NMR Unveils Activity Mechanism of Linear Spider Venom Peptide Fragments Selected by Neural Networks Against Staphylococci Including MRSA
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptide Synthesis
2.2. Database Assembly
2.3. High Resolution NMR Spectroscopy
2.4. Wide-Line 31P-NMR Spectroscopy
2.5. NMR Diffusion Measurements
2.6. Testing Biological Activity of Peptides and Confocal Microscopy Measurements
3. Results
3.1. LP Fragment Library Construction, Neural Network Training, and Activity Prediction
3.2. High-Resolution NMR Study of Peptide IX
3.3. Interaction of Peptide IX with Phospholipid Membranes by Wide-Line 31P-NMR
3.4. Hemolytic Activity of AMP
3.5. Mechanism of Antibacterial Action of Peptide XIII
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 16-DSA | 16-doxylstearic acid |
| AMP | Antimicrobial Peptide |
| BCECF | 2′,7′-Bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein |
| CLSM | Confocal Laser Scanning Microscopy |
| D8PG | Dioctanoyl Phosphatidylglycerol |
| DOPC | Dioleoyl Phosphatidylcholine |
| DOPG | Dioleoyl Phosphatidylglycerol |
| DOPE | Dioleoyl Phosphatidylethanolamine |
| LP | Linear Peptide |
| LPPG | 1-palmitoyl-2-hydroxy-sn-glycero-3-phospho-(1′-rac-glycerol) |
| MIC | Minimum Inhibitory Concentration |
| MHB | Mueller-Hinton Broth |
| MLV | Multilamellar Vesicle |
| MRSA | Methicillin Resistant S. aureus |
| PC | Phosphatidyl Choline |
| PG | Phosphatidyl Glycerol |
| PI | Propidium Iodide |
| RCI | Random Coil Index |
References
- Miller, W.R.; Arias, C.A. ESKAPE Pathogens: Antimicrobial Resistance, Epidemiology, Clinical Impact and Therapeutics. Nat. Rev. Microbiol. 2024, 22, 598–616. [Google Scholar] [CrossRef]
- Magana, M.; Pushpanathan, M.; Santos, A.L.; Leanse, L.; Fernandez, M.; Ioannidis, A.; Giulianotti, M.A.; Apidianakis, Y.; Bradfute, S.; Ferguson, A.L.; et al. The Value of Antimicrobial Peptides in the Age of Resistance. Lancet Infect. Dis. 2020, 20, e216–e230. [Google Scholar] [CrossRef]
- Narayana, J.L.; Mishra, B.; Lushnikova, T.; Golla, R.M.; Wang, G. Modulation of Antimicrobial Potency of Human Cathelicidin Peptides against the ESKAPE Pathogens and In Vivo Efficacy in a Murine Catheter-Associated Biofilm Model. Biochim. Biophys. Acta Biomembr. 2019, 1861, 1592–1602. [Google Scholar] [CrossRef] [PubMed]
- Mechesso, A.F.; Su, Y.; Xie, J.; Wang, G. Enhanced Antimicrobial Screening Sensitivity Enabled the Identification of an Ultrashort Peptide KR-8 for Engineering of LL-37mini to Combat Drug-Resistant Pathogens. ACS Infect. Dis. 2023, 9, 2215–2225. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Hanke, M.L.; Mishra, B.; Lushnikova, T.; Heim, C.E.; Chittezham Thomas, V.; Bayles, K.W.; Kielian, T. Transformation of Human Cathelicidin LL-37 into Selective, Stable, and Potent Antimicrobial Compounds. ACS Chem. Biol. 2014, 9, 1997–2002. