DOPE/CHEMS-Based EGFR-Targeted Immunoliposomes for Docetaxel Delivery: Formulation Development, Physicochemical Characterization and Biological Evaluation on Prostate Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of pH-Sensitive Liposomes
2.1.1. Box–Behnken Design
2.1.2. Functionalization of pH Sensitive Liposome with Cetuximab (Immunoliposome)
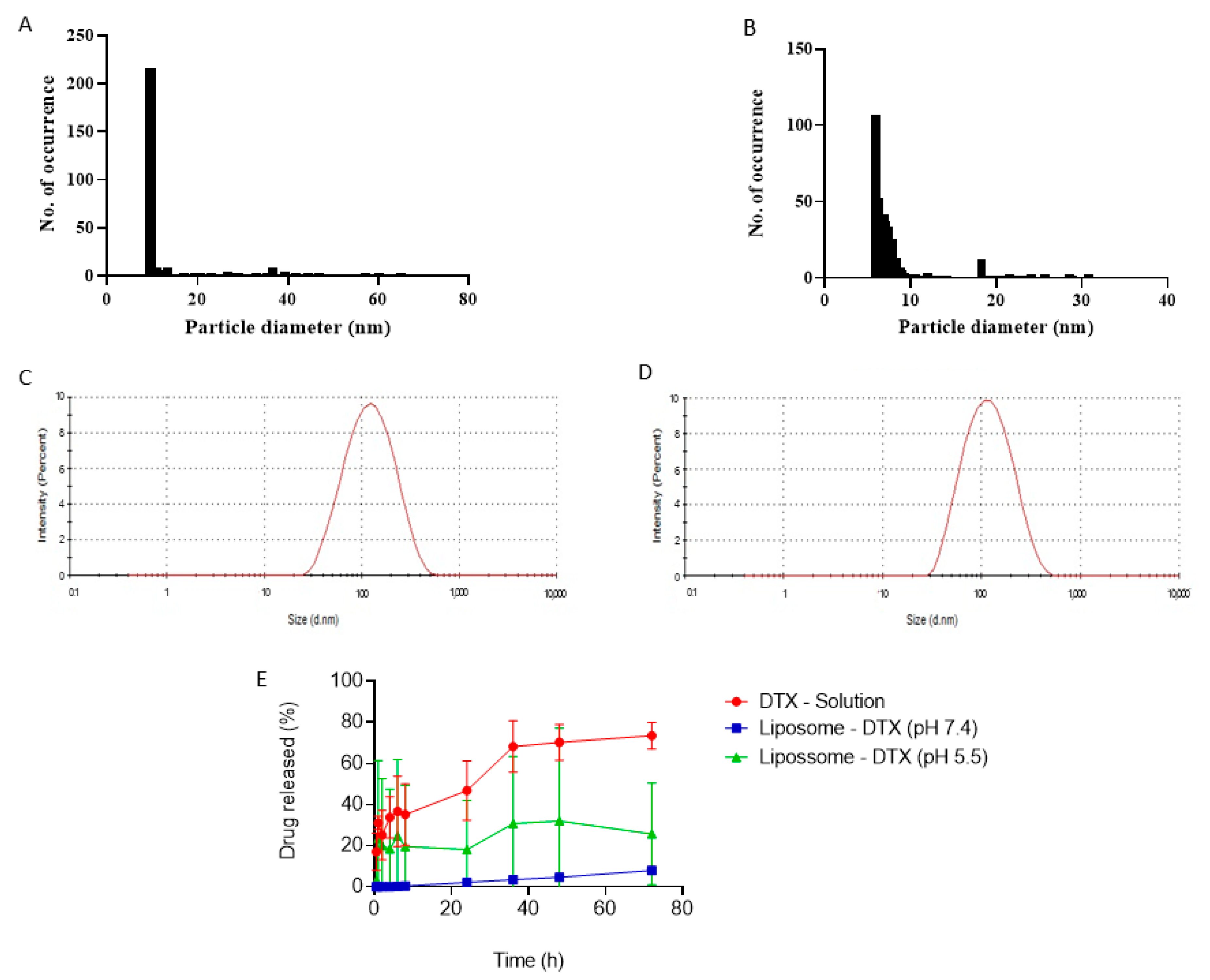
