Innovative Strategies for the Targeted Degradation of Viral Proteins: Paving the Way for Next-Generation Therapeutics
Abstract
1. Introduction
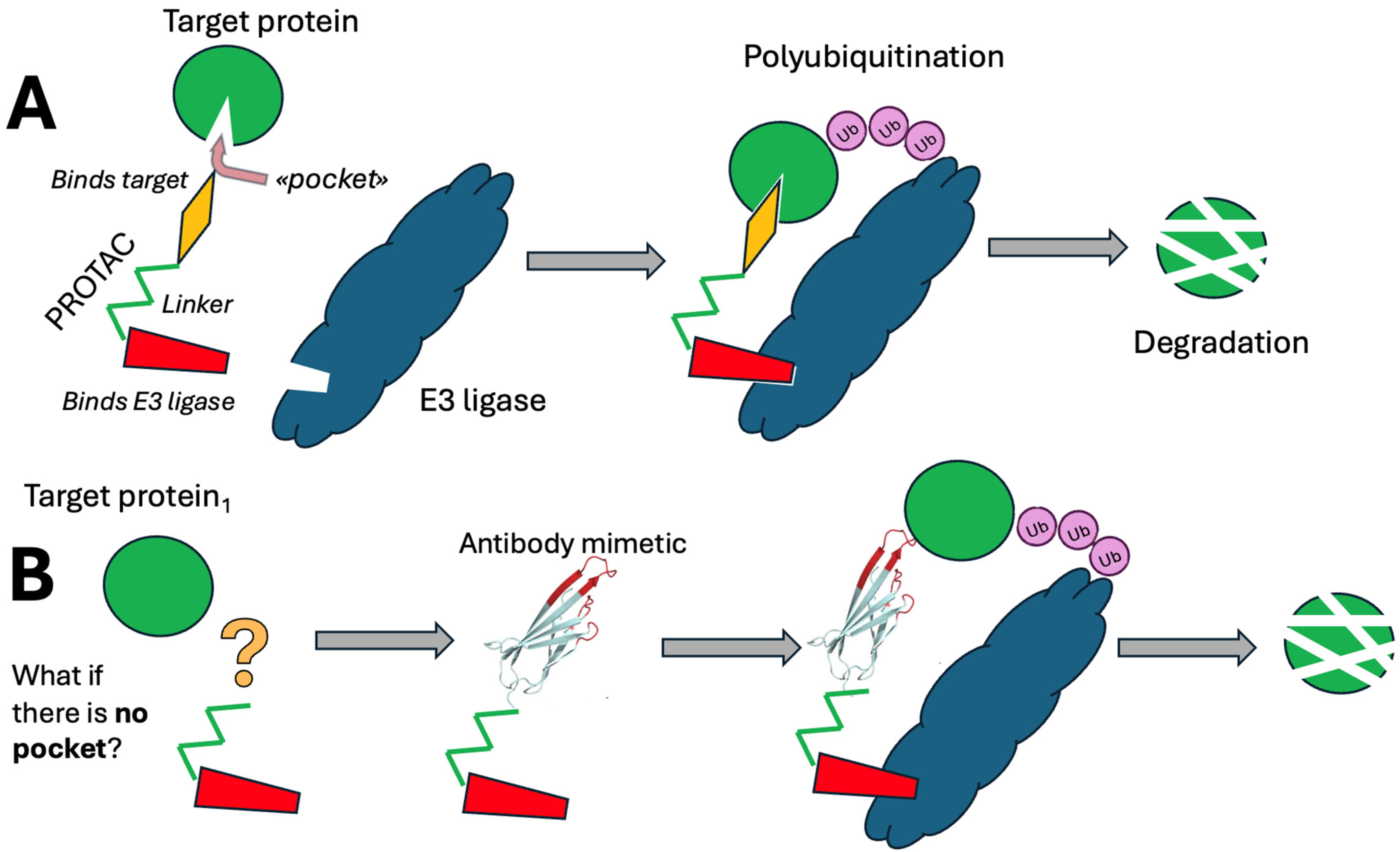
2. PROTACs
3. Diving Antibodies
- The Cell Entry Module: Facilitates entry into cells via receptor-mediated endocytosis or containing cell-penetrating peptides, either specifically or non-specifically.
- The Endosomal Escape Module: Enables escape from endocytotic vesicles into the hyaloplasm.
