Abstract
The study presents data on the anti-inflammatory effects of a combination of sodium dichloroacetate and sodium valproate (DCA–VPA) on the expression of inflammation- and immune response-related genes in T lymphocytes of SARS-CoV-2 patients. The study aimed to assess the effects of DCA–VPA on the genes of cytokine activity, chemokine-mediated signaling, neutrophil chemotaxis, lymphocyte chemotaxis, T-cell chemotaxis, and regulation of T-cell proliferation pathways. The study included 21 patients with SARS-CoV-2 infection and pneumonia: 9 male patients with a mean age of 68.44 ± 15.32 years and 12 female patients with a mean age of 65.42 ± 15.74 years. They were hospitalized between December 2022 and March 2023. At the time of testing, over 90% of sequences analyzed in Lithuania were found to be of the omicron variant of SARS-CoV-2. The T lymphocytes from patients were treated with 5 mmol DCA and 2 mmol VPA for 24 h in vitro. The effect of the DCA–VPA treatment on gene expression in T lymphocytes was analyzed via gene sequencing. The study shows that DCA–VPA has significant anti-inflammatory effects and apparent sex-related differences. The effect is more potent in T cells from male patients with SARS-CoV-2 infection and pneumonia than in females.
1. Introduction
The discovery of novel therapeutic indications for authorized medicines and the development of new medicinal products by a combination of known drugs for treating severe infections are areas of significant interest in medicine. The research presents data from the anti-inflammatory effects of sodium dichloroacetate (DCA) and sodium valproate (VPA) salt combination (DCA–VPA) on SARS-CoV-2 infection patients’ T lymphocytes in vitro. VPA and DCA are well-established medicines with a therapeutic experience and known safety profile, dosage, and blood levels. Therefore, they are attractive for studies to explore potential new therapeutic indications for these medicines or their combination. DCA–VPA has anti-inflammatory effects in mouse thymocytes, significantly affecting the expression of inflammation-related genes involved in thymocyte inflammation-related cytokine activity, the inflammatory response, and the IL17 signaling pathways [1].
Severe acute respiratory syndrome coronavirus 2 disease (SARS-CoV-2 infection; COVID-19) disrupts the immune response, leading to high levels of cytokine and other inflammation molecules released in the blood. However, cytokine-inhibitor therapies have been unsuccessful and can even aggravate the patient’s condition in some cases [2,3]. Severe infections are a sign of an inflammatory and immune response to infection and a manifestation of severe mitochondrial metabolic disturbances [4,5,6], which can lead to an unfavorable outcome and death [7,8,9]. Therefore, treating metabolic disorders is a vital target for treating serious SARS-CoV-2 [10].
The meta-analysis shows that the risk of SARS-CoV-2 infection is the same in men and women. Men with COVID-19 are three times more frequently admitted to the intensive care unit and also more often dying [11,12,13,14]. In contrast to women, a higher mortality in COVID-19 men is found in all age groups [13,15]. Male gender is a decisive predisposing factor, identified by multiple analyses of the patient, for severe SARS-CoV-2 infection [11].
DCA and VPA are investigational drugs exhibiting anti-inflammatory effects in viral and bacterial infections, and these effects are associated with the reversal of mitochondrial dysregulation and impaired immune responses [16,17]. Regulatory guidelines for investigational medicines research call for the importance of the impact of sex on the assessment of drug efficacy, and it is essential to provide relevant information on the sex-specific efficacy of drugs under development [18,19]. Female and male cells have different responses to chemicals. Studies on the sex-specific effects of drugs must be addressed in preclinical and clinical trials [20,21].
DCA has been studied as an investigational medicine for over 50 years. It is a preparation found to lower blood glucose levels; it works as an inhibitor of pyruvate dehydrogenase kinase (PDK) activity, increasing the activity of pyruvate dehydrogenase (PDH) and its complex (PDHC); and it is commonly used to treat diseases associated with mitochondrial defects and increased congenital or acquired lactic acid production [22]. The inactivation of the PDHA1 subunit is associated with metabolic reprogramming and the overproduction of lactic acid, a pro-inflammatory metabolite that increases the production of pro-inflammatory cytokines [23]. PDH inactivation during severe infectious inflammation and sepsis is detected in peripheral blood mononuclear cells, macrophages, and vascular endothelial cells [24,25]. As a modulator of mitochondria, DCA is also a potential agent for inhibiting inflammation. Elevated blood lactate concentration is a marker of the severity of SARS-CoV-2 infection and an independent predictor of poor prognosis in COVID-19, sepsis, and other infections [26,27].
VPA may reduce the expression of the ACE2 receptor and the transmembrane serine protease 2 (TMPRSS2) receptor, which requires co-expression for SARS-CoV-2 viral entry into a cell [28,29]. Male cells have a single ACE2 allele, as the ACE2 gene is encoded on the X chromosome [30,31]. The female cell’s mosaicism of the X chromosome is associated with the heterogeneity of the ACE2 allele, which may encode partial resistance to COVID-19 infection in women [32]. Female tissues have lower ACE2 expression than males [33,34,35]. Older women have lower ACE2 activity than younger women, which is uncommon in men [35]. Testosterone increases ACE2 levels, while estrogens suppress ACE2 expression [36,37,38]. No sex-related differences in TMPRSS2 expression were found in human and mouse lungs [39]. Pharmacological intervention with an androgen receptor antagonist significantly inhibited ACE2 expression in the lungs of male mice [39]. TMPRSS2 is potentially the most promising target for SARS-CoV-2 infection therapy as its specific expression is in alveolar cells [40,41]; TMPRSS2 inhibitors are effective against SARS-CoV-2 [28]. VPA downregulates TMPRSS2 expression [41,42]. SARS-CoV-2 infection was reduced by more than 75% in patients treated with VPA monotherapy compared to the general population [43]. Based on clinical and laboratory endpoints, VPA-treated patients develop less severe COVID-19 than control patients [44].
Bioinformatics approaches allow a broader analysis of the potential effects of registered drugs, thus opening up the possibility of their use in treating SARS-CoV-2 infection [45]. One such drug is VPA, a histone deacetylase (HDAC) inhibitor [46]. The opening of the SARS-CoV-2 spike trimer receptor binding domains is blocked by VPA and coenzyme A conjugate [47]. The X-ray crystal structure of SARS-CoV-2 has shown that central protease (Mpro, 3CLpro) is a crucial protein in the viral life cycle [48,49,50]. HDAC inhibitors bind tightly to the active site of the crystallographic structure of viral Mpro [51]. Viral Mpro is a target for antiviral drugs [52,53]. The SARS-CoV-2 protease NSP5 interacts with HDAC2. It is thought that NSP5 may inhibit HDAC2 translocation into the nucleus and affect HDAC2 potency and mechanisms of inflammation [54,55]. HDAC2 binding to promoters was lower in females than males [56]. VPA reduced HDAC2 levels in the frontal cortex tissue of male rats, but there was no such effect in females [57]. HDAC2 protein levels are sensitive to the selective effects of VPA [58]. VPA has been investigated in treating various viral infections [45,59]. VPA is being investigated as an immunomodulator [45,60].
Elevated glucose levels favor the progression of SARS-CoV-2 infection [61]; glycolysis is essential for the rapid growth and proliferation of virus-activated T cells [62,63,64]. Treatment with DCA or VPA monotherapy reduces blood glucose levels [22,65,66].
The novelty of the DCA–VPA research is based on the mechanisms of synergistic effects of these agents. VPA activates the SLC5A8 gene through DNA demethylation by inhibiting HDAC activity [67]. The SLC5A8 carries short-chain fatty acids and DCA into cells [68,69]. SLC5A8, participating in the mitochondrial β-oxidation pathway, enhances the transport of DCA into the cell, thereby altering the regulation of mitochondrial metabolism [70,71]. SLC5A8 is well expressed in cells in the kidney [72] and lung [73] tissues, which are targets of SARS-CoV-2. Thus, the synergistic effect of VPA and DCA may be pharmacologically relevant to investigate the efficacy of SARS-CoV-2 infection treatment.
Investigating sex-specific differences in treating SARS-CoV-2 infection may lead to a new approach to the personalized treatment of the disease. This study aimed to evaluate DCA–VPA as a potential investigational drug for the treatment of SARS-CoV-2 infection, to elucidate the possible biological pathways of the pathogenesis of the disease, to examine the impact of sex-specific pharmacological mechanisms of SARS-CoV-2 infection, and to explore the possibility of an individualized treatment of the disease.
The study aimed to investigate the effect of the DCA–VPA treatment on the expression of inflammation- and immune response-related genes in T lymphocytes of SARS-CoV-2-infected patients with pneumonia. The objectives of the study were to determine the effect of the DCA–VPA treatment on cytokine activity, chemokine-mediated signaling, neutrophil chemotaxis, lymphocyte chemotaxis, T-cell chemotaxis, and regulation of T-cell proliferation pathways, and to detect sex-related differences in the treatment efficacy.
The study showed that the treatment of T lymphocytes from SARS-CoV-2-infected patients with pneumonia with DCA–VPA in vitro had an anti-inflammatory effect, manifested by inhibiting pro-inflammatory gene expression in T lymphocytes. The impact on cytokine activity, chemokine-mediated signaling, neutrophil chemotaxis, lymphocyte chemotaxis, and regulation of T-cell proliferation pathways was superior in male T lymphocytes than in female T lymphocytes, and gender-related differences in treatment effect were identified.
2. Materials and Methods
2.1. The Investigational Medicinal Product
The investigational medicinal product combines sodium dichloroacetate (DCA; Sigma-Aldrich, Steinheim, Germany) and sodium valproate (VPA; Sigma-Aldrich, Steinheim, Germany). The combination of these medicinal products is a pending patent filed by us that covers DCA–VPA as a new medicinal product for the treatment of viral and bacterial infections (National patent application No. LT2023 532; 22 August 2023), as well as for the treatment of cancer (Official bulletin of the state patent bureau of the Republic of Lithuania, No. 6874, filling date 17 April 2020, https://vpb.lrv.lt/uploads/vpb/documents/files/VPB-OB-Nr23-2021-12-10-1d.pdf (accessed on 2 February 2024)); a European patent application has been submitted (European patent application No. 21168796.7, filing date 16 April 2021, https://register.epo.org/application?number=EP21168796 (accessed on 2 February 2024)).
2.2. Patients with SARS-CoV-2 Infection and Pneumonia
The study included 21 patients (9 males and 12 females) hospitalized in an infectious diseases unit due to a serious condition caused by COVID-19 infection and pneumonia. The study was approved by the Kaunas Regional Biomedical Research Ethics Committee (Authorization to conduct biomedical research 2021-12-28; No. BE-2-140). Patients were hospitalized at the Department of Infectious Diseases of the Lithuanian University of Health Sciences Kaunas Hospital (Kaunas, Lithuania). Blood was collected from the patients, and T lymphocytes were isolated. Patients were enrolled according to established inclusion and exclusion criteria. Participants could be included in the study only if all criteria were applied. Adult males and females who independently gave informed consent to participate in the study were selected.
Inclusion criteria were as follows: (1) adult male and female patients, older than 18 years; (2) patients who understand the essence of the study and can independently give written informed consent; (3) RT-PCR detected a positive SARS-CoV-2 sample (naso–pharyngeal swab); (4) patients with new pneumonia diagnosed via X-ray and according to the criteria of COVID-19 guidance; (5) no active malignancy; (6) no history of pancreatitis; and (7) no prior valproic acid use.
The exclusion criteria were as follows: (1) patients with acute myocardial infarction or stroke; (2) patients with severe hepatic or renal impairment; (3) known HIV positivity, active, uncontrolled infection (e.g., hepatitis A, B, or C infection); (4) patients taking immunosuppressants; (5) cancer patients; (6) patients undergoing blood purification therapy (hemodialysis, hemofiltration, hemadsorption, plasma exchange, plasma adsorption, or peritoneal dialysis).
Criteria for severe illness were as indicated in the guidelines (https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum (accessed on 2 February 2024) when pneumonia was associated with at least one of the following symptoms: individuals who have a SpO2 < 94% on room air at sea level, a ratio of arterial partial pressure of oxygen to fraction of inspired oxygen (PaO2/FiO2) < 300 mm Hg, a respiratory rate > 30 breaths/min, or lung infiltrates > 50%.