2.2. Physicochemical Characterization of Liposomes and Immunoliposomes
2.2.1. Particle Size, Polydispersity and Zeta Potential
2.2.2. Encapsulation Efficiency
2.2.3. Atomic Force Microscopy
2.2.4. Fourier Transform Infrared Spectroscopy (FTIR)
2.2.5. Thermal Analysis
2.2.6. Powder X-ray Diffractometry
2.2.7. In Vitro Release Study
2.3. Antibody Functionalization Characterization
2.3.1. Electrophoresis
2.3.2. Thermophoresis
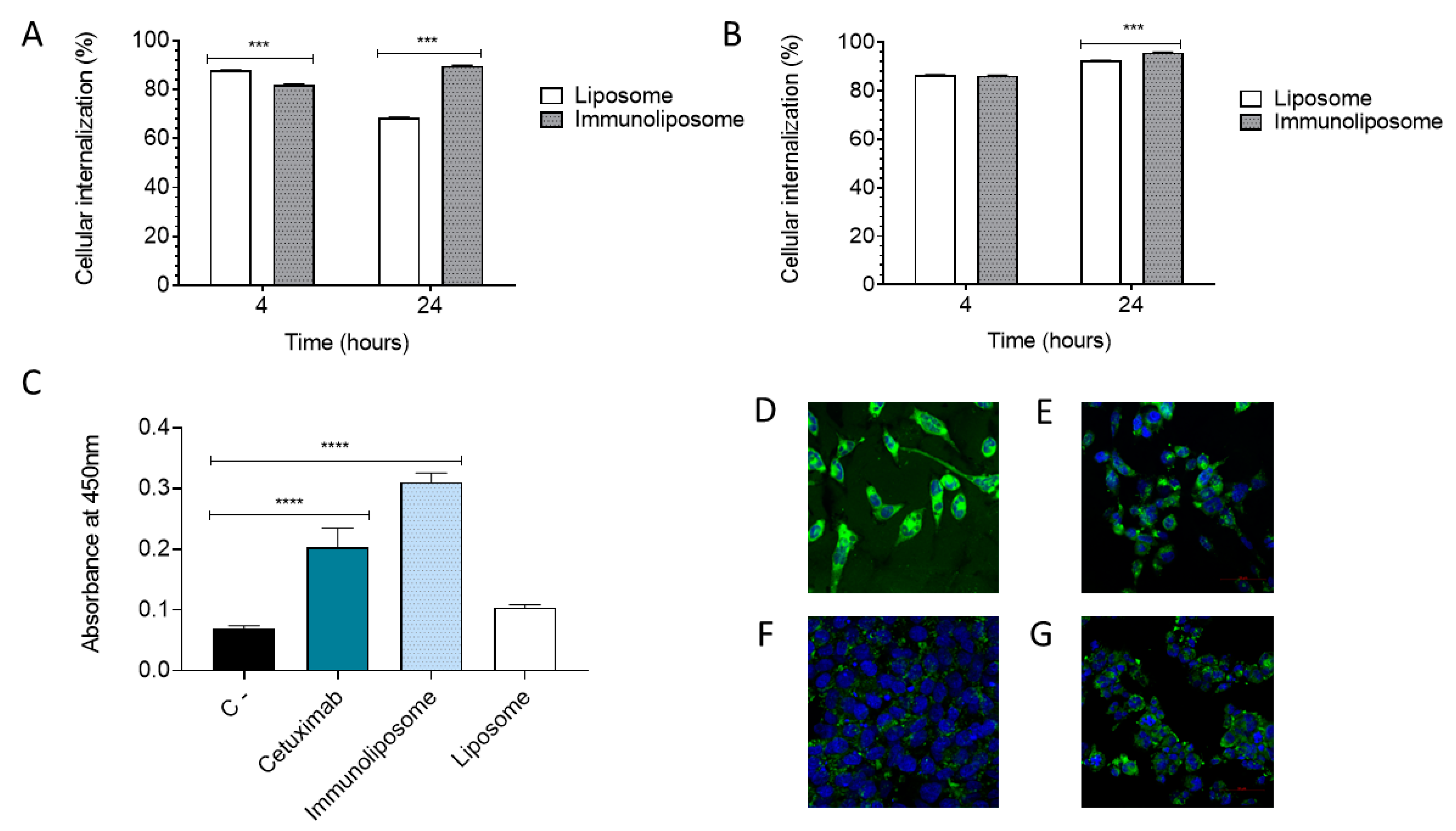
2.3.3. Indirect ELISA
2.4. Cell Studies
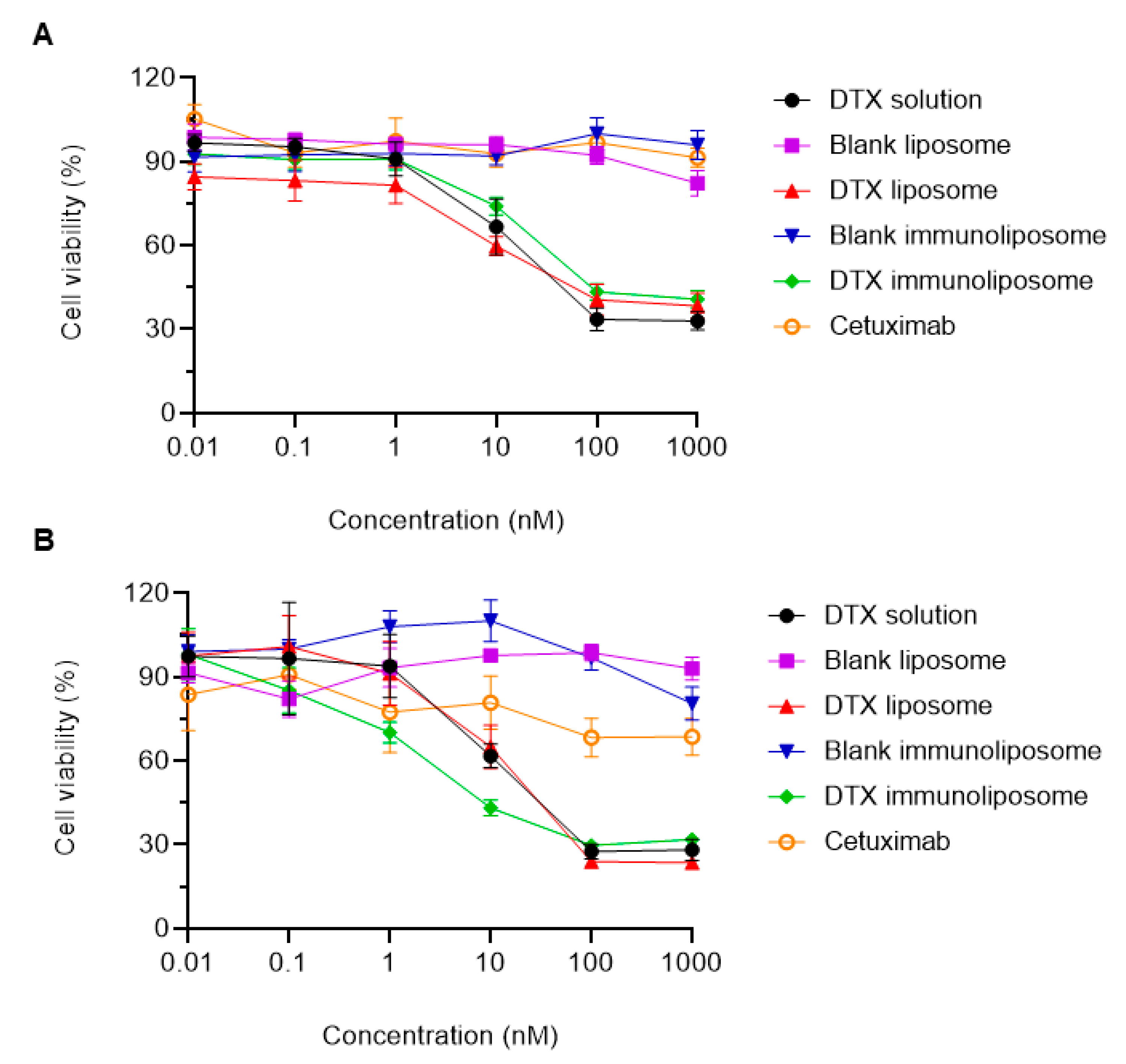
2.4.1. Cellular Viability Assay
2.4.2. Cellular Internalization
2.5. Statistical Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Cancer Observatory. Estimated Number of Prevalent Cases (1-Year) World, Both Sexes, All Ages. 2020. Available online: https://gco.iarc.fr/today (accessed on 7 February 2023).
