAM 1, or Del PAM 2 | 0 | No | Chr6:113,076,075–113,076,096 | + | 113,076,090 | 20.51 |
| GGGATTCTCCCAGGCCCAGGGCGG—hit GNGATTCTCCCAGGCCCAGGGNGG—query | Ins 19 | 0 | Yes | Chr6:113,076,074–113,076,097 | + | 113,076,091 | 0.83 |
| GGGATTCTCCCAGGCCCAGGGCGG—hit GGNATTCTCCCAGGCCCAGGGNGG—query | Ins 18 | 0 | Yes | Chr6:113,076,074–113,076,097 | + | 113,076,091 | 0.85 |
| GGGATTCTCCCAGGCCCAGGGCGG—hit GGANTTCTCCCAGGCCCAGGGNGG—query | Ins 17 | 1 | Yes | Chr6:113,076,074–113,076,097 | + | 113,076,091 | 1.02 |
| GGATTCTCCCAGGCCCAGGGCGGT—hit GGATTCTCCCAGGCCCAGGGNNGG—query | Ins PAM 2, or Ins PAM 3 | 1 | No | Chr6:113,076,075–113,076,098 | + | 113,076,092 | 40.7 |
| GGATTCTCCCAGGCCCAGGGCGGT—hit GGATTCTCCCAGGCCCAGGGNGNG—query | Ins PAM 1 | 1 | No | Chr6:113,076,075–113,076,098 | + | 113,076,092 | 40.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monteagudo, V.; Flores, L.C.B.; Lopes, M.; Fachel, F.N.S.; Martins, G.; Siebert, M.; Carniel, W.d.S.; Garcez, T.N.A.; Teixeira, H.F.; Matte, U.; et al. Newborn Intravenous Injection of Liposomal CRISPR/Cas9 Complex Has No Incidence of Off-Targets or Tumors in Mice. Pharmaceutics 2025, 17, 656. https://doi.org/10.3390/pharmaceutics17050656
Monteagudo V, Flores LCB, Lopes M, Fachel FNS, Martins G, Siebert M, Carniel WdS, Garcez TNA, Teixeira HF, Matte U, et al. Newborn Intravenous Injection of Liposomal CRISPR/Cas9 Complex Has No Incidence of Off-Targets or Tumors in Mice. Pharmaceutics. 2025; 17(5):656. https://doi.org/10.3390/pharmaceutics17050656
Chicago/Turabian StyleMonteagudo, Vinícius, Larissa Cristina Barbosa Flores, Melaine Lopes, Flavia Nathiely Silveira Fachel, Giselle Martins, Marina Siebert, Willian da Silva Carniel, Tuane Nerissa Alves Garcez, Helder Ferreira Teixeira, Ursula Matte, and et al. 2025. "Newborn Intravenous Injection of Liposomal CRISPR/Cas9 Complex Has No Incidence of Off-Targets or Tumors in Mice" Pharmaceutics 17, no. 5: 656. https://doi.org/10.3390/pharmaceutics17050656
APA StyleMonteagudo, V., Flores, L. C. B., Lopes, M., Fachel, F. N. S., Martins, G., Siebert, M., Carniel, W. d. S., Garcez, T. N. A., Teixeira, H. F., Matte, U., Giugliani, R., Baldo, G., Poletto, É., & Schuh, R. S. (2025). Newborn Intravenous Injection of Liposomal CRISPR/Cas9 Complex Has No Incidence of Off-Targets or Tumors in Mice. Pharmaceutics, 17(5), 656. https://doi.org/10.3390/pharmaceutics17050656