- The Ubiquitination Module: Contains an amino acid sequence that binds E3 ubiquitin ligase, promoting ubiquitination of the target protein.
- The Targeting Module: Incorporates an antiviral protein-specific antibody mimetic, with an added hydrolysis site by acid proteases between the targeting and ubiquitination modules and the rest of the molecule for controlled degradation within endocytotic compartments.
- The Subcellular Localization Module is essential for directing the molecule to specific subcellular compartments beyond the hyaloplasm.
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DAb | diving antibody |
| E3LBP | E3-ligase binding peptide |
| HCV | hepatitis C virus |
| Mpro | SARS-CoV-2 main protease |
| PROTACs | PROteolysis TArgeting Chimeras |
| TRIM | tripartite motif protein |
| VHL | von Hippel–Lindau Cullin RING E3 ligase |
References
- Bulatov, E.; Ciulli, A. Targeting Cullin-RING E3 Ubiquitin Ligases for Drug Discovery: Structure, Assembly and Small-Molecule Modulation. Biochem. J. 2015, 467, 365–386. [Google Scholar] [CrossRef] [PubMed]
- Heaton, S.M.; Borg, N.A.; Dixit, V.M. Ubiquitin in the Activation and Attenuation of Innate Antiviral Immunity. J. Exp. Med. 2016, 213, 1–13. [Google Scholar] [CrossRef]
- Wang, S.; Chen, Y.; Li, C.; Wu, Y.; Guo, L.; Peng, C.; Huang, Y.; Cheng, G.; Qin, F.X.F. TRIM14 Inhibits Hepatitis C Virus Infection by SPRY Domain-Dependent Targeted Degradation of the Viral NS5A Protein. Sci. Rep. 2016, 6, 32336. [Google Scholar] [CrossRef]
- Van Gent, M.; Sparrer, K.M.J.; Gack, M.U. TRIM Proteins and Their Roles in Antiviral Host Defenses. Annu. Rev. Virol. 2018, 5, 385–405. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, J.; Pang, X.; Liu, Z.; Li, Q.; Yi, D.; Zhang, Y.; Fang, X.; Zhang, T.; Zhou, R.; et al. An Anti-Influenza A Virus Microbial Metabolite Acts by Degrading Viral Endonuclease PA. Nat. Commun. 2022, 13, 2079. [Google Scholar] [CrossRef]
- Chatterjee, P.; Ponnapati, M.; Kramme, C.; Plesa, A.M.; Church, G.M.; Jacobson, J.M. Targeted Intracellular Degradation of SARS-CoV-2 via Computationally Optimized Peptide Fusions. Commun. Biol. 2020, 3, 715. [Google Scholar] [CrossRef]
- Martinez-Ortiz, W.; Zhou, M.M. Could PROTACs Protect Us From COVID-19? Drug Discov. Today 2020, 25, 1894–1896. [Google Scholar] [CrossRef]
- Nandave, M.; Jain, P. (Eds.) PROTAC-Mediated Protein Degradation: A Paradigm Shift in Cancer Therapeutics; Springer: Singapore, 2024; ISBN 9789819750771. [Google Scholar]
- Békés, M.; Langley, D.R.; Crews, C.M. PROTAC Targeted Protein Degraders: The Past Is Prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef]
- Reboud-Ravaux, M.; El Amri, C. COVID-19 Therapies: Protease Inhibitions and Novel Degrader Strategies. Front. Drug Discov. 2022, 2, 892057. [Google Scholar] [CrossRef]
- Desantis, J.; Goracci, L. Proteolysis Targeting Chimeras in Antiviral Research. Future Med. Chem. 2022, 14, 459–462. [Google Scholar] [CrossRef] [PubMed]
- de Wispelaere, M.; Du, G.; Donovan, K.A.; Zhang, T.; Eleuteri, N.A.; Yuan, J.C.; Kalabathula, J.; Nowak, R.P.; Fischer, E.S.; Gray, N.S.; et al. Small Molecule Degraders of the Hepatitis C Virus Protease Reduce Susceptibility to Resistance Mutations. Nat. Commun. 2019, 10, 3468. [Google Scholar] [CrossRef]
- Zhou, H.; Wu, S.; Xu, Z. A Kind of Oseltamivir PROTAC Compound and Its Preparation Method and Application in Anti-Influenza Virus Drug. CN 112592331 B, 22 October 2021. [Google Scholar]
- Xu, Z.; Liu, X.; Ma, X.; Zou, W.; Chen, Q.; Chen, F.; Deng, X.; Liang, J.; Dong, C.; Lan, K.; et al. Discovery of Oseltamivir-Based Novel PROTACs as Degraders Targeting Neuraminidase to Combat H1N1 Influenza Virus. Cell Insight 2022, 1, 100030. [Google Scholar] [CrossRef]
- Li, H.; Wang, S.; Ma, W.; Cheng, B.; Yi, Y.; Ma, X.; Xiao, S.; Zhang, L.; Zhou, D. Discovery of Pentacyclic Triterpenoid PROTACs as a Class of Effective Hemagglutinin Protein Degraders. J. Med. Chem. 2022, 65, 7154–7169. [Google Scholar] [CrossRef] [PubMed]
- Schäffner, E.; Costard, J. Platform-Derived Broad-Spectrum Antivirals A Progress Report on the SPRIND Challenge “Broad-Spectrum Antivirals”. 2023. Available online: https://cms.system.sprind.org/uploads/Platform_derived_broad_spectrum_antivirals_449f78c65b.pdf (accessed on 19 September 2025).