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; van den Broeck, T.; Cumberbatch, M.G.; de Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. European Urology. 2020, 2, 243–262. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Saleem, S.; Chaudhuri, A.; Ali, J.; Baboota, S. Docetaxel: An update on its molecular mechanisms, therapeutic trajectory and nanotechnology in the treatment of breast, lung and prostate cancer. J. Drug Deliv. Sci. Technol. 2020, 60, 101959. [Google Scholar] [CrossRef]
- Abdel-Rahman, O. Efficacy and toxicity outcomes of elderly castrate-resistant prostate cancer patients treated with docetaxel—A pooled analysis of 3 randomized studies. Urol. Oncol. Semin. Orig. Investig. 2020, 38, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Du, C.; Guo, N.; Teng, Y.; Meng, X.; Sun, H.; Li, S.; Yu, P.; Galons, H. Composition design and medical application of liposomes. Eur. J. Med. Chem. 2019, 164, 640–653. [Google Scholar] [CrossRef]
- Guimarães, D.; Cavaco-Paulo, A.; Nogueira, E. Design of liposomes as drug delivery system for therapeutic applications. Int. J. Pharm. 2021, 161, 120571. [Google Scholar] [CrossRef]
- Nsairat, H.; Khater, D.; Sayed, U.; Odeh, F.; al Bawab, A.; Alshaer, W. Liposomes: Structure, composition, types, and clinical applications. Heliyon 2022, 8, e09394. [Google Scholar] [CrossRef]
- Corrie, L.; Kaur, J.; Awasthi, A.; Vishwas, S.; Gulati, M.; Saini, S.; Kumar, B.; Pandey, N.K.; Gupta, G.; Dureja, H.; et al. Multivariate Data Analysis and Central Composite Design-Oriented Optimization of Solid Carriers for Formulation of Curcumin-Loaded Solid SNEDDS: Dissolution and Bioavailability Assessment. Pharmaceutics 2022, 14, 2395. [Google Scholar] [CrossRef]
- Jain, A.; Hurkat, P.; Jain, S.K. Development of liposomes using formulation by design: Basics to recent advances. Chem. Phys. Lipids 2019, 224, 104764. [Google Scholar] [CrossRef]
- Vardhan, H.; Mittal, P.; Adena, S.K.R.; Mishra, B. Long-circulating polyhydroxybutyrate-co-hydroxyvalerate nanoparticles for tumor targeted docetaxel delivery: Formulation, optimization and in vitro characterization. Eur. J. Pharm. Sci. 2017, 99, 85–94. [Google Scholar] [CrossRef]
- Eloy, J.O.; Ruiz, A.; de Lima, F.T.; Petrilli, R.; Raspantini, G.; Nogueira, K.A.B.; Santos, E.; de Oliveira, C.S.; Borges, J.C.; Marchetti, J.M.; et al. EGFR-targeted immunoliposomes efficiently deliver docetaxel to prostate cancer cells. Colloids Surf. B Biointerfaces 2020, 194, 111185. [Google Scholar] [CrossRef]
- Ashrafizadeh, M.; Delfi, M.; Zarrabi, A.; Bigham, A.; Sharifi, E.; Rabiee, N.; Paiva-Santos, A.C.; Kumar, A.P.; Tan, S.C.; Hushmandi, K.; et al. Stimuli-responsive liposomal nanoformulations in cancer therapy: Pre-clinical & clinical approaches. J. Control. Release 2022, 351, 50–80. [Google Scholar] [PubMed]
- Belfiore, L.; Saunders, D.N.; Ranson, M.; Thurecht, K.J.; Storm, G.; Vine, K.L. Towards clinical translation of ligand-functionalized liposomes in targeted cancer therapy: Challenges and opportunities. J. Control. Release 2018, 277, 1–13. [Google Scholar] [CrossRef]
- Eloy, J.O.; Petrilli, R.; Trevizan, L.N.F.; Chorilli, M. Immunoliposomes: A review on functionalization strategies and targets for drug delivery. Colloids Surf. B Biointerfaces 2017, 159, 454–467. [Google Scholar] [CrossRef] [PubMed]
- Merino, M.; Zalba, S.; Garrido, M.J. Immunoliposomes in clinical oncology: State of the art and future perspectives. J. Control. Release 2018, 275, 162–176. [Google Scholar] [CrossRef] [PubMed]
- Rezaiemanesh, A.; Majidi, J.; Baradaran, B.; Movasaghpour, A.; Nakhlband, A.; Barar, J.; Omidi, Y. Impacts of anti-EGFR monoclonal antibody in prostate cancer PC3 cells. Hum. Antibodies 2010, 19, 63–70. [Google Scholar] [CrossRef]
- Da Silva Santos, E.; Nogueira, K.A.B.; Fernandes, L.C.C.; Martins, J.R.P.; Reis, A.V.F.; de Brito Vieira Neto, J.; da Silva Júnior, I.J.; Pessoa, C.; Petrilli, R.; Eloy, J.O. EGFR targeting for cancer therapy: Pharmacology and immunoconjugates with drugs and nanoparticles. Int. J. Pharm. 2021, 592, 120082. [Google Scholar] [CrossRef] [PubMed]
- Bangham, A.D.; Standish, M.M.; Watkins, J.C. Diffusion of univalent ions across the lamellae of swollen phospholipids. J. Mol. Biol. 1965, 13, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Eloy, J.O.; Petrilli, R.; Topan, J.F.; Antonio, H.M.R.; Barcellos, J.P.A.; Chesca, D.L.; Serafini, L.N.; Tiezzi, D.G.; Lee, R.J.; Marchetti, J.M. Co-loaded paclitaxel/rapamycin liposomes: Development, characterization and in vitro and in vivo evaluation for breast cancer therapy. Colloids Surf. B Biointerfaces 2016, 141, 74–82. [Google Scholar] [CrossRef]
- Solanki, A.B.; Parikh, J.R.; Parikh, R.H. Formulation and Optimization of Piroxicam Proniosomes by 3-Factor, 3-Level Box-Behnken Design. 2007. Available online: http://www.aapspharmscitech.org (accessed on 9 January 2023).