T lymphocytes were obtained from 9 men aged 68.44 ± 15.32 years and 12 women aged 65.42 ± 15.74 years hospitalized from December 2022 to March 2023. At that time, the omicron variant of SARS-CoV-2 was detected in >90% of sequences tested in Lithuania (https://ourworldindata.org/covid-cases (accessed on 2 February 2024). In the female group, the time between a positive SARS-CoV-2 PCR test and blood sampling was 3.27 ± 2.00 days, and in the male group, it was 4.38 ± 3.28 days. The blood glucose concentration on the day of blood sampling was 6.38 ± 1.36 mM/L in males and 9.01 ± 3.12 mM/L in females; the female blood glucose concentration was slightly elevated due to the presence of 5 patients with type 2 diabetes mellitus in the study group.
2.3. Isolation of T Lymphocytes from Human Venous Blood by Ficoll-Paque™ Density Gradient and Treatment
Patient blood was taken using standard venipuncture in 10 mL vacuum tubes with anticoagulant KEDTA. Mononuclear cells were purified using the Ficoll-Paque™ gradient centrifugation method according to the manufacturer’s instructions (Cytiva, Uppsala, Sweden). Commercial PBS GIBCO, a phosphate-buffered saline (LifeTechnologies, The Hague, The Netherlands) and HBSS, a balanced salt solution (Sigma-Aldrich, Dorset, UK), were purchased. All reagents were prewarmed to room temperature before use (18–20 °C). The blood was diluted 1 to 1 in PBS. In total, 20 mL (4 mL per tube) of Ficoll-Paque™ was added to another 5 centrifuge tubes (15 mL). The diluted blood was then slowly layered onto the Ficoll-Paque™ layer using a Pasteur pipette. The tubes were then centrifuged at 532× g for 15 min at room temperature. The cell-containing interface layer was then drawn into new 15 mL centrifuge tubes. The cell washing procedure was repeated twice. The cells were suspended carefully using a sterile Pasteur pipette in approximately 3 volumes (depending on the estimated volume of cells in a tube) of HBSS and centrifuged at 354× g for 5 min at room temperature. The cells were resuspended in HBSS (approximately 3 volumes) and centrifuged at room temperature at 244 g for 5 min. The cells were then resuspended in HBSS. Cell viability was assessed with trypan blue dye using the Evos XL Core Imaging System (ThermoFisherScientific, Carlsbad, CA, USA). The T lymphocytes isolated from patients were treated with DCA–VPA (5 mM DCA and 2 mM VPA) for 24 h in vitro. Due to the synergistic effect of DCA and VPA, the dose of sodium dichloroacetate chosen for the combination was half that of our monotherapy to treat the other cells [74].
2.4. Total RNA Extraction and Next-Generation Sequencing
A commercial PureLink™ RNA Purification Kit (Invitrogen™, Thermo Fisher Scientific, Bleiswijk, The Netherlands) was used to extract total RNA from lymphocytes. After extraction, total RNA was quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), and the RNA integrity number (RIN) was evaluated on the Agilent 2100 capillary electrophoresis system (Agilent Technologies, Santa Clara, CA, USA) using an RNA 6000 Nano kit (Agilent Technologies, Santa Clara, CA, USA). Samples with RIN > 8.5 were used in downstream next-generation sequencing (NGS) experiments. According to the manufacturer’s manual, NGS library preparation was performed using the QIAseq targeted RNA Inflammation and Immunity Transcriptome panel kit (Qiagen, Hilden, Germany). As a starting RNA input, 400 ng of RNA was used for each sample. The final PCR amplification stage was performed in a 22-cycle setting. The size ranges of NGS libraries were measured with a High Sensitivity DNA 1000 kit (Agilent Technologies, Santa Clara, CA, USA) on an Agilent 2100 device. For final library quantification, the Qubit™ High Sensitivity dsDNA Quantification Assay kit (Invitrogen™, Thermo Fisher Scientific, Bleiswijk, The Netherlands) was applied, and fluorescence measurements were taken on a Qubit 4.0 fluorometer (Invitrogen™, Thermo Fisher Scientific, Bleiswijk, The Netherlands). Libraries were denatured and diluted according to the NextSeq library denaturation and dilution guide. A sequencing run was performed on the Illumina NextSeq 550 sequencer (Illumina, San Diego, CA, USA) using the 150-cycle Illumina High Output Kit v2.5 (Illumina, San Diego, CA, USA) under default conditions. T lymphocyte samples from males and females were sequenced on the same flow cell.
2.5. Bioinformatic Analysis
The quality of the data was assessed using MultiQC v1.13 [75]. Adapter sequences with 3 nucleotides and sequences shorter than 15 nucleotides, as well as sequences with a quality score lower than 25, were removed using Cutadapt v1.9.1 [76]. The human genome (GRCh38.p13) was downloaded from the Ensembl database [77]. The remaining sequences were aligned to the human genome using the STAR 2.1.3 tool [78]. The gene expression matrix was obtained using FeatureCounts v3.15 [79]. The sequence expression was normalized using the upper quartile method, and genes with a total expression across samples less than 50 were removed. A differential gene expression analysis was performed using DESeq2 v3.15 [80], and p-values were adjusted using the Benjamin–Hochberg method. An enrichment analysis of biological pathways was conducted using the DAVID server [81,82]. This normalization allows for a fair comparison between gene sets of different sizes. Data from treated T lymphocytes were compared with untreated cells. The gene expression data from treated and control conditions were normalized and logarithmically transformed. The enrichment analysis results from the gene set enrichment analysis (GSEA) were analyzed using the Enrichplot v1.2 and ClusterProfiler v4.8.2 R packages [83]. GSEA was performed using predefined algorithms to calculate enrichment scores and p-values for each dominant pathway or gene set. Gene set annotations were obtained from gene ontology (GO), KEGG, and Reactome databases. NES is the primary statistic for identifying gene sets significantly overrepresented in a ranked list of genes. It is derived from the ES (Enrichment Score) but is normalized for the size of each gene set. The impact of the DCA–VPA treatment was calculated by comparing the gene expression in treated and control T cells. Significant changes in gene expression were determined when p < 0.05.
3. Results
3.1. Data on the Effect of the DCA–VPA Treatment on the Expression of Genes Related to Inflammation and Immune Response in T Lymphocytes from Men with SARS-CoV-2 Infection and Pneumonia
The treatment with DCA–VPA of T lymphocytes of male patients suffering from SARS-CoV-2 infection and pneumonia significantly reduced the expression of 40 genes related to inflammation and immune response and involved in cytokine activity, chemokine mediated signaling, neutrophil chemotaxis, lymphocyte chemotaxis, and the regulation of T-cell proliferation pathogenic mechanisms after 24 h of treatment. Table 1 shows the male patients’ T lymphocyte gene expression data (Log2 counts average of control and treated cells), Log2 fold change compared to control and treated cells data, and p-value for each gene in the tested group.

Table 1.
Gene expression data for T lymphocytes in male patients with SARS-CoV-2 infection in the study groups.
The DCA–VPA treatment significantly downregulated the expression of ACKR3, CCL22, CCL24, CCL4, CCR1, CCR2, CCRL2, CD209, CMKLR1, CSF1, CSF2, CSF3, CSF3R, CX3CR1, CXCL1, CXCL13, CXCL2, CXCL3, CXCL5, CXCL6, CXCR2, CXCR3, EBI3, IL10, IL12RB1, IL19, IL1A, IL1B, IL1RN, IL24, IL23A, IL27, IL27RA, IL2RA, IL6, ITGB2, JAK3, LIF, OSM, and TNFRSF14 genes of T lymphocytes of males suffering from SARS-CoV-2 infection with pneumonia. The strength of the effect of DCA–VPA suppression on gene expression is shown in Figure 1.

Figure 1.
Differentially expressed genes in men suffering from SARS-CoV-2 infection with pneumonia: T lymphocytes following DCA–VPA treatment.
The bar chart represents the fold change in gene expression on a logarithmic scale (log2FoldChange). The direction of the data, as indicated by the negative values on the x-axis, suggests that the genes listed are downregulated following treatment with DCA–VPA in T lymphocytes. In this context, a negative log2 fold change means that the expression of these genes is lower in the treated cells compared to a control group. The greater the absolute value of the log2 fold change, the more significant the downregulation. The p-values provided alongside each gene indicate the statistical significance of the downregulation, with lower p-values denoting higher statistical significance.
GO analysis revealed that differentially expressed changes in control and DCA–VPA-treated T lymphocyte genes of men with SARS-CoV-2 infection are involved in pathogenic mechanisms of inflammation pathways of interest. Table 2 shows the DCA–VPA treatment’s effects on T lymphocyte genes involved in cytokine activity, chemokine-mediated signaling, neutrophil chemotaxis, lymphocyte chemotaxis, and regulation of T-cell proliferation pathways in men suffering from SARS-CoV-2 infection with pneumonia.

Table 2.
Effect of DCA–VPA treatment on men suffering from SARS-CoV-2 infection with pneumonia: T lymphocyte gene expression in the inflammatory pathways of interest.
The treatment of male T lymphocytes with DCA–VPA significantly reduced the gene expression of 22 pro-inflammatory cytokines involved in the cytokine activity pathway. These genes are CCL22, CCL24, CSF1, CSF2, CSF3, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL13, EBI3, IL1A, IL1B, IL1RN, IL6, IL10, IL19, IL24, IL27, LIF, and OSM. Twenty-two genes were affected in the cytokine activity pathway, with an enrichment score of −0.82 and an NES of −2.73, indicating a substantial underrepresentation of these genes following DCA–VPA treatment. The p-value of 1.00 × 10−10 signifies a very high statistical significance.
DCA–VPA-treated male T lymphocytes showed a significantly decreased expression of 17 pro-inflammatory genes, namely, ACKR3, CCL4, CCL22, CCL24, CCR1, CCR2, CCRL2, CMKLR1, CX3CR1, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL13, CXCR2, and CXCR3, which are involved in the chemokine-mediated signaling pathway. This pathway had 17 genes downregulated, with an enrichment score of −0.68, an NES of −2.08, and a p-value of 6.110−5, reinforcing these changes’ statistical significance. The affected genes are mainly chemokine receptors and ligands, suggesting a reduced signaling activity in chemotaxis.
The DCA–VPA treatment significantly reduced the expression of the pro-inflammatory chemokine in the neutrophil chemotaxis pathway genes CCL4, CCL22, CCL24, CSF3R, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL13, and ITGB2 in men’s T lymphocytes. There were 11 genes with decreased expression in this category, with an enrichment score of −0.63, an NES of −1.86, and a p-value of 0.0015, implying a diminished recruitment or movement of neutrophils.
When analyzing the expression of genes in the lymphocyte chemotaxis pathway, it was found that the DCA–VPA treatment reduced the expression of five pro-inflammatory chemokine genes in male T lymphocytes. The genes affected by the treatment were CCL4, CCL22, CCL24, CXCL13, and CXCR3. A smaller set of five genes showed downregulation in this pathway, with an enrichment score and an NES of −0.68 and −1.53, respectively. The p-value of 0.036 suggests a statistically significant change, indicating that the treatment may hinder lymphocyte migration.
The DCA–VPA treatment significantly reduced the gene expression of the regulation of T-cell proliferation pathway factors. These genes were CCR2, CD209, EBI3, IL1A, IL1B, IL2RA, IL6, IL10, IL12RB1, IL23A, IL27, IL27RA, JAK3, and TNFRSF14. The expression of 15 genes related to T-cell proliferation was considerably decreased, as indicated by an enrichment score of −0.47, an NES of −1.52, and a p-value of 0.048. Figure 2 shows the results of the gene set enrichment analysis (GSEA) illustrating the suppressing effect of the DCA–VPA treatment on specific sets of inflammation- and immune response-related genes in cytokine activity, chemokine mediated signaling, neutrophil chemotaxis, lymphocyte chemotaxis, and regulation of T-cell proliferation pathways in T lymphocytes from men with SARS-CoV-2 infection.
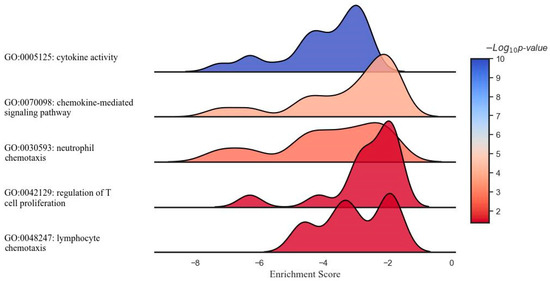
Figure 2.
Ridge plot representing the gene set enrichment analysis of inflammation- and immune-related response modulation through DCA–VPA treatment in male patients with SARS-CoV-2 infection and pneumonia.