- Zhang, K.; Röske, J.; Buddrus, L.; Maple, H.; Moloney, A.; Marsh, G.; Cooper, M.; Karadogan, B.; Chen, Y.-T.; Rox, K.; et al. MproTAC—New Approach for Development of SARS-CoV-2 Antiviral Drugs. 2024. Available online: https://www.xtal-concepts.com/wp-content/uploads/2024-05-31-Kaixuan-poster.pdf (accessed on 19 September 2025).
- Alugubelli, Y.R.; Xiao, J.; Khatua, K.; Kumar, S.; Sun, L.; Ma, Y.; Ma, X.R.; Vulupala, V.R.; Atla, S.; Blankenship, L.R.; et al. Discovery of First-in-Class PROTAC Degraders of SARS-CoV-2 Main Protease. J. Med. Chem. 2024, 67, 6495–6507. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Mountford, S.J.; Kiso, M.; Anderson, D.E.; Papadakis, G.; Jarman, K.E.; Tilmanis, D.R.; Maher, B.; Tran, T.; Shortt, J.; et al. Targeted Protein Degraders of SARS-CoV-2 Mpro Are More Active than Enzymatic Inhibition Alone with Activity against Nirmatrelvir Resistant Virus. Commun. Med. 2025, 5, 140. [Google Scholar] [CrossRef]
- Hahn, F.; Hamilton, S.T.; Wangen, C.; Wild, M.; Kicuntod, J.; Brückner, N.; Follett, J.E.L.; Herrmann, L.; Kheimar, A.; Kaufer, B.B.; et al. Development of a PROTAC-Based Targeting Strategy Provides a Mechanistically Unique Mode of Anti-Cytomegalovirus Activity. Int. J. Mol. Sci. 2021, 22, 12858. [Google Scholar] [CrossRef]
- Grifagni, D.; Lenci, E.; De Santis, A.; Orsetti, A.; Barracchia, C.G.; Tedesco, F.; Bellini Puglielli, R.; Lucarelli, F.; Lauriola, A.; Assfalg, M.; et al. Development of a GC-376 Based Peptidomimetic PROTAC as a Degrader of 3-Chymotrypsin-like Protease of SARS-CoV-2. ACS Med. Chem. Lett. 2024, 15, 250–257. [Google Scholar] [CrossRef]
- Luo, D.; Luo, R.; Wang, W.; Deng, R.; Wang, S.; Ma, X.; Pu, C.; Liu, Y.; Zhang, H.; Yu, S.; et al. Discovery of L15 as a Novel Vif PROTAC Degrader with Antiviral Activity against HIV-1. Bioorg. Med. Chem. Lett. 2024, 111, 129880. [Google Scholar] [CrossRef]
- Mukerjee, N.; Mukherjee, D. PROTAC-Based Therapeutics for Targeting HPV Oncoproteins in Head and Neck Cancers. Nano TransMed 2025, 4, 100071. [Google Scholar] [CrossRef]
- Mukerjee, N.; Maitra, S.; Sharma, R. Proteolysis—Targeting Chimeras in Antiviral Therapy: Leveraging Influenza Virus and Exosome—Mediated Delivery for Targeted Protein Degradation and Therapeutic Advancements. Drug Dev. Res. 2024, 85, e22145. [Google Scholar] [CrossRef]
- Mukerjee, N.; Maitra, S.; Ghosh, A.; Sengupta, T.; Alexiou, A.; Subramaniyan, V.; Anand, K. Synergizing Proteolysis-Targeting Chimeras and Nanoscale Exosome-Based Delivery Mechanisms for HIV and Antiviral Therapeutics. ACS Appl. Nano Mater. 2024, 7, 3499–3514. [Google Scholar] [CrossRef]
- Sobolev, A.S. The Delivery of Biologically Active Agents into the Nuclei of Target Cells for the Purposes of Translational Medicine. Acta Naturae 2020, 12, 47–56. [Google Scholar] [CrossRef]