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, R.; Esmaeili Gouvarchin Ghaleh, H.; Ranjbar, R. Antibiofilm Effect of Melittin Alone and in Combination with Conventional Antibiotics toward Strong Biofilm of MDR-MRSA and -Pseudomonas aeruginosa. Front. Microbiol. 2023, 14, 1030401. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, P.; Xiao, L.; Liu, Y.; Wu, K.; Ni, G.; Li, H.; Wang, T.; Wu, X.; Chen, G.; et al. Caerin 1.1 and 1.9 Peptides from Australian Tree Frog Inhibit Antibiotic-Resistant Bacteria Growth in a Murine Skin Infection Model. Microbiol. Spectr. 2021, 9, e0005121. [Google Scholar] [CrossRef]
- Memariani, H.; Memariani, M.; Pourmand, M.R. Venom-Derived Peptide Mastoparan-1 Eradicates Planktonic and Biofilm-Embedded Methicillin-Resistant Staphylococcus aureus Isolates. Microb. Pathog. 2018, 119, 72–80. [Google Scholar] [CrossRef]
- Kuhn-Nentwig, L.; Lischer, H.E.L.; Pekár, S.; Langenegger, N.; Albo, M.J.; Isaia, M.; Nentwig, W. Linear Peptides-A Combinatorial Innovation in the Venom of Some Modern Spiders. Front. Mol. Biosci. 2021, 8, 705141. [Google Scholar] [CrossRef]
- Santos, D.M.; Verly, R.M.; Piló-Veloso, D.; de Maria, M.; de Carvalho, M.A.R.; Cisalpino, P.S.; Soares, B.M.; Diniz, C.G.; Farias, L.M.; Moreira, D.F.F.; et al. LyeTx I, a Potent Antimicrobial Peptide from the Venom of the Spider Lycosa erythrognatha. Amino Acids 2010, 39, 135–144. [Google Scholar] [CrossRef]
- Kozlov, S.A.; Vassilevski, A.A.; Feofanov, A.V.; Surovoy, A.Y.; Karpunin, D.V.; Grishin, E.V. Latarcins, Antimicrobial and Cytolytic Peptides from the Venom of the Spider Lachesana tarabaevi (Zodariidae) That Exemplify Biomolecular Diversity. J. Biol. Chem. 2006, 281, 20983–20992. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Vassilevski, A.A.; Kozlov, S.A.; Feofanov, A.V.; Grishin, E.V.; Efremov, R.G. Latarcins: Versatile Spider Venom Peptides. Cell. Mol. Life Sci. 2015, 72, 4501–4522. [Google Scholar] [CrossRef]
- Wadhwani, P.; Sekaran, S.; Strandberg, E.; Bürck, J.; Chugh, A.; Ulrich, A.S. Membrane Interactions of Latarcins: Antimicrobial Peptides from Spider Venom. Int. J. Mol. Sci. 2021, 22, 10156. [Google Scholar] [CrossRef]
- Vassilevskii, A.A.; Kozlov, S.A.; Zhmak, M.N.; Kudelina, I.A.; Dubovskii, P.V.; Shaturskii, O.I.; Arseniev, A.S.; Grishin, E.V. Synthetic analogues of antimicrobial peptides from the venom of the Central Asian spider Lachesana tarabaevi. Russ. J. Bioorg. Chem. 2007, 33, 376–382. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Volynsky, P.E.; Polyansky, A.A.; Karpunin, D.V.; Chupin, V.V.; Efremov, R.G.; Arseniev, A.S. Three-Dimensional Structure/Hydrophobicity of Latarcins Specifies Their Mode of Membrane Activity. Biochemistry 2008, 47, 3525–3533. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate Structure Prediction of Biomolecular Interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The Antimicrobial Peptide Database as a Tool for Research and Education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [PubMed]