- Ferreira, S.L.C.; Bruns, R.E.; Ferreira, H.S.; Matos, G.D.; David, J.M.; Brandão, G.C.; da Silva, E.G.P.; Portugal, L.A.; dos Reis, P.S.; Souza, A.S.; et al. Box-Behnken design: An alternative for the optimization of analytical methods. Anal. Chim. Acta 2007, 597, 179–186. [Google Scholar] [CrossRef]
- Bastogne, T. Quality-by-design of nanopharmaceuticals—A state of the art. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 2151–2157. [Google Scholar] [CrossRef]
- Abdelbary, A.A.; Aboughaly, M.H.H. Design and optimization of topical methotrexate loaded niosomes for enhanced management of psoriasis: Application of Box-Behnken design, in-vitro evaluation and in-vivo skin deposition study. Int. J. Pharm. 2015, 485, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Wagner, J.R.; Mount, E.M.; Giles, H.F. Design of Experiments. In Extrusion; Elsevier: Amsterdam, The Netherlands, 2014; pp. 291–308. Available online: https://linkinghub.elsevier.com/retrieve/pii/B9781437734812000259 (accessed on 15 February 2023).
- Petrilli, R.; Eloy, J.O.; Saggioro, F.P.; Chesca, D.L.; de Souza, M.C.; Dias, M.V.S.; daSilva, L.L.P.; Lee, R.J.; Lopez, R.F.V. Skin cancer treatment effectiveness is improved by iontophoresis of EGFR-targeted liposomes containing 5-FU compared with subcutaneous injection. J. Control. Release 2018, 283, 151–162. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, Y.S.; Oliveira, A.C.; Ayala, A.P. Mechanochemically induced solid state transformations: The case of raloxifene hydrochloride. Eur. J. Pharm. Sci. 2018, 114, 146–154. [Google Scholar] [CrossRef]
- De Pinho Pessoa Nogueira, L.; de Oliveira, Y.S.; de CFonseca, J.; Costa, W.S.; Raffin, F.N.; Ellena, J.; Ayala, A.P. Crystalline structure of the marketed form of Rifampicin: A case of conformational and charge transfer polymorphism. J. Mol. Struct. 2018, 1155, 260–266. [Google Scholar] [CrossRef]
- Claro De Souza, M.; Marotta-Oliveira, S.S.A.; Rocha, N.H.S.; Eloy, J.O.; Marchetti, J.M. Development of a Method to Evaluate the Release Profile of Tamoxifen from Pegylated Hybrid Micelles. J. Liq. Chromatogr. Relat. Technol. 2015, 38, 1223–1229. [Google Scholar] [CrossRef]
- Kulhari, H.; Pooja, D.; Shrivastava, S.; Telukutala, S.R.; Barui, A.K.; Patra, C.R.; Naidu Vegi, G.M.; Adams, D.J.; Sistla, R. Cyclic-RGDfK peptide conjugated succinoyl-TPGS nanomicelles for targeted delivery of docetaxel to integrin receptor over-expressing angiogenic tumours. Nanomedicine 2015, 11, 1511–1520. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, U.S.; Straubinger, R.M. Paclitaxel-liposomes for intracavitary therapy of intraperitoneal P388 leukemia. Cancer Lett. 1996, 107, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Sheng, Z.; Liu, Z.; Zhang, X.; Xiao, Y.; Xie, J.; Zhang, Y.; Xu, T. CMTM5-v1 inhibits cell proliferation and migration by downregulating oncogenic EGFR signaling in prostate cancer cells. J. Cancer 2020, 11, 3762–3770. [Google Scholar] [CrossRef]
- Silva, R.; Ferreira, H.; Little, C.; Cavaco-Paulo, A. Effect of ultrasound parameters for unilamellar liposome preparation. Ultrason. Sonochem. 2010, 17, 628–632. [Google Scholar] [CrossRef]
- Kanamala, M.; Palmer, B.D.; Jamieson, S.M.F.; Wilson, W.R.; Wu, Z. Dual pH-sensitive liposomes with low pH-triggered sheddable PEG for enhanced tumor-targeted drug delivery. Nanomedicine 2019, 14, 1971–1989. [Google Scholar] [CrossRef]
- Lafi, Z.; Alshaer, W.; Hatmal, M.M.; Zihlif, M.; Alqudah, D.A.; Nsairat, H.; Azzam, H.; Aburjai, T.; Bustanji, Y.; Awidi, A. Aptamer-functionalized pH-sensitive liposomes for a selective delivery of echinomycin into cancer cells. RSC Adv. 2021, 11, 29164–29177. [Google Scholar] [CrossRef]
- Isalomboto Nkanga, C.; Murhimalika Bapolisi, A.; Ikemefuna Okafor, N.; Werner Maçedo Krause, R. General Perception of Liposomes: Formation, Manufacturing and Applications. In Liposomes—Advances and Perspectives; IntechOpen: London, UK, 2019. [Google Scholar]
- Lee, Y.; Thompson, D.H. Stimuli-responsive liposomes for drug delivery. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2017, 9, e1450. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, A.; Mohamed, H.; Mustafa, A.; Ali, H.; Haythum, A.R.B.; Izzeldeen, N.; Abdelgadir, N.; Saeed, A.A. PH-sensitive Liposomes and Application. World J. Pharm. Pharm. Sci. SJIF Impact Factor 2020, 9, 212–228. Available online: www.wjpps.com (accessed on 11 February 2023).