Figure 2 shows the results of a GSEA illustrating the modulatory effect of the DCA–VPA treatment on specific sets of inflammation- and immune response-related genes in T lymphocytes from men with SARS-CoV-2 infection. Each plot represents a unique gene ontology (GO) term associated with distinct aspects of response. From top to bottom, the gene sets represent suppressing effects of cytokine activity, chemokine-mediated signaling, neutrophil chemotaxis, regulation of T cell proliferation, and lymphocyte chemotaxis biological pathways. The x-axis denotes the ES, which reflects the degree to which a set of genes is overrepresented at the extremes (top or bottom) of the entire ranked list of genes. The y-axis represents the gene sets, organized by inflammation- and immune-related function categories. The color intensity within each plot corresponds to the −Log10 p-value, indicating the level of statistical significance, with blue indicating a higher significance level. The peaks shifting towards the left of the 0 mark suggest the downregulation of the corresponding gene sets post treatment with DCA–VPA, which may imply a modulating effect on these inflammation/immune mechanisms, potentially leading to a suppression of responses. Table 3 shows the changes in the expression of genes involved in cytokine activity, chemokine-mediated signaling, neutrophil chemotaxis, lymphocyte chemotaxis, and lymphocyte chemotaxis in T lymphocytes from women with SARS-CoV-2 infection, which were significantly altered in the 24 h after treatment with DCA–VPA when comparing the untreated to the treated group.

Table 3.
Gene expression data for T lymphocytes in female patients with SARS-CoV-2 infection and pneumonia in the study groups.
3.2. Data on the Effect of the DCA–VPA Treatment on the Expression of Genes Related to Inflammation and Immune Response in T Lymphocytes from Females with SARS-CoV-2 Infection with Pneumonia
The DCA–VPA treatment of SARS-CoV-2-infected women’s T lymphocytes significantly altered the expression of 21 genes, including 11 genes—ACKR3, CCL2, CCL7, CCL24, CCR1, CCR3, CSF3R, CXCL1, CXCL5, CXCL6, and CXCR2—whose expression was decreased, and 10 genes—BMP6, CCL20, CXCL2, CXCL8, IL11, IL1A, IL1B, IL23A, IL24, and NODAL—whose expression was increased. The DCA–VPA treatment’s efficacy in gene expression changes is illustrated in Figure 3.
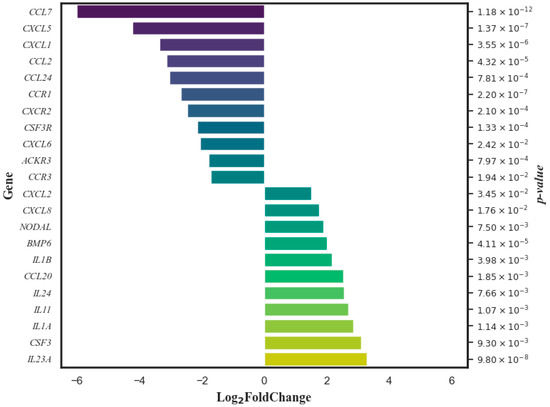
Figure 3.
Genes whose expression in women’s T lymphocytes was significantly changed by DCA–VPA treatment in patients suffering from SARS-CoV-2 infection with pneumonia.
The Log2 fold change data are organized in such a way that genes with reduced expression appear in descending order of change magnitude, and those with increased expression are in ascending order. The p-value on the right assesses the statistical significance of these changes.
Table 4 extends these findings to the GO analysis of DCA–VPA-treated female T lymphocytes, identifying genes in the biological pathways of cytokine activity, chemokine-mediated signaling, neutrophil chemotaxis, and lymphocyte chemotaxis. The table details the number of genes involved, the enrichment score, the NES, the effects on gene expression, and the significance of these effects as denoted by the p-values.

Table 4.
Effect of DCA–VPA treatment on gene expression in biological pathways in the T lymphocytes of women suffering from SARS-CoV-2 infection with pneumonia.
The DCA–VPA treatment altered the expression of 11 genes in the cytokine activity pathway. CSF3 was downregulated, while BMP6, CCL20, CXCL2, CXCL8, IL11, IL1A, IL1B, IL23A, IL24, and NODAL were significantly upregulated. An enrichment score of 0.41, an NES of 1.74, and a p-value of 0.015 indicate a significant shift toward enhanced cytokine activity.
Within the chemokine-mediated signaling pathway, the expression of 10 genes was significantly decreased: ACKR3, CCL2, CCL24, CCL7, CCR1, CCR3, CXCL1, CXCL5, CXCL6, and CXCR2. This is supported by an enrichment score of −0.61, an NES of −1.89, and a p-value of 0.025, indicating a notable suppression of chemokine-mediated signaling.
For the neutrophil chemotaxis pathway, the expression of CCL2, CCL24, CCL7, CSF3R, CXCL1, CXCL5, CXCL6, and CXCR2 was significantly downregulated, as shown by an enrichment score of −0.66, an NES of −1.97, and a p-value of 0.001, denoting a significant reduction in neutrophil chemotaxis by the treatment.
The treatment’s effects in the lymphocyte chemotaxis pathway were evident in the downregulation of three genes, CCL2, CCL7, and CCL24, with an enrichment score of −0.72, an NES of −1.55, and a p-value of 0.04, indicating diminished lymphocyte chemotaxis.
The GO analysis indicated no significant effect of the DCA–VPA treatment on the regulation of T-cell proliferation pathway in women with SARS-CoV-2 infection, suggesting that the treatment under the conditions studied did not impact this particular aspect of T-cell function. Figure 4 shows the GSEA results, illustrating the effect of the DCA–VPA treatment on specific sets of inflammation- and immune response-related genes in cytokine activity, chemokine-mediated signaling, neutrophil chemotaxis, and lymphocyte chemotaxis pathways in T lymphocytes from women with SARS-CoV-2 infection and pneumonia.
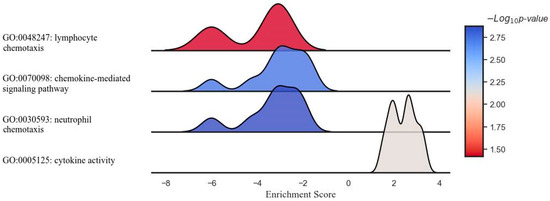
Figure 4.
Ridge plot representing the gene set enrichment analysis of inflammation- and immune-related response modulation in pathways of interest by DCA–VPA treatment in female patients with SARS-CoV-2 infection and pneumonia.
In a study elucidating the molecular impact of the DCA–VPA treatment in female patients with SARS-CoV-2 infection, a ridge plot was employed to visualize the gene set enrichment analysis across several key immunological pathways. The image presents a ridge plot illustrating the GSEA for four different gene ontology (GO) pathways in the context of female patients with SARS-CoV-2 infection treated with DCA–VPA. The plot visualizes the distribution of enrichment scores across the four pathways. It correlates these scores with the statistical significance of the enrichment, as indicated by the negative logarithm of the p-value (−Log10 p-value). The enrichment scores on the horizontal axis revealed no strong up- or downregulation in the cytokine activity pathway, with the associated p-values suggesting minimal statistical significance. A slight negative shift in the neutrophil chemotaxis pathway hinted at a potential gene downregulation, though the significance remained low. The leftward distribution in the chemokine-mediated signaling pathway was more pronounced, indicating a more substantial downregulation of genes, with a corresponding moderate increase in statistical significance. Most notably, the lymphocyte chemotaxis pathway demonstrated a marked negative enrichment, signaling a robust downregulation of gene activity, corroborated by the highest statistical significance among the evaluated pathways. This comprehensive GSEA underscores the nuanced and differential gene expression modulation enacted by the DCA–VPA treatment, offering insights into the therapeutic alteration of inflammation- and immune response-related pathways of SARS-CoV-2 infection.
3.3. Sex-Specific Gene Expression Responses to DCA-VPA Treatment in T Lymphocytes of SARS-CoV-2-Infected Patients with Pneumonia
The analysis of DCA–VPA-treated patients’ T lymphocytes revealed distinct gene expression profiles, highlighting a differential sex-specific response to the treatment in biological pathways of concern. The DCA–VPA treatment significantly altered the expression of 22 genes in male T lymphocytes and 11 genes in the female cells’ cytokine activity pathway. In the chemokine-mediated signaling pathway, the expressions of 17 genes expressions were altered in males and 10 in females; 11 genes were changed in neutrophil chemotaxis in males and 8 in females; 5 genes were altered in lymphocyte chemotaxis in males; and 3 genes were changed in T lymphocytes in females. These figures also indicate sex-related differences. The GO analysis showed that the DCA–VPA treatment did not significantly affect the regulation of T-cell proliferation pathway in SARS-CoV-2-infected women. In contrast, substantial changes in 14 genes were detected in male cells. Figure 5 represents the bubble plot impact of the DCA–VPA therapy on inflammation- and immune-related gene sets in the investigated biological pathways.
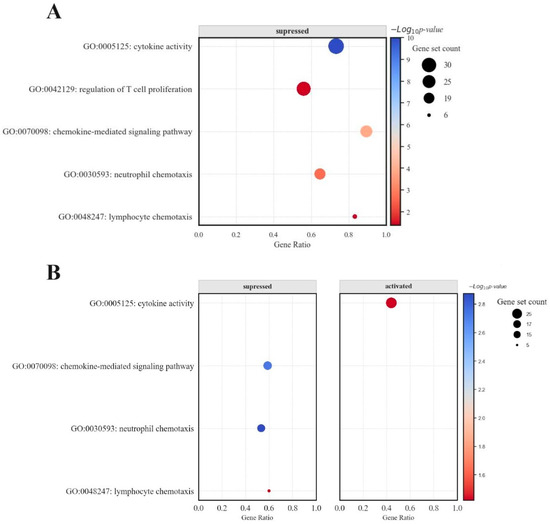
Figure 5.
Bubble plot representing the impact of DCA–VPA therapy on inflammation- and immune-related gene sets (A) in males with SARS-CoV-2 infection and pneumonia patients, and (B) in females with SARS-CoV-2 infection and pneumonia patients.
In male patients, gene sets associated with cytokine activity and chemokine-mediated signaling pathways were downregulated, as evidenced by negative enrichment scores in GSEA. Conversely, female patients exhibited a bimodal distribution in the enrichment scores for lymphocyte chemotaxis, indicating a nuanced regulatory effect of the treatment on this pathway. While still affected, the chemokine-mediated signaling pathway showed a less pronounced response than male cells. Additionally, the bubble (dot) plots for females demonstrated a balance between upregulated and downregulated genes, a pattern not as evident in male patients. The bar chart data for women showed a more evenly distributed expression change across the examined genes, including significant upregulation in specific genes—a response not observed in the male cohort.
4. Discussion
The study on the effects of DCA–VPA showed sex-related differences in cytokine activity, chemokine signaling, the regulation of neutrophil chemotaxis and T lymphocyte proliferation, and in the expression of inflammation- and immunity-associated genes in the T lymphocytes of men and women with SARS-CoV-2 infection with pneumonia.
4.1. DCA–VPA Effects on Cytokine Activity and Sex-Related Differences
Notably, the bar charts revealed a considerable downregulation in genes related to cytokine activity in males, with 22 genes showing substantial log2 fold changes. This pattern was not as prominent in females, where the changes in gene expression were more varied and included notable upregulation instances. Treatment with DCA–VPA significantly reduced the expression of IL1A, IL1B, IL1RN, IL6, IL10, IL19, IL24, IL27, OSM, and LIF in the T lymphocytes of male patients with infection. The DCA–VPA treatment of T lymphocytes from male SARS-CoV-2-infected patients with pneumonia was probably most effective in suppressing the expression of the IL6 gene, a key cytokine of the infection. IL6 levels are significantly elevated in the blood of patients with severe COVID-19 [84]. IL6 has been identified as a cause of the cytokine release syndrome in SARS-CoV-2 infection [85]. The S protein of SARS-CoV-2 increased IL6 mRNA expression in macrophages [86]. In COVID-19 patients, a decrease in IL6 mRNA expression indicates a reduction in inflammatory reactions and gradual recovery [87]. IL6 inhibitors are being used to treat severe COVID-19 [85]. DCA–VPA inhibited OSM (oncostatin M) and LIF gene expression in male T lymphocytes of SARS-CoV-2 patients. The positive regulator of IL6 signaling, OSM, was upregulated in lethal COVID-19 patients and SARS-CoV-2-infected patients with moderate compared to mild pneumonia [88]. Monocytes and neutrophils produced OSM, stimulating IL6 production in fibroblasts and endothelial cells. The level of circulating OSM is positively correlated with the severity of COVID-19 [89]. LIF cytokine, a member of the IL6 family, is highly expressed in lung cells of mild and severe COVID-19 patients but not in healthy subjects. LIF is elevated in the lungs of ARDS patients [90]. EBI3 (Epstein–Barr virus-induced gene 3) is a subunit of the composite cytokines IL27 and IL35; EBI3 activity was linked to its capacity to mediate IL6 trans-signaling, and by mediating trans-signaling, it can promote pro-inflammatory IL6 functions [91].