- Pirtskhalava, M.; Amstrong, A.A.; Grigolava, M.; Chubinidze, M.; Alimbarashvili, E.; Vishnepolsky, B.; Gabrielian, A.; Rosenthal, A.; Hurt, D.E.; Tartakovsky, M. DBAASP v3: Database of Antimicrobial/Cytotoxic Activity and Structure of Peptides as a Resource for Development of New Therapeutics. Nucleic Acids Res. 2021, 49, D288–D297. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Kang, X.; Dong, F.; Liu, Y.; Zhu, N.; Hu, Y.; Xu, H.; Lao, X.; Zheng, H. DRAMP 3.0: An Enhanced Comprehensive Data Repository of Antimicrobial Peptides. Nucleic Acids Res. 2022, 50, D488–D496. [Google Scholar] [CrossRef]
- Dubovskii, P.V. Unusual Titration of the Membrane-Bound Artificial Hemagglutinin Fusion Peptide. Eur. Biophys. J. 2012, 41, 1077–1084. [Google Scholar] [CrossRef]
- Shen, Y.; Bax, A. Protein Structural Information Derived from NMR Chemical Shift with the Neural Network Program TALOS-N. Methods Mol. Biol. 2015, 1260, 17–32. [Google Scholar] [CrossRef]
- Güntert, P. Automated NMR Structure Calculation with CYANA. Methods Mol. Biol. 2004, 278, 353–378. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Lesovoy, D.M.; Dubinnyi, M.A.; Utkin, Y.N.; Arseniev, A.S. Interaction of the P-Type Cardiotoxin with Phospholipid Membranes. Eur. J. Biochem. 2003, 270, 2038–2046. [Google Scholar] [CrossRef]
- Dubinnyi, M.A.; Lesovoy, D.M.; Dubovskii, P.V.; Chupin, V.V.; Arseniev, A.S. Modeling of 31P-NMR Spectra of Magnetically Oriented Phospholipid Liposomes: A New Analytical Solution. Solid State Nucl. Magn. Reson. 2006, 29, 305–311. [Google Scholar] [CrossRef]
- Zheng, G.; Stait-Gardner, T.; Anil Kumar, P.G.; Torres, A.M.; Price, W.S. PGSTE-WATERGATE: An STE-Based PGSE NMR Sequence with Excellent Solvent Suppression. J. Magn. Reson. 2008, 191, 159–163. [Google Scholar] [CrossRef]
- Chou, J.J.; Baber, J.L.; Bax, A. Characterization of Phospholipid Mixed Micelles by Translational Diffusion. J. Biomol. NMR 2004, 29, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Tokuyama, M.; Oppenheim, I. Dynamics of Hard-Sphere Suspensions. Phys. Rev. E 1994, 50, R16–R19. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Ignatova, A.A.; Volynsky, P.E.; Ivanov, I.A.; Zhmak, M.N.; Feofanov, A.V.; Efremov, R.G. Improving Therapeutic Potential of Antibacterial Spider Venom Peptides: Coarse-Grain Molecular Dynamics Guided Approach. Future Med. Chem. 2018, 10, 2309–2322. [Google Scholar] [CrossRef]
- Du, Z.; Ding, X.; Xu, Y.; Li, Y. UniDL4BioPep: A Universal Deep Learning Architecture for Binary Classification in Peptide Bioactivity. Brief. Bioinform. 2023, 24, bbad135. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Shin, M.K.; Yoo, J.S.; Jang, W.; Sung, J.-S. Identifying Novel Antimicrobial Peptides from Venom Gland of Spider Pardosa astrigera by Deep Multi-Task Learning. Front. Microbiol. 2022, 13, 971503. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Sutherland, D.; Hammond, S.A.; Yang, C.; Taho, F.; Bergman, L.; Houston, S.; Warren, R.L.; Wong, T.; Hoang, L.M.N.; et al. AMPlify: Attentive Deep Learning Model for Discovery of Novel Antimicrobial Peptides Effective against WHO Priority Pathogens. BMC Genom. 2022, 23, 77. [Google Scholar] [CrossRef]