- Khramtsov, Y.V.; Ulasov, A.V.; Slastnikova, T.A.; Rosenkranz, A.A.; Lupanova, T.N.; Georgiev, G.P.; Sobolev, A.S. Modular Nanotransporters Delivering Biologically Active Molecules to the Surface of Mitochondria. Pharmaceutics 2023, 15, 2687. [Google Scholar] [CrossRef] [PubMed]
- Khramtsov, Y.V.; Ulasov, A.V.; Rosenkranz, A.A.; Slastnikova, T.A.; Lupanova, T.N.; Georgiev, G.P.; Sobolev, A.S. Modular Nanotransporters Deliver Anti-Keap1 Monobody into Mouse Hepatocytes, Thereby Inhibiting Production of Reactive Oxygen Species. Pharmaceutics 2024, 16, 1345. [Google Scholar] [CrossRef] [PubMed]
- Khramtsov, Y.V.; Rosenkranz, A.A.; Ulasov, A.V.; Slastnikova, T.A.; Lupanova, T.N.; Alieva, R.T.; Georgiev, G.P.; Sobolev, A.S. Modular Nanotransporters Containing Keap1 Monobodies Are Capable of Reducing the Toxic Effect of Acetaminophen on the Liver of Mice. Dokl. Biochem. Biophys. 2025, 521, 174–177. [Google Scholar] [CrossRef]
- Lupanova, T.N.; Ulasov, A.V.; Khramtsov, Y.V.; Rozenkranz, A.A.; Georgiev, G.P.; Sobolev, A.S. Intracellular Delivery of an Antibody-Like Molecule Capable of Inhibiting c-Myc. Dokl. Biochem. Biophys. 2023, 509, 70–72. [Google Scholar] [CrossRef]
- Khramtsov, Y.V.; Ulasov, A.V.; Lupanova, T.N.; Georgiev, G.P.; Sobolev, A.S. Quantitative Description of the N-Protein of the SARS-CoV-2 Virus Degradation in Cells Stably Expressing It under the Influence of New Modular Nanotransporters. Dokl. Biochem. Biophys. 2024, 513, S63–S66. [Google Scholar] [CrossRef] [PubMed]
- Khramtsov, Y.V.; Ulasov, A.V.; Lupanova, T.N.; Slastnikova, T.A.; Rosenkranz, A.A.; Bunin, E.S.; Georgiev, G.P.; Sobolev, A.S. Intracellular Degradation of SARS-CoV-2 N-Protein Caused by Modular Nanotransporters Containing Anti-N-Protein Monobody and a Sequence That Recruits the Keap1 E3 Ligase. Pharmaceutics 2024, 16, 4. [Google Scholar] [CrossRef]
- Khramtsov, Y.V.; Ulasov, A.V.; Lupanova, T.N.; Georgiev, G.P.; Sobolev, A.S. Modular Nanotransporters Capable of Causing Intracellular Degradation of the N-Protein of the SARS-CoV-2 Virus in A549 Cells with Temporary Expression of This Protein Fused with a Fluorescent Protein MRuby3. Dokl. Biochem. Biophys. 2024, 513, S60–S62. [Google Scholar] [CrossRef]
- Khramtsov, Y.V.; Lupanova, T.N.; Rosenkranz, A.A.; Georgiev, G.P.; Sobolev, A.S. Optimization of A549 Cell Transfection Efficiency with a Plasmid Encoding the N-Protein of the SARS-CoV-2 Virus. Dokl. Biochem. Biophys. 2025, 521, 157–159. [Google Scholar] [CrossRef]
- Khramtsov, Y.V.; Ulasov, A.V.; Lupanova, T.N.; Georgiev, G.P.; Sobolev, A.S. Modular Nanotransporters Capable of Binding to SARS-CoV-2 Virus Nucleocapsid Protein in Target Cells. Dokl. Biochem. Biophys. 2023, 510, 87–90. [Google Scholar] [CrossRef]
- Hantschel, O.; Biancalana, M.; Koide, S. Monobodies as Enabling Tools for Structural and Mechanistic Biology. Curr. Opin. Struct. Biol. 2020, 60, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Dai, T.; Qin, Z.; Pan, T.; Chu, F.; Lou, L.; Zhang, L.; Yang, B.; Huang, H.; Lu, H.; et al. Targeting Liquid–Liquid Phase Separation of SARS-CoV-2 Nucleocapsid Protein Promotes Innate Antiviral Immunity by Elevating MAVS Activity. Nat. Cell Biol. 2021, 23, 718–732. [Google Scholar] [CrossRef] [PubMed]