DCA–VPA suppressed IL1 in male SARS-CoV-2 patients’ T lymphocytes. IL1 is essential in developing severe SARS-CoV-2 infection associated with mortality [92]. IL1A is expressed in lung cells [93], and it predicts a bad prognosis for COVID-19 [94]. IL1A and IL1B are associated with a susceptibility to pandemic A/H1N1 influenza virus [95]. Although the overall effect of DCA–VPA on the cytokine release pathway is inhibitory (see Figure 3), the fact that the combination also inhibits the expression of the IL1RN may make the effect of our treatment ambiguous. IL1RN modulates the COVID-19 cytokine release syndrome via endogenous “anti-inflammatory” pathways. IL1RN variants modulate the severity of SARS-CoV-2 infection: the IL1RN CTA haplotype and the rs419598 CC single nucleotide variant are associated with lower levels of pro-inflammatory IL1B, IL6, and IL2, together with higher levels of anti-inflammatory IL-10. They are associated with a significant reduction in male mortality. We did not identify genetic variants in IL1RN, which does not allow us to determine the genetic regulation of the inflammatory pathway by IL1RN variants. However, other studies suggest that IL1 is an essential target for further studies in severe SARS-CoV-2 infection [92].
The suppressive effect of DCA–VPA on the gene expression of the IL10 family of cytokines in male T lymphocytes would also indicate an inhibitory effect on SARS-CoV-2 infection. Patients with severe COVID-19 have elevated plasma levels of the IL10 family of cytokines, IL19, and IL24, which decrease during recovery [96]. Airway epithelial cells secrete IL19 in response to stimulation with various pro-inflammatory cytokines [97]. The plasma and saliva levels of IL19 significantly increase in patients with severe COVID-19 [98]. To generalize regarding IL10 gene expression change and the characterization of the treatment’s effects on male T lymphocytes, it is crucial to emphasize that the gene set enrichment analysis (GSEA) showed significant downregulation in cytokine gene expression, including both pro- and anti-inflammatory cytokines, with a notable negative net enrichment score (NES) of −2.73 (p-value 1.00 × 10−10). This broad spectrum modulation suggests the treatment’s complex impact on cytokine dynamics rather than a unilateral effect. Specifically, including IL10, a traditionally anti-inflammatory cytokine, in the list of downregulated genes highlights the intricate balance the drug achieves in cytokine regulation. The therapeutic potential of the treatment, as suggested by our analysis, may stem from its capacity to attenuate excessive inflammatory responses, a key pathological feature of severe COVID-19, rather than simply augmenting or suppressing individual cytokines. The nuanced understanding of cytokine interactions, including the pleiotropic effects of cytokines like IL10, supports our conclusion of the treatment’s net anti-inflammatory effect, contributing to its efficacy in managing COVID-19-related inflammation.
The data that DCA–VPA suppresses IL27 expression in male T lymphocytes may also be important for treating infection. IL27 at the time of admission was strongly associated with patients with severe/critical COVID-19 pneumonia and is, therefore, an excellent predictor of adverse prognosis [94], although there is conflicting evidence that IL27 levels were significantly higher in survivors of the severe form of COVID-19 than in those who died from the severe form [99].
DCA–VPA significantly downregulated CSF1, CSF2, and CSF3 expression in the cytokine activity pathway. High granulocyte colony-stimulating factor (G-CSF) levels are detected in parallel with elevated serum levels of IL6 and IL10 in severe COVID-19 [100,101]. T cells can activate monocytes via the pro-inflammatory CSF2 and CSF1 receptors and induce a cytokine storm [102]. CSF3 is the most highly upregulated gene in SARS-CoV-2 infection, indicating that CSF3 may be a candidate target for drug therapy [103].
The treatment of male T lymphocytes with DCA–VPA significantly reduced the expression of chemokines genes as CCL4, CCL22, CCL24, CCR1, CCR2, CCRL2, CX3CR1, CXCL1, CXCL13, CXCL2, CXCL3, CXCL5, CXCL6, CXCR2, and CXCR3. Chemokines are critical mediators of inflammation, which remove pathogens. However, the overproduction of chemokines promotes inflammation: chemokines are a direct contributor to acute respiratory syndrome, which is fatal in around 40% of severe cases. Chemokines are involved in various stages of SARS-CoV-2 infection [104]. CCL22 is a biomarker of severe COVID-19; CCL22 controls immunity by promoting the communication of regulatory T cells to dendritic cells, which is essential for recruiting Th2 cells in the respiratory tissues [105]. COVID-19 patients who progressed showed persistent Th2 inflammation, which was strongly affected by CCL24 [106]. The genes for the vital pro-inflammatory factors CXCL1, CXCL2, CXCL3, CXCL5, and CSF2 are targets of SARS-CoV-2 infection [107]. Studies in mice have shown that the deactivation of ACE2 leads to an increase in the release of pro-inflammatory CXCL1 and CXCL5 and an increase in neutrophil infiltration with inflammatory lung injury [84]. MERS-CoV or SARS-CoV-2 viruses modulate the expression of CXCL5 and CXCL6 in lung cells. These genes encode proteins that affect lymphocyte and neutrophil function [108,109]. Deleting the CXCL5 in a mouse model reduced lung inflammation, indicating that CXCL5 inhibition may be a target to control lung inflammation [109]. As COVID-19 patients progressed, a statistically significant dysregulation of CXCL5 was found [110]. The CXCL6 exerts its chemotactic effect by interacting with receptors CXCR1 and CXCR2 to attract neutrophil granulocytes [108]. Pro-inflammatory CXCL13 has a high specificity for disease progression: it was significantly higher in ICU than in non-ICU patients. CXCL13 is implicated in pulmonary fibrosis and the regulation of B-cell response [111].
In the T lymphocytes of SARS-CoV-2-infected women with pneumonia, the DCA–VPA treatment changed the expression of 11 genes: it increased the expression of IL11, IL1A, IL1B, IL23A, IL24, CCL20, CXCL2, CXCL8, BMP6, and NODAL, and decreased the expression of CSF3. IL11, a member of the IL6 cytokine family, is defined as an “epithelial interleukin”. High levels of IL11 are linked to prolonged inflammation [112]. IL23A is one of two subunits of cytokine IL23. IL23 is produced by dendritic cells and macrophages and is an integral part of the inflammatory response [113].
The CCL20 chemokine binds to the CCR6 receptor, which determines the chemotaxis of dendritic cells, T lymphocytes, and B cells [114]. Increased CCL20 expression is correlated with increased circulating CCL20 levels during the prolonged course of COVID-19 and with prolonged viral clearance [115]. CXCL8 is a prognostic marker for the severity of SARS and MERS infection. During SARS-CoV-2 infection, CXCL8 levels are increased in blood and alveoli; the increase is also associated with bacterial infection [104]. The increased CXCL8 expression is associated with anti-inflammatory effects. The CXCL8 enhances T helper cell function [116].
BMP6 (bone morphogenetic protein 6) is involved in antiviral response, enhances the antiviral activity of IFNs, and regulates the expression of critical antiviral effectors [117,118]. The NODAL gene encodes the TGF-β (transforming growth factor-β) protein. TGF-β is essential in the control of immunity and inflammation mechanisms [119].
The comparison of the enrichment analysis of cytokine activity in the T lymphocytes of male and female SARS-CoV-2-infected patients treated with DCA–VPA shows sex differences in the modulatory effect. The enrichment score is shifted in opposite directions, with males to the left and females to the right of the midline (0 value); the Log10 p-value indicates a higher level of statistical significance for female T lymphocytes. In male T lymphocytes, 22 genes associated with inflammation are suppressed, whereas in female T lymphocytes, only CSF3 is suppressed (as in male T lymphocytes). BMP6, CXCL20, CXCL8, IL1A, IL1B, IL11, IL23A, IL24, and NODAL were found to be activated in female cells. The increased BMP6, CXCL8 and CXCL20 expressions are associated with anti-inflammatory effects. Whether the increase in IL11 levels is pathogenic or a natural host response to restore homeostasis in disease remains unanswered [112]. NODAL is important in controlling immunity and inflammation mechanisms, and its expression could be linked to female-specific immune responses. The same could be interpreted as the determined opposite effect of DCA–VPA in the increase of IL1A, IL1B, IL23A, IL24, and CXCL2 expression after the treatment of female cells. The difference in female immune mechanisms from those of males in SARS-CoV-2 infection is reported [120]. Our studies have shown that the effect of DCA–VPA on male T lymphocytes is significantly associated with the suppression of IL6 and related cytokines genes, which is not the case in female cells. Here, we examine the sex-specific anti-inflammatory effects of DCA–VPA by assessing T lymphocyte gene expression changes in the chemokine-mediated signaling, neutrophil chemotaxis, lymphocyte chemotaxis, and regulation of T-cell proliferation pathways.
4.2. DCA–VPA Effects on Chemokine-Mediated Signaling, Neutrophil Chemotaxis, Lymphocyte Chemotaxis, and Regulation of T-Cell Proliferation Pathways and Sex-Related Differences
The suppression of CCL22, CCL24, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, and CXCL13 in male T lymphocytes coincides with the overlap between cytokine-activated and chemokine-mediated signaling pathways, suggesting that the DCA–VPA treatment is associated with an anti-inflammatory effect via both of these pathways. No overlap of genes inhibited by the DCA–VPA treatment in the cytokine activity and chemokine-mediated signaling pathways was determined in female T lymphocytes.
In men and women patients with SARS-2-CoV-2 infection, the treatment of T lymphocytes with DCA–VPA significantly decreased the expression of the same genes—ACKR3, CCL24, CCR1, CXCL1, CXCL5, and CXCL6—in the chemokine-mediated signaling pathway. ACKR3 (Atypical Chemokine Receptor 3) in lymphocytes separates two phenotypically, transcriptionally, and functionally distinct populations of mouse MZ B cells [121]. Its relevance to inflammation and SARS-CoV-2 infection has not been investigated. SARS-CoV-2 upregulates CCR1 and CCR2 in human thoracic dorsal root ganglia, indicating an inflammatory mediator effect on pulmonary sensory neurons. Thus, the pharmacological inhibition of receptors would suggest that it could potentially suppress hyperinflammation in critically ill COVID-19 patients [104].
In male T lymphocytes, sex-specific effects of the DCA–VPA treatment, which significantly suppressed CCL4, CCL22, CCR2, CCRL2, CMKLR1, CX3CR1, CXCL2, CXCL3, CXCL13, and CXCR3 expression, were revealed in the chemokine-mediated signaling pathway. Severe COVID-19 patients over-expressed CCL4, and lung macrophages showed an over-expression of the gene [122]. CCRL2 mRNA and protein were detected in monocytes, macrophages, neutrophils, CD4- and CD8-positive T lymphocytes, B cells, monocyte-derived dendritic cells, and CD34-positive cells [123]. CCRL2 was increased by proinflammatory stimuli, such as LPS or TNF-α alone or in combination with IFN-γ or GM-CSF [123,124].
The clinical relevance of CMKLR1 as a marker of lung inflammation in ARDS was confirmed using RNA sequencing data, which showed that CMKLR1 expression is significantly increased in lung monocytes and macrophages in COVID-19 patients. CMKLR1-targeted PET is essential for monitoring the dynamics of lung inflammation and response to anti-inflammatory therapy [125]. The pro-inflammatory chemokine receptors CCR1, CCR2, CXCR3, and CX3CR1 mediate the immune response in the lung and are present in myeloid cells, T cells, and NK cells [126]. CXCR3 is the receptor for CXCL10. The CXCL10-CXCR3 signaling axis is in the pathogenesis of severe COVID-19 infection, making it a potential therapeutic target [127].