- Li, C.; Warren, R.L.; Birol, I. Models and Data of AMPlify: A Deep Learning Tool for Antimicrobial Peptide Prediction. BMC Res. Notes 2023, 16, 11. [Google Scholar] [CrossRef]
- Adedeji-Olulana, A.F.; Wacnik, K.; Lafage, L.; Pasquina-Lemonche, L.; Tinajero-Trejo, M.; Sutton, J.A.F.; Bilyk, B.; Irving, S.E.; Portman Ross, C.J.; Meacock, O.J.; et al. Two Codependent Routes Lead to High-Level MRSA. Science 2024, 386, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Charoenkwan, P.; Kanthawong, S.; Schaduangrat, N.; Li’, P.; Moni, M.A.; Shoombuatong, W. SCMRSA: A New Approach for Identifying and Analyzing Anti-MRSA Peptides Using Estimated Propensity Scores of Dipeptides. ACS Omega 2022, 7, 32653–32664. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Vorontsova, O.V.; Utkin, Y.N.; Arseniev, A.S.; Efremov, R.G.; Feofanov, A.V. Cobra Cytotoxins: Determinants of Antibacterial Activity. Mendeleev Commun. 2015, 25, 70–71. [Google Scholar] [CrossRef]
- Jiang, Z.; Vasil, A.I.; Hale, J.D.; Hancock, R.E.W.; Vasil, M.L.; Hodges, R.S. Effects of Net Charge and the Number of Positively Charged Residues on the Biological Activity of Amphipathic Alpha-Helical Cationic Antimicrobial Peptides. Biopolymers 2008, 90, 369–383. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- de Souza, A.N.; Cardoso, G.d.A.; Nunes, L.O.; Aisenbrey, C.; Salnikov, E.; de Souza, K.R.; Saad, A.; de Lima, M.E.; Resende, J.M.; Bechinger, B.; et al. Comparative Structural and Biophysical Investigation of Lycosa erythrognatha Toxin I (LyeTx I) and Its Analog LyeTx I-b. Antibiotics 2025, 14, 66. [Google Scholar] [CrossRef]
- Reis, P.V.M.; Boff, D.; Verly, R.M.; Melo-Braga, M.N.; Cortés, M.E.; Santos, D.M.; Pimenta, A.M.d.C.; Amaral, F.A.; Resende, J.M.; de Lima, M.E. LyeTxI-b, a Synthetic Peptide Derived From Lycosa erythrognatha Spider Venom, Shows Potent Antibiotic Activity in Vitro and in Vivo. Front. Microbiol. 2018, 9, 667. [Google Scholar] [CrossRef]
- Danielsson, J.; Jarvet, J.; Damberg, P.; Gräslund, A. Translational Diffusion Measured by PFG-NMR on Full Length and Fragments of the Alzheimer Aβ(1–40) Peptide. Determination of Hydrodynamic Radii of Random Coil Peptides of Varying Length. Magn. Reson. Chem. 2002, 40, 89–97. [Google Scholar] [CrossRef]
- García de la Torre, J.; Huertas, M.L.; Carrasco, B. HYDRONMR: Prediction of NMR Relaxation of Globular Proteins from Atomic-Level Structures and Hydrodynamic Calculations. J. Magn. Reson. 2000, 147, 138–146. [Google Scholar] [CrossRef]
- Kemmink, J.; Creighton, T.E. Local Conformations of Peptides Representing the Entire Sequence of Bovine Pancreatic Trypsin Inhibitor and Their Roles in Folding. J. Mol. Biol. 1993, 234, 861–878. [Google Scholar] [CrossRef]