- Paliwal, S.R.; Paliwal, R.; Vyas, S.P. A review of mechanistic insight and application of pH-sensitive liposomes in drug delivery. Drug Deliv. 2015, 22, 231–242. [Google Scholar] [CrossRef] [PubMed]
- Rawal, M.; Singh, A.; Amiji, M.M. Quality-by-Design Concepts to Improve Nanotechnology-Based Drug Development. Pharm. Res. 2019, 36, 153. [Google Scholar] [CrossRef]
- Singh, B.; Sharma, T.; Saini, S.; Kaur, R.; Jain, A.; Raza, K.; Beg, S. Systematic Development of Drug Nanocargos Using Formulation by Design (FbD): An Updated Overview. 2014. Available online: www.begellhouse.com (accessed on 11 February 2023).
- Magalhães, J.; Chaves, L.L.; Vieira, A.C.; Santos, S.G.; Pinheiro, M.; Reis, S. Optimization of rifapentine-loaded lipid nanoparticles using a Quality-by-Design strategy. Pharmaceutics 2020, 12, 75. [Google Scholar] [CrossRef]
- Khuri, A.I.; Mukhopadhyay, S. Response surface methodology. Wiley Interdiscip. Rev. Comput. Stat. 2010, 3, 128–149. [Google Scholar] [CrossRef]
- Tamizharasan, T.; Senthilkumar, N.; Selvakumar, V.; Dinesh, S. Taguchi’s methodology of optimizing turning parameters over chip thickness ratio in machining P/M AMMC. SN Appl. Sci. 2019, 1, 160. [Google Scholar] [CrossRef]
- Das Kurmi, B.; Paliwal, R.; Paliwal, S. Dual cancer targeting using estrogen functionalized chitosan nanoparticles loaded with doxorubicin-estrone conjugate: A quality by design approach. Int. J. Biol. Macromol. 2020, 164, 2881–2894. [Google Scholar] [CrossRef]
- Monteiro, L.O.F.; Malachias, Â.; Pound-Lana, G.; Magalhães-Paniago, R.; Mosqueira, V.C.F.; Oliveira, M.C.; de Barros, A.L.B.; Leite, E.A. Paclitaxel-Loaded pH-Sensitive Liposome: New Insights on Structural and Physicochemical Characterization. Langmuir 2018, 34, 5728–5737. [Google Scholar] [CrossRef]
- Bahari, L.A.S.; Hamishehkar, H. The impact of variables on particle size of solid lipid nanoparticles and nanostructured lipid carriers; A comparative literature review. Adv. Pharm. Bull. 2016, 6, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Hasanzadeh Davarani, F.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of particle size and polydispersity index on the clinical applications of lipidic nanocarrier systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef]
- Chugh, H.; Sood, D.; Chandra, I.; Tomar, V.; Dhawan, G.; Chandra, R. Role of gold and silver nanoparticles in cancer nano-medicine. Artif. Cells Nanomed. Biotechnol. 2018, 46, 1210–1220. [Google Scholar] [CrossRef]
- Gaumet, M.; Vargas, A.; Gurny, R.; Delie, F. Nanoparticles for drug delivery: The need for precision in reporting particle size parameters. Eur. J. Pharm. Biopharm. 2008, 69, 1–9. [Google Scholar] [CrossRef]
- Badran, M. Formulation and In Vitro Evaluation of Flufenamic Acid Loaded Deformable Liposomes for Improved Skin Delivery. Dig. J. Nanomater. Biostruct. 2014, 9, 83–91. [Google Scholar]
- Chen, M.; Liu, X.; Fahr, A. Skin penetration and deposition of carboxyfluorescein and temoporfin from different lipid vesicularsystems: In vitro study with finite and infinite dosage application. Int. J. Pharm. 2011, 408, 223–234. [Google Scholar] [CrossRef]
- Putri, D.C.A.; Dwiastuti, R.; Marchaban, M.; Nugroho, A.K. Optimization of mixing temperature and sonication duration in liposome preparation. J. Pharm. Sci. Community 2017, 14, 79–85. [Google Scholar] [CrossRef]