In female T lymphocytes, sex-specific effects of the DCA–VPA treatment, which involved the significant suppression of CCL24, CCL7, CCR3, and CXCL1 were determined in the chemokine-mediated signaling pathway. Severe COVID-19 patients were reported to over-express CCL7 in lung macrophages [128,129]. The CCR3 receptor is involved in migrating macrophages, NK cells, and monocytes and is upregulated in early post-SARS-CoV-2 infection [104].
In the neutrophil chemotaxis pathway, the DCA–VPA treatment significantly suppressed the expression of CCL24, CCL7, CSF3R, CXCL1, CXCL5, and CXCL6 in both male and female T lymphocytes. The following sex-related difference was found: only in men was the expression of CCL4, CCL22, CXCL2, CXCL3, CXCL13, and ITGB2 additionally suppressed, while in women, the expression of CCL2, CCL7, and CXCR2 was significantly suppressed sex-specifically.
In the regulation of the T-cell proliferation pathway, the significant downregulation of pro-inflammatory CCR2, CD209, EBI3, IL1A, IL1B, IL2RA, IL6, IL10, IL12RB1, IL23A, IL27, IL27RA, JAK3, and TNFRSF14 was determined in T lymphocytes of men suffering from SARS-CoV-2 infection. However, no reliable gene therapy effect was found in female T lymphocytes in regulating the T-cell proliferation pathway. In addition to the genes mentioned above in the article, in male T cells, the DCA–VPA treatment significantly inhibited IL12RB1 (interleukin 12 receptor beta 1 subunit), which is linked to a significant association between susceptibility to SARS-CoV-2 infection [130], IL27RB1, whose expression is required for CD4+ and CD8+ T-cell differentiation in humans [131], JAK3, which has a cell type-specific role in IL-2-induced proliferative signal transduction [132], and TNFRSF14 (tumor necrosis factor-related cytokine LIGHT (TNFSF14), which has proinflammatory activity, with multifaceted roles in stimulating T cells [133]. In turn, suppressing CD209 by the medicinal product could inhibit the receptor for SARS-CoV-2 from attachment onto host cells [134]. Given the complexity of T-cell proliferation processes, sex-related differences in the effects of the DCA–VPA treatment on the regulation of the T-cell proliferation pathway can be discussed in the context of the overall impact on T-cell proliferation [135]. T lymphocytes proliferate, and their functional responses depend on metabolism through T-cell differentiation processes and on the inflammatory microenvironment in which they are located [135]. The inhibition of pro-inflammatory cytokine generation may be linked to the effect of not only DCA but also the inhibition of glycolysis, which may influence the T-cell proliferation pathway in T lymphocytes.
This study faces a situation where the effect of DCA–VPA is not controlled by an anti-inflammatory agent that modulates inflammatory immune responses via mitochondrial metabolic mechanisms. This could be seen as a limitation of the study. However, this happens because no analogous anti-inflammatory preparation corrects metabolic disturbances. Nevertheless, it is considered that the efficacy of DCA–VPA has been sufficiently demonstrated in the treated cells compared to untreated controls. Another limitation of the study could be that post-menopausal women were studied. As sex-related differences were found, it would be essential to study the T lymphocytes of young women. A pharmacovigilance alert has been issued that patients with COVID-19 should avoid treatment with the anti-inflammatory ibuprofen as it may worsen the condition [136]. Ibuprofen is a specific SLC5A8 blocker [137]. Potentially promising research data have been reported for the treatment of COVID-19 with anti-inflammatory small molecule Ebselen derivative compounds, which are inhibitors of SARS-CoV-2 protease Mpro, papain-like protease (PLpro), and nsp14 guanine N7-methyltransferase [138]. The actuality and the rationale for applying the anti-inflammatory effects of the DCA–VPA combination study is that VPA activates the SLC5A8 via DNA demethylation [139]. SLC5A8 carries short-chain fatty acids and transports DCA into cells [68,69]. SLC5A8 transports DCA into the cell [22,70,71], correcting mitochondrial metabolism in tissue cells, and the DCA–VPA treatment should attenuate the inflammation and tissue damage and protect against disease progression. DCAs and VPAs have sex-specific metabolism and biological effects [16,140]. Therefore, further research on the sex-specific efficacy of the preparation is relevant.
5. Conclusions
The gene sequencing analysis revealed a significant effect of DCA–VPA on changes in the expression of the genes in cytokine activity, chemokine-mediated signaling, neutrophil chemotaxis, lymphocyte chemotaxis, T-cell chemotaxis, and regulation of T-cell proliferation pathways. DCA–VPA exerts anti-inflammatory effects, and there were apparent sex-related differences: the anti-inflammatory effect was more strongly expressed in T lymphocytes from male patients than female patients with SARS-CoV-2 infection with pneumonia. To further clarify sex differences, it would be appropriate to determine whether these differences are influenced by data on women with diabetes.
The sex-specific modulation of inflammation/immune response by the DCA–VPA treatment in the T lymphocytes of SARS-CoV-2 patients with pneumonia underscores the potential for personalized therapeutic approaches. The distinct immune profiles warrant further investigation into the molecular mechanisms driving these differences and could provide insights into the optimal management of SARS-CoV-2 infection in diverse patient populations.
6. Patents
The combination of these medicinal products is a pending patent filed by us that covers DCA–VPA as a new medicinal product for the treatment of viral and bacterial infections (National patent application No. LT2023 532; 22 August 2023), as well as for the treatment of cancer (Official bulletin of the state patent bureau of the Republic of Lithuania, No. 6874, filling date 17 April 2020, https://vpb.lrv.lt/uploads/vpb/documents/files/VPB-OB-Nr23-2021-12-10-1d.pdf (accessed on 2 February 2024); a European patent application has been submitted (European patent application No. 21168796.7, filing date 16 April 2021, https://register.epo.org/application?number=EP21168796 (accessed on 2 February 2024).
Author Contributions
Conceptualization, V.L. and D.S.; Data curation, A.M., T.T. and D.G.; Bioinformatic analysis, L.K.; Formal analysis D.G., I.B., A.V., T.T. and R.U.; Funding acquisition, V.L. and D.S.; Investigation, D.G., L.K., A.M., T.T. and V.T.; Methodology; D.G., L.K., R.U. and V.T.; Project administration, V.L. and D.S.; Resources, V.L. and D.G.; Software D.G., R.U. and L.K.; Supervision, V.L.; Validation, D.G., L.K. and V.T.; Visualization L.K.; Writing—original draft D.S., L.K., V.T., D.G. and V.L.; Writing—review and editing, All authors. All authors have read and agreed to the published version of the manuscript.
Funding
This project has received funding from the European Regional Development Fund (project No 13.1.1-LMT-L-718-05-0030) under a grant agreement with the Research Council of Lithuania (LMTLT), funded as the European Union’s measure in response to the COVID-19 pandemic.
Institutional Review Board Statement
Authorization to conduct biomedical research was granted by Kaunas Regional Biomedicine Research Ethics Committee (Lithuania) 28-12-2021 No. BE-2-140.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Stakišaitis, D.; Kapočius, L.; Kilimaitė, E.; Gečys, D.; Šlekienė, L.; Balnytė, I.; Palubinskienė, J.; Lesauskaitė, V. Preclinical Study in Mouse Thymus and Thymocytes: Effects of Treatment with a Combination of Sodium Dichloroacetate and Sodium Valproate on Infectious Inflammation Pathways. Pharmaceutics 2023, 15, 2715. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, K.; Ferrer-Espada, R.; Martinez-de-Tejada, G.; Nehls, C.; Fukuoka, S.; Mauss, K.; Weindl, G.; Garidel, P. A Comparison between SARS-CoV-2 and Gram-Negative Bacteria-Induced Hyperinflammation and Sepsis. Int. J. Mol. Sci. 2023, 24, 15169. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Qin, S.; Li, Z.; Gao, W.; Tang, M.; Dong, X. Early immune system alterations in patients with septic shock. Front. Immunol. 2023, 14, 1126874. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, C.; Scherag, A.; Adhikari, N.K.J.; Hartog, C.S.; Tsaganos, T.; Schlattmann, P.; Angus, D.C.; Reinhart, K. International Forum of Acute Care Trialists, Assessment of Global Incidence and Mortality of Hospital-treated Sepsis. Current Estimates and Limitations. Am. J. Respir. Crit. Care Med. 2016, 193, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.-L.; Bakker, J. Blood lactate levels in sepsis: In 8 questions. Curr. Opin. Crit. Care 2021, 27, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Shoraka, S.; Samarasinghe, A.E.; Ghaemi, A.; Mohebbi, S.R. Host mitochondria: More than an organelle in SARS-CoV-2 infection. Front. Cell Infect. Microbiol. 2023, 13, 1228275. [Google Scholar] [CrossRef] [PubMed]
- Langley, R.J.; Tsalik, E.L.; van Velkinburgh, J.C.; Glickman, S.W.; Rice, B.J.; Wang, C.; Chen, B.; Carin, L.; Suarez, A.; Mohney, R.P.; et al. An integrated clinico-metabolomic model improves prediction of death in sepsis. Sci. Transl. Med. 2013, 5, 195ra95. [Google Scholar] [CrossRef]
- Kotas, M.E.; Medzhitov, R. Homeostasis, inflammation, and disease susceptibility. Cell 2015, 816–827. [Google Scholar] [CrossRef]
- Soares, M.P.; Teixeira, L.; Moita, L.F. Disease tolerance and immunity in host protection against infection. Nat. Rev. Immunol. 2017, 17, 83–96. [Google Scholar] [CrossRef]
- Bhowal, C.; Ghosh, S.; Ghatak, D.; De, R. Pathophysiological involvement of host mitochondria in SARS-CoV-2 infection that causes COVID-19: A comprehensive evidential insight. Mol. Cell Biochem. 2023, 478, 1325–1343. [Google Scholar] [CrossRef]
- Vardavas, C.I.; Mathioudakis, A.G.; Nikitara, K.; Stamatelopoulos, K.; Georgiopoulos, G.; Phalkey, R.; Leonardi-Bee, J.; Fernandez, E.; Carnicer-Pont, D.; Vestbo, J.; et al. Prognostic factors for mortality, intensive care unit and hospital admission due to SARS-CoV-2: A systematic review and meta-analysis of cohort studies in Europe. Eur. Respir. Rev. 2022, 31, 220098. [Google Scholar] [CrossRef]