- Kemmink, J.; van Mierlo, C.P.; Scheek, R.M.; Creighton, T.E. Local Structure Due to an Aromatic-Amide Interaction Observed by 1H-Nuclear Magnetic Resonance Spectroscopy in Peptides Related to the N Terminus of Bovine Pancreatic Trypsin Inhibitor. J. Mol. Biol. 1993, 230, 312–322. [Google Scholar] [CrossRef]
- Platzer, G.; Okon, M.; McIntosh, L.P. pH-Dependent Random Coil (1)H, (13)C, and (15)N Chemical Shifts of the Ionizable Amino Acids: A Guide for Protein pKa Measurements. J. Biomol. NMR 2014, 60, 109–129. [Google Scholar] [CrossRef]
- Epand, R.M.; Rotem, S.; Mor, A.; Berno, B.; Epand, R.F. Bacterial Membranes as Predictors of Antimicrobial Potency. J. Am. Chem. Soc. 2008, 130, 14346–14352. [Google Scholar] [CrossRef] [PubMed]
- Lipfert, J.; Columbus, L.; Chu, V.B.; Lesley, S.A.; Doniach, S. Size and Shape of Detergent Micelles Determined by Small-Angle X-Ray Scattering. J. Phys. Chem. B 2007, 111, 12427–12438. [Google Scholar] [CrossRef]
- Pyrkov, T.V.; Chugunov, A.O.; Krylov, N.A.; Nolde, D.E.; Efremov, R.G. PLATINUM: A Web Tool for Analysis of Hydrophobic/Hydrophilic Organization of Biomolecular Complexes. Bioinformatics 2009, 25, 1201–1202. [Google Scholar] [CrossRef]
- Strandberg, E.; Sparrman, T.; Lindblom, G. Phase Diagrams of Systems with Cationic Alpha-Helical Membrane-Spanning Model Peptides and Dioleoylphosphatidylcholine. Adv. Colloid Interface Sci. 2001, 89–90, 239–261. [Google Scholar] [CrossRef]
- Pinheiro, T.J.; Watts, A. Lipid Specificity in the Interaction of Cytochrome c with Anionic Phospholipid Bilayers Revealed by Solid-State 31P NMR. Biochemistry 1994, 33, 2451–2458. [Google Scholar] [CrossRef]
- Claessens, M.M.A.E.; Leermakers, F.A.M.; Hoekstra, F.A.; Stuart, M.A.C. Opposing Effects of Cation Binding and Hydration on the Bending Rigidity of Anionic Lipid Bilayers. J. Phys. Chem. B 2007, 111, 7127–7132. [Google Scholar] [CrossRef]
- Burnell, E.E.; Cullis, P.R.; de Kruijff, B. Effects of Tumbling and Lateral Diffusion on Phosphatidylcholine Model Membrane 31P-NMR Lineshapes. Biochim. Biophys. Acta 1980, 603, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Taraschi, T.F.; De Kruijff, B.; Verkleij, A.; Van Echteld, C.J. Effect of Glycophorin on Lipid Polymorphism. A 31P-NMR Study. Biochim. Biophys. Acta 1982, 685, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Batenburg, A.M.; Bougis, P.E.; Rochat, H.; Verkleij, A.J.; de Kruijff, B. Penetration of a Cardiotoxin into Cardiolipin Model Membranes and Its Implications on Lipid Organization. Biochemistry 1985, 24, 7101–7110. [Google Scholar] [CrossRef]
- Dubovskii, P.V.; Volynsky, P.E.; Polyansky, A.A.; Chupin, V.V.; Efremov, R.G.; Arseniev, A.S. Spatial Structure and Activity Mechanism of a Novel Spider Antimicrobial Peptide. Biochemistry 2006, 45, 10759–10767. [Google Scholar] [CrossRef]