- Bertrand, N.; Wu, J.; Xu, X.; Kamaly, N.; Farokhzad, O.C. Cancer nanotechnology: The impact of passive and active targeting in the era of modern cancer biology. Adv. Drug Deliv. Rev. 2014, 66, 2–25. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Tang, M. Safety of novel liposomal drugs for cancer treatment: Advances and prospects. Chem. Biol. Interact. 2018, 295, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Paxton, J.W.; Wu, Z. Enhanced pH-responsiveness, cellular trafficking, cytotoxicity and long-circulation of PE gylated liposomes with post-insertion technique using gemcitabine as a model drug. Pharm. Res. 2015, 32, 2428–2438. [Google Scholar] [CrossRef]
- Zhang, J.X.; Zalipsky, S.; Mullah, N.; Pechar, M.; Allen, T.M. Pharmaco attributes of dioleoylphosphatidylethanolamine/cholesterylhemisuccinate liposomes containing different types of cleavable lipopolymers. Pharmacol. Res. 2004, 49, 185–198. [Google Scholar] [CrossRef]
- Sudhakar, B.; Krishna, M.C.; Murthy, K.V.R. Factorial design studies of antiretroviral drug-loaded stealth liposomal injectable: PEGylation, lyophilization and pharmacokinetic studies. Appl. Nanosci. 2016, 6, 43–60. [Google Scholar] [CrossRef]
- Corrie, L.; Gulati, M.; Awasthi, A.; Vishwas, S.; Kaur, J.; Khursheed, R.; Kumar, R.; Kumar, A.; Imran, M.; Chellappan, D.K.; et al. Polysaccharide, fecal microbiota, and curcumin-based novel oral colon-targeted solid self-nanoemulsifying delivery system: Formulation, characterization, and in-vitro anticancer evaluation. Mater Today Chem. 2022, 26, 101165. [Google Scholar] [CrossRef]
- Palaniappan, S.P.; Muthukumara, K.; Sabariraj, R.V.; Kumar, S.D.; Sathish, T. CNC turning process parameters optimization on Aluminium 6082 alloy by using Taguchi and ANOVA. Mater. Today Proc. 2020, 21, 1013–1021. [Google Scholar] [CrossRef]
- Liu, K.C.; Arivajiagane, A.; Wu, S.J.; Tzou, S.C.; Chen, C.Y.; Wang, Y.M. Development of a novel thermal-sensitive multifunctional liposome with antibody conjugation to target EGFR-expressing tumors. Nanomedicine 2019, 15, 285–294. [Google Scholar] [CrossRef]
- Jain, S.; Deore, S.V.; Ghadi, R.; Chaudhari, D.; Kuche, K.; Katiyar, S.S. Tumor microenvironment responsive VEGF-antibody functionalized pH sensitive liposomes of docetaxel for augmented breast cancer therapy. Mater. Sci. Eng. C 2021, 121, 111832. [Google Scholar] [CrossRef]
- Mamot, C.; Drummond, D.C.; Greiser, U.; Hong, K.; Kirpotin, D.B.; Marks, J.D.; Park, J.W.; Krishnada, A.; Onyuksel, H. Epidermal growth factor receptor (EGFR)-targeted immunoliposomes mediate specific and efficient drug delivery to EGFR- and EGFRvIII-overexpressing tumor cells. Women’s Oncol. Rev. 2004, 4, 101–103. [Google Scholar]
- Asmari, M.; Ratih, R.; Alhazmi, H.A.; el Deeb, S. Thermophoresis for characterizing biomolecular interaction. Methods 2018, 146, 107–119. [Google Scholar] [CrossRef]
- Marasini, R.; Nguyen, T.D.T.; Rayamajhi, S.; Aryal, S. Synthesis and characterization of a tumor-seeking LyP-1 peptide integrated lipid-polymer composite nanoparticle. Mater. Adv. 2020, 1, 469–480. [Google Scholar] [CrossRef]
- Maya, S.; Kumar, L.G.; Sarmento, B.; Sanoj Rejinold, N.; Menon, D.; Nair, S.V.; Jayakumar, R. Cetuximab conjugated O-carboxymethyl chitosan nanoparticles for targeting EGFR overexpressing cancer cells. Carbohydr. Polym. 2013, 93, 661–669. [Google Scholar] [CrossRef]