- Peckham, H.; de Gruijter, N.M.; Raine, C.; Radziszewska, A.; Ciurtin, C.; Wedderburn, L.R.; Rosser, E.C.; Webb, K.; Deakin, C.T. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020, 11, 6317. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, C.; Regitz-Zagrosek, V.; Neuhauser, H.K.; Morgan, R.; Klein, S.L. Impact of sex and gender on COVID-19 outcomes in Europe. Biol. Sex. Differ. 2020, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Williamson, E.J.; Walker, A.J.; Bhaskaran, K.; Bacon, S.; Bates, C.; Morton, C.E.; Curtis, H.J.; Mehrkar, A.; Evans, D.; Inglesby, P.; et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature 2020, 584, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Grasselli, G.; Zangrillo, A.; Zanella, A.; Antonelli, M.; Cabrini, L.; Castelli, A.; Cereda, D.; Coluccello, A.; Foti, G.; Fumagalli, R.; et al. Baseline Characteristics and Outcomes of 1591 Patients Infected With SARS-CoV-2 Admitted to ICUs of the Lombardy Region, Italy. JAMA 2020, 323, 1574–1581. [Google Scholar] [CrossRef] [PubMed]
- Stakišaitis, D.; Kapočius, L.; Valančiūtė, A.; Balnytė, I.; Tamošuitis, T.; Vaitkevičius, A.; Sužiedėlis, K.; Urbonienė, D.; Tatarūnas, V.; Kilimaitė, E.; et al. SARS-CoV-2 Infection; Differences, S.-R. and a Possible Personalized Treatment Approach with Valproic Acid: A Review. Biomedicines 2022, 10, 962. [Google Scholar] [CrossRef] [PubMed]
- Mainali, R.; Zabalawi, M.; Long, D.; Buechler, N.; Quillen, E.; Key, C.-C.; Zhu, X.; Parks, J.S.; Furdui, C.; Stacpoole, P.W.; et al. Dichloroacetate reverses sepsis-induced hepatic metabolic dysfunction. Elife 2021, 10, e64611. [Google Scholar] [CrossRef] [PubMed]
- Cavagnaro, J.A. Preclinical safety evaluation of biotechnology-derived pharmaceuticals. Nat. Rev. Drug Discov. 2002, 1, 469–475. [Google Scholar] [CrossRef]
- U.S. Food and Drug Administration. Guideline for the study and evaluation of gender differences in the clinical evaluation of drugs; notice. Fed. Regist. 1993, 58, 39406–39416. [Google Scholar]
- Clayton, J.A.; Collins, F.S. Policy: NIH to balance sex in cell and animal studies. Nature 2014, 509, 282–283. [Google Scholar] [CrossRef]
- de Vries, S.T.; Starokozhko, V.; Schellens, I.M.M.; Wijnans, L.; Enzmann, H.; Cavaleri, M.; Mol, P.G.M. Attention for sex in COVID-19 trials: A review of regulatory dossiers. BMJ Glob. Health 2022, 7, e008173. [Google Scholar] [CrossRef]
- Schoenmann, N.; Tannenbaum, N.; Hodgeman, R.M.; Raju, R.P. Regulating mitochondrial metabolism by targeting pyruvate dehydrogenase with dichloroacetate, a metabolic messenger. Biochim. Biophys. Acta Mol. Basis Dis. 2023, 1869, 166769. [Google Scholar] [CrossRef]
- Nuzzo, E.; Berg, K.M.; Andersen, L.W.; Balkema, J.; Montissol, S.; Cocchi, M.N.; Liu, X.; Donnino, M.W. Pyruvate Dehydrogenase Activity Is Decreased in the Peripheral Blood Mononuclear Cells of Patients with Sepsis. A Prospective Observational Trial. Ann. Am. Thorac. Soc. 2015, 12, 1662–1666. [Google Scholar] [CrossRef]
- Icard, P.; Lincet, H.; Wu, Z.; Coquerel, A.; Forgez, P.; Alifano, M.; Fournel, L. The key role of Warburg effect in SARS-CoV-2 replication and associated inflammatory response. Biochimie 2021, 180, 169–177. [Google Scholar] [CrossRef]
- Yang, K.; Fan, M.; Wang, X.; Xu, J.; Wang, Y.; Tu, F.; Gill, P.S.; Ha, T.; Liu, L.; Williams, D.L.; et al. Lactate promotes macrophage HMGB1 lactylation, acetylation, and exosomal release in polymicrobial sepsis. Cell Death Differ. 2022, 29, 133–146. [Google Scholar] [CrossRef]
- Colegio, O.R.; Chu, N.-Q.; Szabo, A.L.; Chu, T.; Rhebergen, A.M.; Jairam, V.; Cyrus, N.; Brokowski, C.E.; Eisenbarth, S.C.; Phillips, G.M.; et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature 2014, 513, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.S. The Lactate and the Lactate Dehydrogenase in Inflammatory Diseases and Major Risk Factors in COVID-19 Patients. Inflammation 2022, 45, 2091–2123. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Pitt, B.; Sutton, N.R.; Wang, Z.; Goonewardena, S.N.; Holinstat, M. Potential repurposing of the HDAC inhibitor valproic acid for patients with COVID-19. Eur. J. Pharmacol. 2021, 898, 173988. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, T.; Suzuki, Y.; Imai, J.; Sugano, S.; Hida, M.; Tanigami, A.; Muroi, S.; Yamada, Y.; Hanaoka, K. Molecular cloning, mRNA expression and chromosomal localization of mouse angiotensin-converting enzyme-related carboxypeptidase (mACE2). DNA Seq. 2002, 13, 217–220. [Google Scholar] [CrossRef]
- Tipnis, S.R.; Hooper, N.M.; Hyde, R.; Karran, E.; Christie, G.; Turner, A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000, 275, 33238–33243. [Google Scholar] [CrossRef]
- Kloc, M.; Ghobrial, R.M.; Kubiak, J.Z. The Role of Genetic Sex and Mitochondria in Response to COVID-19 Infection. Int. Arch. Allergy Immunol. 2020, 181, 629–634. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, Z.; Wang, Y.; Zhou, Y.; Ma, Y.; Zuo, W. Single-Cell RNA Expression Profiling of ACE2, the Receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020, 202, 756–759. [Google Scholar] [CrossRef]
- Patel, S.K.; Velkoska, E.; Burrell, L.M. Emerging markers in cardiovascular disease: Where does angiotensin-converting enzyme 2 fit in? Clin. Exp. Pharmacol. Physiol. 2013, 40, 551–559. [Google Scholar] [CrossRef]
- Fernández-Atucha, A.; Izagirre, A.; Fraile-Bermúdez, A.B.; Kortajarena, M.; Larrinaga, G.; Martinez-Lage, P.; Echevarría, E.; Gil, J. Sex differences in the aging pattern of renin-angiotensin system serum peptidases. Biol. Sex. Differ. 2017, 8, 5. [Google Scholar] [CrossRef]
- Bukowska, A.; Spiller, L.; Wolke, C.; Lendeckel, U.; Weinert, S.; Hoffmann, J.; Bornfleth, P.; Kutschka, I.; Gardemann, A.; Isermann, B.; et al. Protective regulation of the ACE2/ACE gene expression by estrogen in human atrial tissue from elderly men. Exp. Biol. Med. 2017, 242, 1412–1423. [Google Scholar] [CrossRef]
- Dalpiaz, P.L.M.; Lamas, A.Z.; Caliman, I.F.; Ribeiro, R.F.; Abreu, G.R.; Moyses, M.R.; Andrade, T.U.; Gouvea, S.A.; Alves, M.F.; Carmona, A.K.; et al. Sex Hormones Promote Opposite Effects on ACE and ACE2 Activity, Hypertrophy and Cardiac Contractility in Spontaneously Hypertensive Rats. PLoS ONE 2015, 10, e0127515. [Google Scholar] [CrossRef]
- Fischer, M.; Baessler, A.; Schunkert, H. Renin angiotensin system and gender differences in the cardiovascular system. Cardiovasc. Res. 2002, 53, 672–677. [Google Scholar] [CrossRef] [PubMed]
- Bartachian, M.; McManus, J.M.; Berk, M.P.; Nakamura, F.; Mukhopadhyay, S.; Xu, W.; Erzurum, S.; Drazba, J.; Peterson, J.; Klain, E.A.; et al. Androgen regulation of pulmonary AR, TMPRSS2 and ACE2 with implications for sex-discordant COVID-19 outcomes. Sci. Rep. 2021, 11, 11130. [Google Scholar] [CrossRef] [PubMed]
- Vaarala, M.H.; Porvari, K.S.; Kellokumpu, S.; Kyllönen, A.P.; Vihko, P.T. Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues. J. Pathol. 2001, 193, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Zarubin, A.; Stepanov, V.; Markov, A.; Kolesnikov, N.; Marusin, A.; Khitrinskaya, I.; Swarovskaya, M.; Litvinov, S.; Ekomasova, N.; Dzhaubermezov, M.; et al. Pharmacogenetic Properties of TMPRSS2 Gene as a Potential Target for COVID-19 Therapy. Genes 2020, 12, 19. [Google Scholar] [CrossRef]
- Fortson, W.S.; Kayarthodi, S.; Fujimura, Y.; Xu, H.; Matthews, R.; Grizzle, W.E.; Rao, V.N.; Bhat, G.K.; Reddy, E.S.P. Histone deacetylase inhibitors, valproic acid and trichostatin-A induce apoptosis and affect acetylation status of p53 in ERG-positive prostate cancer cells. Int. J. Oncol. 2011, 39, 111–119. [Google Scholar] [CrossRef]
- Moreno-Pérez, O.; Merino, E.; Ramos, J.M.; Rodríguez, J.C.; Diaz, C.; Mas, P.; Reus, S.; Sánchez-Martínez, R.; Boix, V.; Chico-Sánchez, P.; et al. Valproic Acid Could Help in the Fight Against COVID-19: A case-control study. Neurologia 2022. [Google Scholar] [CrossRef]
- Collazos, J.; Domingo, P.; Fernández-Araujo, N.; Asensi-Díaz, E.; Vilchez-Rueda, H.; Lalueza, A.; Roy-Vallejo, E.; Blanes, R.; Raya-Cruz, M.; Sanz-Cánovas, J.; et al. Valproic Acid in COVID-19 Study Group, Exposure to valproic acid is associated with less pulmonary infiltrates and improvements in diverse clinical outcomes and laboratory parameters in patients hospitalized with COVID-19. PLoS ONE 2022, 17, e0262777. [Google Scholar] [CrossRef]
- Naasani, I.; Analysis, C.O.P.R. A Bioinformatic Approach to Accelerate Drug Repurposing against Covid-19 and Other Emerging Epidemics. SLAS Discov. 2021, 26, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Phiel, C.J.; Zhang, F.; Huang, E.Y.; Guenther, M.G.; Lazar, M.A.; Klein, P.S. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 2001, 276, 36734–36741. [Google Scholar] [CrossRef]
- Maschietto, F.; Qiu, T.; Wang, J.; Shi, Y.; Allen, B.; Lisi, G.P.; Lolis, E.; Batista, V.S. Valproate-coenzyme A conjugate blocks opening of receptor binding domains in the spike trimer of SARS-CoV-2 through an allosteric mechanism. Comput. Struct. Biotechnol. J. 2023, 21, 1066–1076. [Google Scholar] [CrossRef]
- Mengist, H.M.; Fan, X.; Jin, T. Designing of improved drugs for COVID-19: Crystal structure of SARS-CoV-2 main protease Mpro. Signal Transduct. Target. Ther. 2020, 5, 67. [Google Scholar] [CrossRef]
- Elfiky, A.A. Anti-HCV nucleotide inhibitors repurposing against COVID-19. Life Sci. 2020, 248, 117477. [Google Scholar] [CrossRef] [PubMed]
- Elfiky, A.A. Ribavirin; Remdesivir; Sofosbuvir; Galidesivir, and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci. 2020, 253, 117592. [Google Scholar] [CrossRef] [PubMed]
- Mamdouh, F.; Mohamed, M.; El-Din, G.; Abuo-Rahma, G.E.-D.; Hayallah, A.M.; Aziz, M.; Nafady, A.; Samir, E. Molecular Docking Study Reveals the Potential Repurposing of Histone Deacetylase Inhibitors Against COVID-19. Int. J. Pharm. Sci. Res. 2020, 11, 4261–4270. [Google Scholar] [CrossRef]