- Budnik, B.A.; Olsen, J.V.; Egorov, T.A.; Anisimova, V.E.; Galkina, T.G.; Musolyamov, A.K.; Grishin, E.V.; Zubarev, R.A. De Novo Sequencing of Antimicrobial Peptides Isolated from the Venom Glands of the Wolf Spider Lycosa singoriensis. J. Mass Spectrom. 2004, 39, 193–201. [Google Scholar] [CrossRef]
- Oh, J.H.; Park, J.; Park, Y. Anti-Biofilm and Anti-Inflammatory Effects of Lycosin-II Isolated from Spiders against Multi-Drug Resistant Bacteria. Biochim. Biophys. Acta Biomembr. 2022, 1864, 183769. [Google Scholar] [CrossRef]
- Park, J.; Kim, H.; Kang, H.-K.; Choi, M.-C.; Park, Y. Lycosin-II Exhibits Antifungal Activity and Inhibits Dual-Species Biofilm by Candida albicans and Staphylococcus aureus. J. Fungi 2022, 8, 901. [Google Scholar] [CrossRef]
- Tan, H.; Wang, J.; Song, Y.; Liu, S.; Lu, Z.; Luo, H.; Tang, X. Antibacterial Potential Analysis of Novel α-Helix Peptides in the Chinese Wolf Spider Lycosa sinensis. Pharmaceutics 2022, 14, 2540. [Google Scholar] [CrossRef]
- Wang, K.; Mwangi, J.; Cao, K.; Wang, Y.; Gao, J.; Yang, M.; Michira, B.B.; Lu, Q.; Li, J. Peptide Toxin Diversity and a Novel Antimicrobial Peptide from the Spider Oxyopes forcipiformis. Toxins 2024, 16, 466. [Google Scholar] [CrossRef]
- Oh, J.W.; Shin, M.K.; Park, H.-R.; Kim, S.; Lee, B.; Yoo, J.S.; Chi, W.-J.; Sung, J.-S. PA-Win2: In Silico-Based Discovery of a Novel Peptide with Dual Antibacterial and Anti-Biofilm Activity. Antibiotics 2024, 13, 1113. [Google Scholar] [CrossRef]
- Peng, Z.; Wei, C.; Cai, J.; Zou, Z.; Chen, J. Characterization of an Antimicrobial Peptide Family from the Venom Gland of Heteropoda venatoria. Toxicon 2024, 241, 107657. [Google Scholar] [CrossRef] [PubMed]
- Fuscaldi, L.L.; de Avelar Júnior, J.T.; Dos Santos, D.M.; Boff, D.; de Oliveira, V.L.S.; Gomes, K.A.G.G.; Cruz, R.d.C.; de Oliveira, P.L.; Magalhães, P.P.; Cisalpino, P.S.; et al. Shortened Derivatives from Native Antimicrobial Peptide LyeTx I: In Vitro and In Vivo Biological Activity Assessment. Exp. Biol. Med. 2021, 246, 414–425. [Google Scholar] [CrossRef]
- Vieira, A.P.G.C.; de Souza, A.N.; Lima, W.G.; Brito, J.C.M.; Simião, D.C.; Gonçalves, L.V.R.; Cordeiro, L.P.B.; de Oliveira Scoaris, D.; Fernandes, S.O.A.; Resende, J.M.; et al. The Synthetic Peptide LyeTx I mn∆K, Derived from Lycosa erythrognatha Spider Toxin, Is Active against Methicillin-Resistant Staphylococcus aureus (MRSA) In Vitro and In Vivo. Antibiotics 2024, 13, 248. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, X.; Song, Y.; Liu, Z.; Tang, X.; Tan, H. LC-AMP-I1, a Novel Venom-Derived Antimicrobial Peptide from the Wolf Spider Lycosa coelestis. Antimicrob. Agents Chemother. 2025, 69, e0042424. [Google Scholar] [CrossRef]
- Fischer, W. Lipoteichoic Acid and Lipids in the Membrane of Staphylococcus aureus. Med. Microbiol. Immunol. 1994, 183, 61–76. [Google Scholar] [CrossRef]
- Wang, G. Determination of Solution Structure and Lipid Micelle Location of an Engineered Membrane Peptide by Using One NMR Experiment and One Sample. Biochim. Biophys. Acta 2007, 1768, 3271–3281. [Google Scholar] [CrossRef]