- Mihalič, F.; Simonetti, L.; Giudice, G.; Sander, M.R.; Lindqvist, R.; Peters, M.B.A.; Benz, C.; Kassa, E.; Badgujar, D.; Inturi, R.; et al. Large-Scale Phage-Based Screening Reveals Extensive Pan-Viral Mimicry of Host Short Linear Motifs. Nat. Commun. 2023, 14, 2409. [Google Scholar] [CrossRef]
- Cermakova, K.; Hodges, H.C. Interaction Modules That Impart Specificity to Disordered Protein. Trends Biochem. Sci. 2023, 48, 477–490. [Google Scholar] [CrossRef]
- Khramtsov, Y.V.; Ulasov, A.V.; Lupanova, T.N.; Georgiev, G.P.; Sobolev, A.S. Among antibody-like molecules, monobodies, able to interact with nucleocapsid protein of SARS-CoV virus, there are monobodies with high affinity to nucleocapsid protein of SARS-CoV-2 virus. Dokl. Biochem. Biophys. 2022, 503, 90–92. [Google Scholar] [CrossRef]
- Hong, K.B.; An, H. Degrader–antibody conjugates: Emerging new modality. J. Med. Chem. 2023, 66, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Khoo, R.; Peh, M.; Teo, J.; Chang, S.C.; Ng, S.; Beilhartz, G.L.; Melnyk, R.A.; Johannes, C.W.; Brown, C.J.; et al. BioPROTACs as Versatile Modulators of Intracellular Therapeutic Targets Including Proliferating Cell Nuclear Antigen (PCNA). Proc. Natl. Acad. Sci. USA 2020, 117, 5791–5800. [Google Scholar] [CrossRef]

| Virus | Target Protein | PROTAC | Reference |
|---|---|---|---|
| Hepatitis C | NS3/4A protease | DGY-08-097 | [12] |
| Influenza A | Neuraminidase | 8e | [13,14] |
| Influenza A | Hemagglutinin | V3 | [15] |
| SARS-CoV-2 | Main protease | MPD2 | [18] |
| SARS-CoV-2 | Main protease | BP-198 | [19] |
| SARS-CoV-2 | 3-chymotrypsin-like protease | PROTAC 1 | [21] |
| Human cytomegalovirus | Cyclin-dependent kinase 9 | THAL-SNS032 | [20] |
| Human immunodeficiency virus-1 | Vif | L15 | [22] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobolev, A.S.; Georgiev, G.P. Innovative Strategies for the Targeted Degradation of Viral Proteins: Paving the Way for Next-Generation Therapeutics. Pharmaceutics 2025, 17, 1420. https://doi.org/10.3390/pharmaceutics17111420
Sobolev AS, Georgiev GP. Innovative Strategies for the Targeted Degradation of Viral Proteins: Paving the Way for Next-Generation Therapeutics. Pharmaceutics. 2025; 17(11):1420. https://doi.org/10.3390/pharmaceutics17111420
Chicago/Turabian StyleSobolev, Alexander S., and Georgii P. Georgiev. 2025. "Innovative Strategies for the Targeted Degradation of Viral Proteins: Paving the Way for Next-Generation Therapeutics" Pharmaceutics 17, no. 11: 1420. https://doi.org/10.3390/pharmaceutics17111420
APA StyleSobolev, A. S., & Georgiev, G. P. (2025). Innovative Strategies for the Targeted Degradation of Viral Proteins: Paving the Way for Next-Generation Therapeutics. Pharmaceutics, 17(11), 1420. https://doi.org/10.3390/pharmaceutics17111420