- Jin, Z.; Du, X.; Xu, Y.; Deng, Y.; Liu, M.; Zhao, Y.; Zhang, B.; Li, X.; Zhang, L.; Peng, C.; et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020, 582, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Pillaiyar, T.; Manickam, M.; Namasivayam, V.; Hayashi, Y.; Jung, S.-H. An Overview of Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3CL Protease Inhibitors: Peptidomimetics and Small Molecule Chemotherapy. J. Med. Chem. 2016, 59, 6595–6628. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. Role of HDAC2 in the pathophysiology of COPD. Annu. Rev. Physiol. 2009, 71, 451–464. [Google Scholar] [CrossRef] [PubMed]
- Dewe, J.M.; Fuller, B.L.; Lentini, J.M.; Kellner, S.M.; Fu, D. TRMT1-Catalyzed tRNA Modifications Are Required for Redox Homeostasis To Ensure Proper Cellular Proliferation and Oxidative Stress Survival. Mol. Cell. Biol. 2017, 37, e00214-17. [Google Scholar] [CrossRef]
- Matsuda, K.I.; Mori, H.; Nugent, B.M.; Pfaff, D.W.; McCarthy, M.M.; Kawata, M. Histone deacetylation during brain development is essential for permanent masculinization of sexual behavior. Endocrinology 2011, 152, 2760–2767. [Google Scholar] [CrossRef] [PubMed]
- Tyler, C.R.S.; Smoake, J.J.W.; Solomon, E.R.; Villicana, E.; Caldwell, K.K.; Allan, A.M. Sex-Dependent Effects of the Histone Deacetylase Inhibitor, Sodium Valproate, on Reversal Learning After Developmental Arsenic Exposure. Front. Genet. 2018, 9, 200. [Google Scholar] [CrossRef]
- Krämer, O.H.; Zhu, P.; Ostendorff, H.P.; Golebiewski, M.; Tiefenbach, J.; Peters, M.A.; Brill, B.; Groner, B.; Bach, I.; Heinzel, T.; et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003, 22, 3411–3420. [Google Scholar] [CrossRef]
- Andreu, S.; Ripa, I.; Bello-Morales, R.; López-Guerrero, J.A. Valproic Acid and Its Amidic Derivatives as New Antivirals against Alphaherpesviruses. Viruses 2020, 12, 1356. [Google Scholar] [CrossRef]
- Michaelis, M.; Doerr, H.W.; Cinatl, J. Valproic acid as anti-cancer drug. Curr. Pharm. Des. 2007, 13, 3378–3393. [Google Scholar] [CrossRef]
- Codo, A.C.; Davanzo, G.G.; de Brito Monteiro, L.; de Souza, G.F.; Muraro, S.P.; Virgilio-da-Silva, J.V.; Prodonoff, J.S.; Carregari, V.C.; de Biagi Junior, C.A.O.; Crunfli, F.; et al. Elevated Glucose Levels Favor SARS-CoV-2 Infection and Monocyte Response through a HIF-1α/Glycolysis-Dependent Axis. Cell Metab. 2020, 32, 437–446.e5. [Google Scholar] [CrossRef]
- Sinclair, L.V.; Rolf, J.; Emslie, E.; Shi, Y.-B.; Taylor, P.M.; Cantrell, D.A. Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation. Nat. Immunol. 2013, 14, 500–508. [Google Scholar] [CrossRef]
- Kidani, Y.; Elsaesser, H.; Hock, M.B.; Vergnes, L.; Williams, K.J.; Argus, J.P.; Marbois, B.N.; Komisopoulou, E.; Wilson, E.B.; Osborne, T.F.; et al. Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat. Immunol. 2013, 14, 489–499. [Google Scholar] [CrossRef]
- Wang, R.; Dillon, C.P.; Shi, L.Z.; Milasta, S.; Carter, R.; Finkelstein, D.; McCormick, L.L.; Fitzgerald, P.; Chi, H.; Munger, J.; et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 2011, 35, 871–882. [Google Scholar] [CrossRef]
- Khan, S.; Jena, G. Valproic Acid Improves Glucose Homeostasis by Increasing Beta-Cell Proliferation, Function, and Reducing its Apoptosis through HDAC Inhibition in Juvenile Diabetic Rat. J. Biochem. Mol. Toxicol. 2016, 30, 438–446. [Google Scholar] [CrossRef]
- Rakitin, A.; Kõks, S.; Haldre, S. Valproate modulates glucose metabolism in patients with epilepsy after first exposure. Epilepsia 2015, 56, e172–e175. [Google Scholar] [CrossRef]
- Mihaylova, M.M.; Vasquez, D.S.; Ravnskjaer, K.; Denechaud, P.-D.; Yu, R.T.; Alvarez, J.G.; Downes, M.; Evans, R.M.; Montminy, M.; Shaw, R.J. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 2011, 145, 607–621. [Google Scholar] [CrossRef]
- Thangaraju, M.; Ananth, S.; Martin, P.M.; Roon, P.; Smith, S.B.; Sterneck, E.; Prasad, P.D.; Ganapathy, V. c/ebpdelta Null mouse as a model for the double knock-out of slc5a8 and slc5a12 in kidney. J. Biol. Chem. 2006, 281, 26769–26773. [Google Scholar] [CrossRef]
- Frank, H.; Gröger, N.; Diener, M.; Becker, C.; Braun, T.; Boettger, T. Lactaturia and loss of sodium-dependent lactate uptake in the colon of SLC5A8-deficient mice. J. Biol. Chem. 2008, 283, 24729–24737. [Google Scholar] [CrossRef]
- Singh, N.; Thangaraju, M.; Prasad, P.D.; Martin, P.M.; Lambert, N.A.; Boettger, T.; Offermanns, S.; Ganapathy, V. Blockade of dendritic cell development by bacterial fermentation products butyrate and propionate through a transporter (Slc5a8)-dependent inhibition of histone deacetylases. J. Biol. Chem. 2010, 285, 27601–27608. [Google Scholar] [CrossRef]
- Zimmerman, M.A.; Singh, N.; Martin, P.M.; Thangaraju, M.; Ganapathy, V.; Waller, J.L.; Shi, H.; Robertson, K.D.; Munn, D.H.; Liu, K. Butyrate suppresses colonic inflammation through HDAC1-dependent Fas upregulation and Fas-mediated apoptosis of T cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G1405–G1415. [Google Scholar] [CrossRef]
- Guo, Z.; Zhong, F.; Hou, M.; Xie, J.; Zhang, A.Z.; Li, X.; Li, Y.; Chang, B.; Yang, J. Key enzyme in charge of ketone reabsorption of renal tubular SMCT1 may be a new target in diabetic kidney disease. Nephrol. Dial. Transplant. 2023, gfad173. [Google Scholar] [CrossRef] [PubMed]
- PubChem, SLC5A8—Solute Carrier Family 5 Member 8 (Human). Available online: https://pubchem.ncbi.nlm.nih.gov/gene/SLC5A8/human (accessed on 10 November 2023).
- Damanskienė, E.; Balnytė, I.; Valančiūtė, A.; Lesauskaitė, V.; Alonso, M.M.; Stakišaitis, D. The Comparative Experimental Study of Sodium and Magnesium Dichloroacetate Effects on Pediatric PBT24 and SF8628 Cell Glioblastoma Tumors Using a Chicken Embryo Chorioallantoic Membrane Model and on Cells In Vitro. Int. J. Mol. Sci. 2022, 23, 10455. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, gkac194. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Sodhi, C.P.; Wohlford-Lenane, C.; Yamaguchi, Y.; Prindle, T.; Fulton, W.B.; Wang, S.; McCray, P.B.; Chappell, M.; Hackam, D.J.; Jia, H. Attenuation of pulmonary ACE2 activity impairs inactivation of des-Arg9 bradykinin/BKB1R axis and facilitates LPS-induced neutrophil infiltration. Am. J. Physiol. Lung Cell. Mol. Physiol. 2018, 314, L17–L31. [Google Scholar] [CrossRef]
- Chen, L.Y.C.; Hoiland, R.L.; Stukas, S.; Wellington, C.L.; Sekhon, M.S. Confronting the controversy: Interleukin-6; the COVID-19 cytokine storm syndrome. Eur. Respir. J. 2020, 56, 2003006. [Google Scholar] [CrossRef]
- Pantazi, I.; Al-Qahtani, A.A.; Alhamlan, F.S.; Alothaid, H.; Matou-Nasri, S.; Sourvinos, G.; Vergadi, E.; Tsatsanis, C. SARS-CoV-2/ACE2 Interaction Suppresses IRAK-M Expression and Promotes Pro-Inflammatory Cytokine Production in Macrophages. Front. Immunol. 2021, 12, 683800. [Google Scholar] [CrossRef]
- Chiu, K.H.-Y.; Yip, C.C.-Y.; Poon, R.W.-S.; Leung, K.-H.; Li, X.; Hung, I.F.-N.; To, K.K.-W.; Cheng, V.C.-C.; Yuen, K.-Y. Correlations of Myeloperoxidase (MPO), Adenosine deaminase (ADA), C-C motif chemokine 22 (CCL22), Tumour necrosis factor alpha (TNFα) and Interleukin-6 (IL-6) mRNA expression in the nasopharyngeal specimens with the diagnosis and severity of SARS-CoV-2 infections. Emerg. Microbes Infect. 2023, 12, 2157338. [Google Scholar] [CrossRef]
- Russell, C.D.; Valanciute, A.; Gachanja, N.N.; Stephen, J.; Penrice-Randal, R.; Armstrong, S.D.; Clohisey, S.; Wang, B.; Al Qsous, W.; Wallace, W.A.; et al. Tissue Proteomic Analysis Identifies Mechanisms and Stages of Immunopathology in Fatal COVID-19. Am. J. Respir. Cell Mol. Biol. 2022, 66, 196–205. [Google Scholar] [CrossRef]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.A.P.M.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tsang, O.T.-Y.; et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef]
- Jorens, P.G.; De Jongh, R.; Bossaert, L.L.; De Backer, W.; Herman, A.G.; Pollet, H.; Bosmans, E.; Taupin, J.L.; Moreau, J.F. High levels of leukaemia inhibitory factor in ARDS. Cytokine 1996, 8, 873–876. [Google Scholar] [CrossRef]
- Chehboun, S.; Labrecque-Carbonneau, J.; Pasquin, S.; Meliani, Y.; Meddah, B.; Ferlin, W.; Sharma, M.; Tormo, A.; Masson, J.-F.; Gauchat, J.-F. Epstein-Barr virus-induced gene. J. Biol. Chem. 2017, 292, 6644–6656. [Google Scholar] [CrossRef]
- Attur, M.; Petrilli, C.; Adhikari, S.; Iturrate, E.; Li, X.; Tuminello, S.; Hu, N.; Chakravarti, A.; Beck, D.; Abramson, S.B. Interleukin-1 receptor antagonist gene ( IL1RN ) variants modulate the cytokine release syndrome and mortality of SARS-CoV-2. medRxiv 2023. preprint. [Google Scholar] [CrossRef]
- Allan, S.M.; Tyrrell, P.J.; Rothwell, N.J. Interleukin-1; neuronal injury. Nat. Rev. Immunol. 2005, 5, 629–640. [Google Scholar] [CrossRef]
- Tamayo-Velasco, Á.; Martínez-Paz, P.; Peñarrubia-Ponce, M.J.; de la Fuente, I.; Pérez-González, S.; Fernández, I.; Dueñas, C.; Gómez-Sánchez, E.; Lorenzo-López, M.; Gómez-Pesquera, E.; et al. IL-1α, and IL-27 Are Robust Biomarkers in Early Severity Stratification of COVID-19 Patients. J. Clin. Med. 2021, 10, 2017. [Google Scholar] [CrossRef]
- Liu, Y.; Li, S.; Zhang, G.; Nie, G.; Meng, Z.; Mao, D.; Chen, C.; Chen, X.; Zhou, B.; Zeng, G. Genetic variants in IL1A and IL1B contribute to the susceptibility to 2009 pandemic H1N1 influenza A virus. BMC Immunol 2013, 14, 37. [Google Scholar] [CrossRef]
- Rajamanickam, A.; Nathella, P.K.; Selvaraj, N.; Manoj, M.; Thangaraj, J.W.V.; Muthusamy, S.K.; Purushothaman, G.K.C.; Bhatnagar, T.; Ponnaiah, M.; Ramasamy, S.; et al. Characterization of IL-10 Family of Cytokines in Acute and Convalescent COVID-19 Individuals. J. Interferon Cytokine Res. 2023, 43, 469–477. [Google Scholar] [CrossRef]
- Huang, F.; Wachi, S.; Thai, P.; Loukoianov, A.; Tan, K.H.; Forteza, R.M.; Wu, R. Potentiation of IL-19 expression in airway epithelia by IL-17A and IL-4/IL-13: Important implications in asthma. J. Allergy Clin. Immunol. 2008, 121, 1415–1421.e3. [Google Scholar] [CrossRef]
- Sharif-Askari, F.S.; Sharif-Askari, N.S.; Hafezi, S.; Goel, S.; Alsayed, H.A.H.; Ansari, A.W.; Mahboub, B.; Al-Muhsen, S.; Temsah, M.-H.; Hamid, Q.; et al. Upregulation of interleukin-19 in saliva of patients with COVID-19. Sci. Rep. 2022, 12, 16019. [Google Scholar] [CrossRef]
- Zamani, B.; Momen-Heravi, M.; Erami, M.; Motedayyen, H.; ArefNezhad, R. Impacts of IL-27 and IL-32 in the pathogenesis and outcome of COVID-19 associated mucormycosis. J. Immunoassay Immunochem. 2023, 44, 242–255. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Yang, X.; Yu, Y.; Xu, J.; Shu, H.; Xia, J.; Liu, H.; Wu, Y.; Zhang, L.; Yu, Z.; Fang, M.; et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: A single-centered, retrospective, observational study. Lancet Respir. Med. 2020, 8, 475–481. [Google Scholar] [CrossRef]