- Jung, D.; Rozek, A.; Okon, M.; Hancock, R.E.W. Structural Transitions as Determinants of the Action of the Calcium-Dependent Antibiotic Daptomycin. Chem. Biol. 2004, 11, 949–957. [Google Scholar] [CrossRef]
- Grein, F.; Müller, A.; Scherer, K.M.; Liu, X.; Ludwig, K.C.; Klöckner, A.; Strach, M.; Sahl, H.-G.; Kubitscheck, U.; Schneider, T. Ca2+-Daptomycin Targets Cell Wall Biosynthesis by Forming a Tripartite Complex with Undecaprenyl-Coupled Intermediates and Membrane Lipids. Nat. Commun. 2020, 11, 1455. [Google Scholar] [CrossRef]
- Tyurin, A.P.; Alferova, V.A.; Paramonov, A.S.; Shuvalov, M.V.; Kudryakova, G.K.; Rogozhin, E.A.; Zherebker, A.Y.; Brylev, V.A.; Chistov, A.A.; Baranova, A.A.; et al. Gausemycins A,B: Cyclic Lipoglycopeptides from Streptomyces sp. Angew. Chem. Int. Ed. 2021, 60, 18694–18703. [Google Scholar] [CrossRef] [PubMed]
- Kravchenko, T.V.; Paramonov, A.S.; Kudzhaev, A.M.; Efimova, S.S.; Khorev, A.S.; Kudryakova, G.K.; Ivanov, I.A.; Chistov, A.A.; Baranova, A.A.; Krasilnikov, M.S.; et al. Gausemycin Antibiotic Family Acts via Ca2+-Dependent Membrane Targeting. J. Nat. Prod. 2024, 87, 664–674. [Google Scholar] [CrossRef]
- Pereira-Dias, L.; Oliveira-Pinto, P.R.; Fernandes, J.O.; Regalado, L.; Mendes, R.; Teixeira, C.; Mariz-Ponte, N.; Gomes, P.; Santos, C. Peptaibiotics: Harnessing the Potential of Microbial Secondary Metabolites for Mitigation of Plant Pathogens. Biotechnol. Adv. 2023, 68, 108223. [Google Scholar] [CrossRef] [PubMed]
- Shenkarev, Z.O.; Balashova, T.A.; Efremov, R.G.; Yakimenko, Z.A.; Ovchinnikova, T.V.; Raap, J.; Arseniev, A.S. Spatial Structure of Zervamicin IIB Bound to DPC Micelles: Implications for Voltage-Gating. Biophys. J. 2002, 82, 762–771. [Google Scholar] [CrossRef]
- Dalla Torre, C.; Sannio, F.; Battistella, M.; Docquier, J.-D.; De Zotti, M. Peptaibol Analogs Show Potent Antibacterial Activity against Multidrug Resistant Opportunistic Pathogens. Int. J. Mol. Sci. 2023, 24, 7997. [Google Scholar] [CrossRef] [PubMed]
- Samsonova, O.V.; Kudryashova, K.S.; Feofanov, A.V. N-Terminal Moiety of Antimicrobial Peptide Ltc1-k Increases Its Toxicity for Eukaryotic Cells. Acta Naturae 2011, 3, 68–78. [Google Scholar] [CrossRef] [PubMed][Green Version]
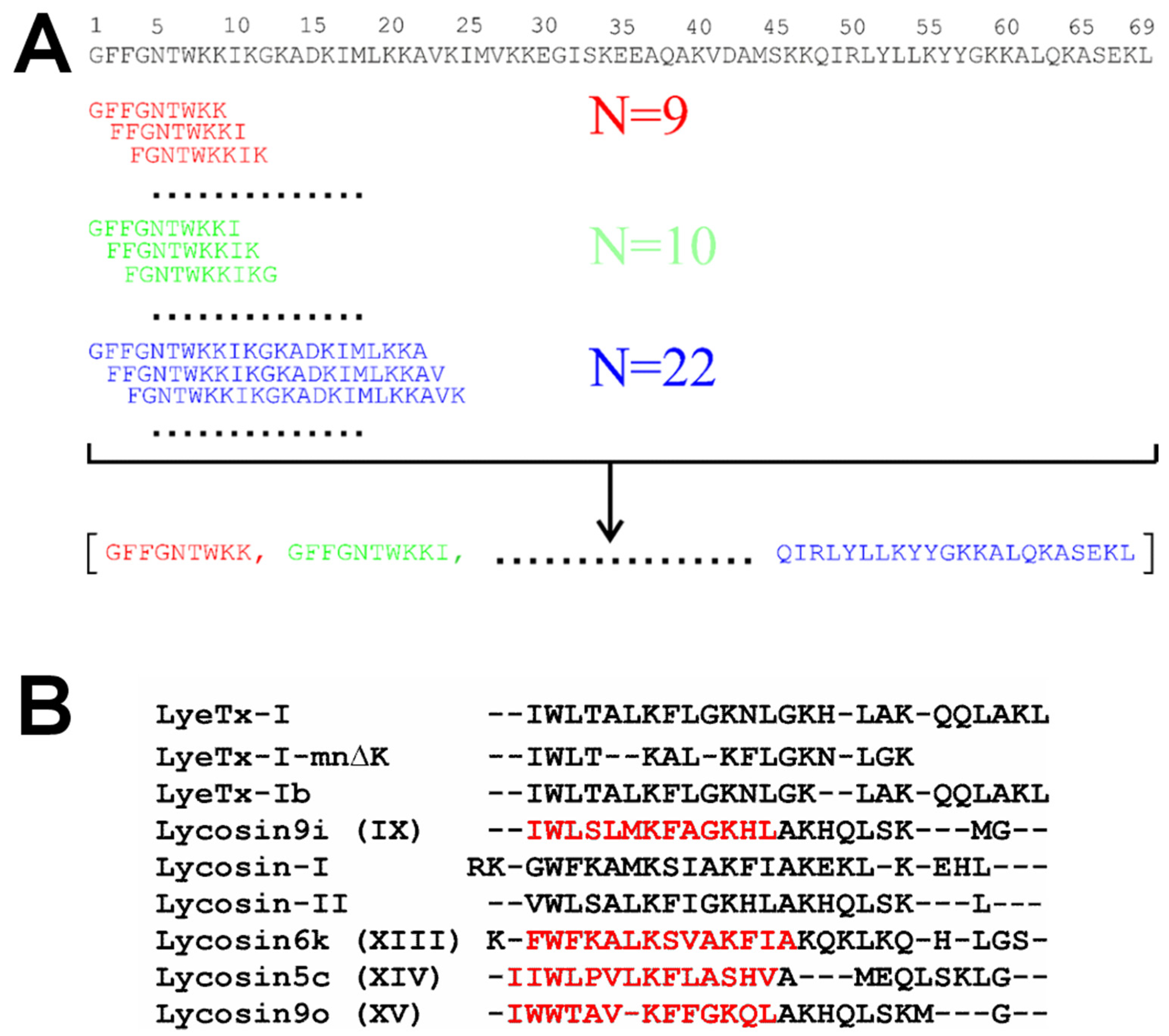

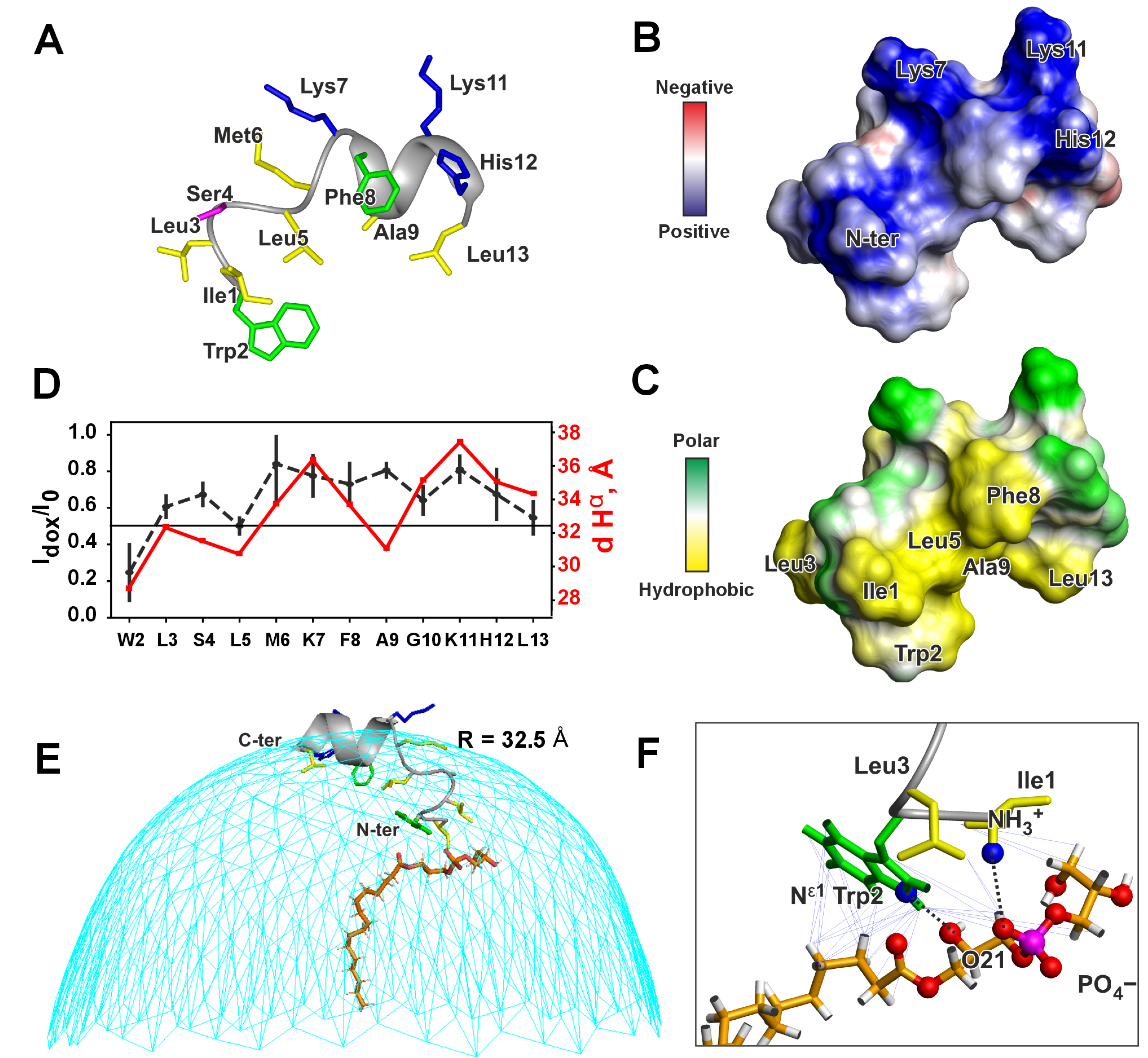
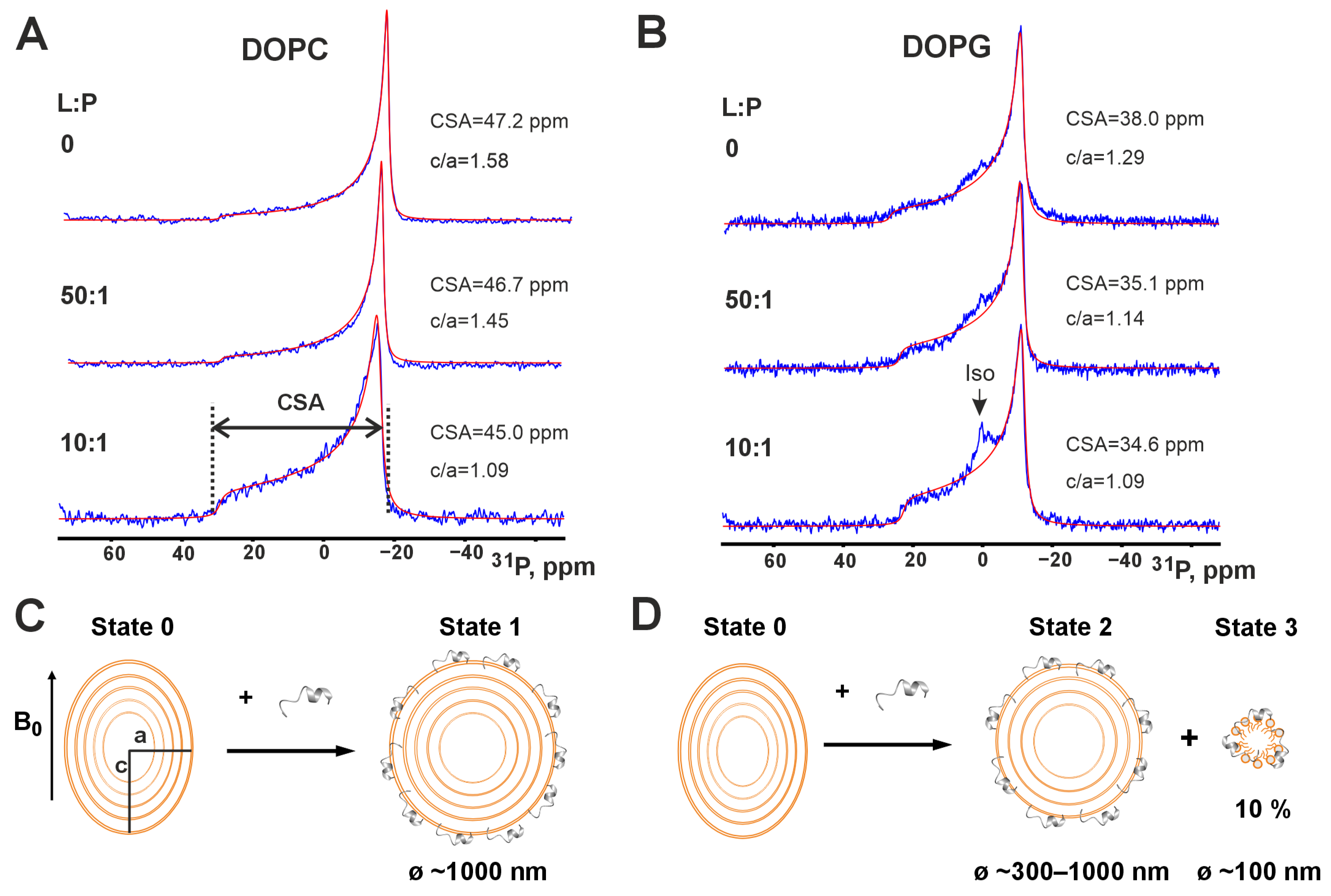



| № | Parent LP and Fragment Sequences 1 | Name | Organism | C-Term 2 | Length | Charge 3 | Gravy 4 | Uniprot ID/GenBank |
|---|---|---|---|---|---|---|---|---|
| I | EAGWMKALKEHVEKLNKTGKLKNLKPPETDTCSFAANAYKALATIRETIDTLKNKLC | Pardosin 13f | Pardosa amentata | – | 20 | 4 | −1.01 | – |
| II | AGLRDFMKRLISKGKIGKEKLVAFIKRVISRVKSR | Pardosin 11d | Pardosa palustris | – | 19 | 5 | 0.05 | A0A8D7ZRV4C/AG6443143.1 |
| III | AIWSSAMQFFIKHLKKENLKKLG | Alopecosin 6c | Alopecosa marikovskyi | – | 16 | 3 | 0.19 | A0A8D7ZRX2/CAG6443179.1 |