- Meidaninikjeh, S.; Sabouni, N.; Marzouni, H.Z.; Bengar, S.; Khalili, A.; Jafari, R. Monocytes and macrophages in COVID-19: Friends and foes. Life Sci. 2021, 269, 119010. [Google Scholar] [CrossRef]
- Nunnari, G.; Sanfilippo, C.; Castrogiovanni, P.; Imbesi, R.; Volti, G.L.; Barbagallo, I.; Musumeci, G.; Di Rosa, M. Network perturbation analysis in human bronchial epithelial cells following SARS-CoV2 infection. Exp. Cell Res. 2020, 395, 112204. [Google Scholar] [CrossRef]
- Khalil, B.A.; Elemam, N.M.; Maghazachi, A.A. Chemokines and chemokine receptors during COVID-19 infection. Comput. Struct. Biotechnol. J. 2021, 19, 976–988. [Google Scholar] [CrossRef]
- Torri, A.; Beretta, O.; Ranghetti, A.; Granucci, F.; Ricciardi-Castagnoli, P.; Foti, M. Gene expression profiles identify inflammatory signatures in dendritic cells. PLoS ONE 2010, 5, e9404. [Google Scholar] [CrossRef]
- Baker, J.R.; Mahdi, M.; Nicolau, D.V.; Ramakrishnan, S.; Barnes, P.J.; Simpson, J.L.; Cass, S.P.; Russell, R.E.K.; Donnelly, L.E.; Bafadhel, M. Early Th2 inflammation in the upper respiratory mucosa as a predictor of severe COVID-19 and modulation by early treatment with inhaled corticosteroids: A mechanistic analysis. Lancet Respir. Med. 2022, 10, 545–556. [Google Scholar] [CrossRef]
- Li, Y.; Han, X. Bioinformatics Approaches to Identify the Comorbidity Complexities of SARS-CoV-2 Infection with Crohn’s Disease. J. Coll. Physicians Surg. Pak. 2023, 33, 1093–1099. [Google Scholar] [CrossRef]
- Jang, Y.; Seo, S.H. Gene expression pattern differences in primary human pulmonary epithelial cells infected with MERS-CoV or SARS-CoV-2. Arch Virol. 2020, 165, 2205–2211. [Google Scholar] [CrossRef]
- Liang, Y.; Li, H.; Li, J.; Yang, Z.-N.; Li, J.-L.; Zheng, H.-W.; Chen, Y.-L.; Shi, H.-J.; Guo, L.; Liu, L.-D. Role of neutrophil chemoattractant CXCL5 in SARS-CoV-2 infection-induced lung inflammatory innate immune response in an in vivo hACE2 transfection mouse model. Zool. Res. 2020, 41, 621–631. [Google Scholar] [CrossRef]
- Sims, J.T.; Krishnan, V.; Chang, C.-Y.; Engle, S.M.; Casalini, G.; Rodgers, G.H.; Bivi, N.; Nickoloff, B.J.; Konrad, R.J.; de Bono, S.; et al. Characterization of the cytokine storm reflects hyperinflammatory endothelial dysfunction in COVID-19. J. Allergy Clin. Immunol. 2021, 147, 107–111. [Google Scholar] [CrossRef]
- Perreau, M.; Suffiotti, M.; Marques-Vidal, P.; Wiedemann, A.; Levy, Y.; Laouénan, C.; Ghosn, J.; Fenwick, C.; Comte, D.; Roger, T.; et al. The cytokines HGF and CXCL13 predict the severity and the mortality in COVID-19 patients. Nat. Commun. 2021, 12, 4888. [Google Scholar] [CrossRef]
- López-Ayllón, B.D.; de Lucas-Rius, A.; Mendoza-García, L.; García-García, T.; Fernández-Rodríguez, R.; Suárez-Cárdenas, J.M.; Santos, F.M.; Corrales, F.; Redondo, N.; Pedrucci, F.; et al. SARS-CoV-2 accessory proteins involvement in inflammatory and profibrotic processes through IL11 signaling. Front. Immunol. 2023, 14, 1220306. [Google Scholar] [CrossRef]
- Oppmann, B.; Lesley, R.; Blom, B.; Timans, J.C.; Xu, Y.; Hunte, B.; Vega, F.; Yu, N.; Wang, J.; Singh, K.; et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity 2000, 13, 715–725. [Google Scholar] [CrossRef]
- Nelson, R.T.; Boyd, J.; Gladue, R.P.; Paradis, T.; Thomas, R.; Cunningham, A.C.; Lira, P.; Brissette, W.H.; Hayes, L.; Hames, L.M.; et al. Genomic organization of the CC chemokine mip-3alpha/CCL20/larc/exodus/SCYA20, showing gene structure, splice variants, and chromosome localization. Genomics 2001, 73, 28–37. [Google Scholar] [CrossRef]
- Hue, S.; Beldi-Ferchiou, A.; Bendib, I.; Surenaud, M.; Fourati, S.; Frapard, T.; Rivoal, S.; Razazi, K.; Carteaux, G.; Delfau-Larue, M.-H.; et al. Uncontrolled Innate and Impaired Adaptive Immune Responses in Patients with COVID-19 Acute Respiratory Distress Syndrome. Am. J. Respir. Crit. Care Med. 2020, 202, 1509–1519. [Google Scholar] [CrossRef]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Helper T Cells and Lymphocyte Activation. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002; Available online: https://www.ncbi.nlm.nih.gov/books/NBK26827/ (accessed on 20 November 2023).
- Jiyarom, B.; Giannakopoulos, S.; Strange, D.P.; Panova, N.; Gale, M.; Verma, S. RIG-I and MDA5 are modulated by bone morphogenetic protein (BMP6) and are essential for restricting Zika virus infection in human Sertoli cells. Front. Microbiol. 2022, 13, 1062499. [Google Scholar] [CrossRef]
- Eddowes, L.A.; Al-Hourani, K.; Ramamurthy, N.; Frankish, J.; Baddock, H.T.; Sandor, C.; Ryan, J.D.; Fusco, D.N.; Arezes, J.; Giannoulatou, E.; et al. Antiviral activity of bone morphogenetic proteins and activins. Nat. Microbiol. 2019, 4, 339–351. [Google Scholar] [CrossRef]
- Miller, D.S.J.; Hill, C.S.; Signaling, F.; Bradshaw, R.A.; Hart, G.W.; Stahl, P.D. Encyclopedia of Cell Biology, 2nd ed.; Academic Press: Oxford, UK, 2023; pp. 46–61. [Google Scholar] [CrossRef]
- Zovi, A.; Ferrara, F.; Langella, R.; Cavallaro, F.; Vitiello, A. Sex affects immune response capacity against COVID-19 infection. Rev. Med. Virol. 2023, 33, e2450. [Google Scholar] [CrossRef]
- GB Atypical Chemokine Receptors ACKR3 & ACKR5. Luxembourg Institute of Health, Luxembourg Institute of Health (n.d.). Available online: https://www.lih.lu/en/event/atypical-chemokine-receptors-ackr3-ackr5/ (accessed on 21 November 2023).
- Zhou, Z.; Ren, L.; Zhang, L.; Zhong, J.; Xiao, Y.; Jia, Z.; Guo, L.; Yang, J.; Wang, C.; Jiang, S.; et al. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe 2020, 27, 883–890.e2. [Google Scholar] [CrossRef]
- Schioppa, T.; Sozio, F.; Barbazza, I.; Scutera, S.; Bosisio, D.; Sozzani, S.; Del Prete, A. Molecular Basis for CCRL2 Regulation of Leukocyte Migration. Front. Cell Dev. Biol. 2020, 8, 615031. [Google Scholar] [CrossRef]
- Galligan, C.L.; Matsuyama, W.; Matsukawa, A.; Mizuta, H.; Hodge, D.R.; Howard, O.M.Z.; Yoshimura, T. Up-regulated expression and activation of the orphan chemokine receptor, CCRL2, in rheumatoid arthritis. Arthritis Rheum. 2004, 50, 1806–1814. [Google Scholar] [CrossRef]
- Mannes, P.Z.; Barnes, C.E.; Biermann, J.; Latoche, J.D.; Day, K.E.; Zhu, Q.; Tabary, M.; Xiong, Z.; Nedrow, J.R.; Izar, B.; et al. Molecular imaging of chemokine-like receptor 1 (CMKLR1) in experimental acute lung injury. Proc. Natl. Acad. Sci. USA 2023, 120, e2216458120. [Google Scholar] [CrossRef] [PubMed]
- Liechti, T.; Iftikhar, Y.; Mangino, M.; Beddall, M.; Goss, C.W.; O’Halloran, J.A.; Mudd, P.A.; Roederer, M. Immune phenotypes that are associated with subsequent COVID-19 severity inferred from post-recovery samples. Nat. Commun. 2022, 13, 7255. [Google Scholar] [CrossRef] [PubMed]
- Blot, M.; Jacquier, M.; Glele, L.-S.A.; Beltramo, G.; Nguyen, M.; Bonniaud, P.; Prin, S.; Andreu, P.; Bouhemad, B.; Bour, J.-B.; et al. Pneumochondrie study group, CXCL10 could drive longer duration of mechanical ventilation during COVID-19 ARDS. Crit. Care 2020, 24, 632. [Google Scholar] [CrossRef]
- Liao, M.; Liu, Y.; Yuan, J.; Wen, Y.; Xu, G.; Zhao, J.; Cheng, L.; Li, J.; Wang, X.; Wang, F.; et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020, 26, 842–844. [Google Scholar] [CrossRef]
- Chua, R.L.; Lukassen, S.; Trump, S.; Hennig, B.P.; Wendisch, D.; Pott, F.; Debnath, O.; Thürmann, L.; Kurth, F.; Völker, M.T.; et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020, 38, 970–979. [Google Scholar] [CrossRef]
- Darbeheshti, F.; Mahdiannasser, M.; Uhal, B.D.; Ogino, S.; Gupta, S.; Rezaei, N. Interindividual immunogenic variants: Susceptibility to coronavirus, respiratory syncytial virus and influenza virus. Rev. Med. Virol. 2021, 31, e2234. [Google Scholar] [CrossRef]
- Miller, H.E.; Robinson, R.T. Early control of Mycobacterium tuberculosis infection requires il12rb1 expression by rag1-dependent lineages. Infect. Immun. 2012, 80, 3828–3841. [Google Scholar] [CrossRef]
- Fujii, H. Cell type-specific roles of Jak3 in IL-2-induced proliferative signal transduction. Biochem. Biophys. Res. Commun. 2007, 354, 825–829. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ward-Kavanagh, L.K.; Lin, W.W.; Šedý, J.R.; Ware, C.F. The TNF Receptor Superfamily in Co-stimulating and Co-inhibitory Responses. Immunity 2016, 44, 1005–1019. [Google Scholar] [CrossRef] [PubMed]
- Anisul, M.; Shilts, J.; Schwartzentruber, J.; Hayhurst, J.; Buniello, A.; Mohammed, E.S.E.; Zheng, J.; Holmes, M.; Ochoa, D.; Carmona, M.; et al. A proteome-wide genetic investigation identifies several SARS-CoV-2-exploited host targets of clinical relevance. Elife 2021, 10, e69719. [Google Scholar] [CrossRef]
- Moro-García, M.A.; Mayo, J.C.; Sainz, R.M.; Alonso-Arias, R. Influence of Inflammation in the Process of T Lymphocyte Differentiation: Proliferative, Metabolic, and Oxidative Changes. Front. Immunol. 2018, 9, 339. [Google Scholar] [CrossRef] [PubMed]
- Day, M. Covid-19: Ibuprofen should not be used for managing symptoms, say doctors and scientists. BMJ 2020, 368, m1086. [Google Scholar] [CrossRef] [PubMed]
- Itagaki, S.; Gopal, E.; Zhuang, L.; Fei, Y.-J.; Miyauchi, S.; Prasad, P.D.; Ganapathy, V. Interaction of ibuprofen and other structurally related NSAIDs with the sodium-coupled monocarboxylate transporter SMCT1 (SLC5A8). Pharm. Res. 2006, 23, 1209–1216. [Google Scholar] [CrossRef] [PubMed]
- Zmudzinski, M.; Rut, W.; Olech, K.; Granda, J.; Giurg, M.; Kaleta, R.; Zgarbova, M.; Kasprzyk, R.; Zhang, L.; Sun, X.; et al. Ebselen derivatives inhibit SARS-CoV-2 replication by inhibition of its essential proteins: PLpro and Mpro proteases, and nsp14 guanine N7-methyltransferase. Sci. Rep. 2023, 13, 1–16. [Google Scholar] [CrossRef]
- Mello, M.L.S. Sodium Valproate-Induced Chromatin Remodeling. Front. Cell. Dev. Biol. 2021, 9, 645518. [Google Scholar] [CrossRef]
- Stakišaitis, D.; Juknevičienė, M.; Damanskienė, E.; Valančiūtė, A.; Balnytė, I.; Alonso, M.M. The Importance of Gender-Related Anticancer Research on Mitochondrial Regulator Sodium Dichloroacetate in Preclinical Studies In Vivo. Cancers 2019, 11, 1210. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).