| IV | GIKDYLKKMLLKLKEKLKSMTS | Peucetin 6 (Peu 6) | Peucetia striata | – | 14 | 3 | −0.22 | A0A8D7ZRT4/CAG6443209.1 |
| V | MILADLIAKLKVRAAKVSG | Trochosin 2l | Trochosa ruricola | NH2 | 14 | 3 | 1.14 | A0A8D8EPX9/CAG6443258.1 |
| VI | IWFSLMKFAGKHLAKHQLSKMG | Lycosin 9l | Lycosa hispanica | NH2 | 19 | 5 | −0.33 | A0A8D8EPN8/CAG6442973.1 |
| VII | IWLSLMKFAGKHLAKHQLSKMG | Lycosin 9i | Lycosa hispanica | – | 16 | 3 | 0.04 | A0A8D7ZRD0/CAG6442955.1 |
| VIII | IWLSLMKFAGKHLAKHQLSKMG | Lycosin 9i | Lycosa hispanica | NH2 | 14 | 4 | 0.03 | A0A8D7ZRD0/CAG6442955.1 |
| IX | IWLSLMKFAGKHLAKHQLSKMG | Lycosin 9i | Lycosa hispanica | NH2 | 13 | 3 | 0.71 | A0A8D7ZRD0/CAG6442955.1 |
| X | IWLSLMKFAGKHLAKHQLSKMG | Lycosin 9i | Lycosa hispanica | – | 13 | 2 | 0.71 | A0A8D7ZRD0/CAG6442955.1 |
| XI | IWLSLMKFAGKHLAKHQLSKMG | Lycosin 9i | Lycosa hispanica | NH2 | 12 | 4 | −0.22 | A0A8D7ZRD0/CAG6442955.1 |
| XII | IWLSLMKFAGKHLAKHQLSKMG | Lycosin 9i | Lycosa hispanica | NH2 | 10 | 4 | −0.83 | A0A8D7ZRD0/CAG6442955.1 |
| XIII | KFWFKALKSVAKFIAKQKLKQHLGSE | Lycosin 6k | Lycosa hispanica | NH2 | 14 | 4 | 0.92 | A0A8D8EQY2/CAG6442961.1 |
| XIV | IIWLPVLKFLASHVAMEQLSKLG | Lycosin 5c | Lycosa hispanica, Alopecosa marikovskyi | NH2 | 14 | 2 | 1.64 | A0A8D7ZUM9/CAG6443133.1 |
| XV | IWWTAVKFFGKQLAKHQLSKMG | Lycosin 9o | Lycosa hispanica | NH2 | 13 | 3 | 0.44 | A0A8D8EQW4/CAG6442974.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mironov, P.A.; Baranova, A.A.; Alferova, V.A.; Egorova, N.S.; Ignatova, A.A.; Feofanov, A.V.; Shenkarev, Z.O.; Dubovskii, P.V. NMR Unveils Activity Mechanism of Linear Spider Venom Peptide Fragments Selected by Neural Networks Against Staphylococci Including MRSA. Pharmaceutics 2025, 17, 1526. https://doi.org/10.3390/pharmaceutics17121526
Mironov PA, Baranova AA, Alferova VA, Egorova NS, Ignatova AA, Feofanov AV, Shenkarev ZO, Dubovskii PV. NMR Unveils Activity Mechanism of Linear Spider Venom Peptide Fragments Selected by Neural Networks Against Staphylococci Including MRSA. Pharmaceutics. 2025; 17(12):1526. https://doi.org/10.3390/pharmaceutics17121526
Chicago/Turabian StyleMironov, Pavel A., Anna A. Baranova, Vera A. Alferova, Natalya S. Egorova, Anastasia A. Ignatova, Alexey V. Feofanov, Zakhar O. Shenkarev, and Peter V. Dubovskii. 2025. "NMR Unveils Activity Mechanism of Linear Spider Venom Peptide Fragments Selected by Neural Networks Against Staphylococci Including MRSA" Pharmaceutics 17, no. 12: 1526. https://doi.org/10.3390/pharmaceutics17121526
APA StyleMironov, P. A., Baranova, A. A., Alferova, V. A., Egorova, N. S., Ignatova, A. A., Feofanov, A. V., Shenkarev, Z. O., & Dubovskii, P. V. (2025). NMR Unveils Activity Mechanism of Linear Spider Venom Peptide Fragments Selected by Neural Networks Against Staphylococci Including MRSA. Pharmaceutics, 17(12), 1526. https://doi.org/10.3390/pharmaceutics17121526