- Vella-Zarb, L.; Dinnebier, R.E.; Baisch, U. The devil is in the detail: A rare H-bonding motif in new forms of docetaxel. Cryst. Growth Des. 2013, 13, 4402–4410. [Google Scholar] [CrossRef]
- Yousefi, A.; Esmaeili, F.; Rahimian, S.; Atyabi, F.; Dinarvand, R. Preparation and in vitro evaluation of a pegylated nano-liposomal formulation containing docetaxel. Sci. Pharm. 2009, 77, 453–464. [Google Scholar] [CrossRef]
- Fan, X.; Chen, J.; Shen, Q. Docetaxel-nicotinamide complex-loaded nanostructured lipid carriers for transdermal delivery. Int. J. Pharm. 2013, 458, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Broström, M.; Enestam, S.; Backman, R.; Mäkelä, K. Condensation in the KCl-NaCl system. Fuel Process. Technol. 2013, 105, 142–148. [Google Scholar] [CrossRef]
- Lee, T.; da Chang, G. Sucrose conformational polymorphism: A jigsaw puzzle with multiple routes to a unique solution. Cryst. Growth Des. 2009, 9, 3551–3561. [Google Scholar] [CrossRef]
- Takechi-Haraya, Y.; Matsuoka, M.; Imai, H.; Izutsu, K.; Sakai-Kato, K. Detection of material-derived differences in the stiffness of egg yolk phosphatidylcholine-containing liposomes using atomic force microscopy. Chem. Phys. Lipids 2020, 233, 104992. [Google Scholar] [CrossRef] [PubMed]
- Onyesom, I.; Lamprou, D.A.; Sygellou, L.; Owusu-Ware, S.K.; Antonijevic, M.; Chowdhry, B.Z.; Douroumis, D. Sirolimus encapsulated liposomes for cancer therapy: Physicochemical and mechanical characterization of sirolimus distribution within liposome bilayers. Mol. Pharm. 2013, 10, 4281–4293. [Google Scholar] [CrossRef]
- Eaton, P.; Quaresma, P.; Soares, C.; Neves, C.; de Almeida, M.P.; Pereira, E.; West, P. A direct comparison of experimental methods to measure dimensions of synthetic nanoparticles. Ultramicroscopy 2017, 182, 179–190. [Google Scholar] [CrossRef]
- Lee, J.M.; Park, H.; Oh, K.T.; Lee, E.S. pH-Responsive hyaluronated liposomes for docetaxel delivery. Int. J. Pharm. 2018, 547, 377–384. [Google Scholar] [CrossRef]
- Zuo, T.; Guan, Y.; Chang, M.; Zhang, F.; Lu, S.; Wei, T.; Shao, W.; Lin, G. RGD(Arg-Gly-Asp) internalized docetaxel-loaded pH sensitive liposomes: Preparation, characterization and antitumor efficacy in vivo and in vitro. Colloids Surf. B Biointerfaces 2016, 147, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Sicard, G.; Paris, C.; Giacometti, S.; Rodallec, A.; Ciccolini, J.; Rocchi, P.; Fanciullino, R. Enhanced antisense oligonucleotide delivery using cationic liposomes grafted with trastuzumab: A proof-of-concept study in prostate cancer. Pharmaceutics 2020, 12, 1166. [Google Scholar] [CrossRef]
- Mack, K.; Rüger, R.; Fellermeier, S.; Seifert, O.; Kontermann, R.E. Dual targeting of tumor cells with bispecific single-chain fv-immunoliposomes. Antibodies 2012, 1, 199–214. [Google Scholar] [CrossRef]
- Yang, Q.; Shi, G.; Chen, X.; Lin, Y.; Cheng, L.; Jiang, Q.; Yan, X.; Jiang, M.; Li, Y.; Zhang, H.; et al. Nanomicelle protects the immune activation effects of Paclitaxel and sensitizes tumors to anti-PD-1 Immunotherapy. Theranostics 2020, 10, 8382–8399. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Q.; Chen, J.; Tang, C.; Yin, C. Enhanced antitumor efficacy of glutathione-responsive chitosan based nanoparticles through co-delivery of chemotherapeutics, genes, and immune agents. Carbohydr. Polym. 2021, 270, 118384. [Google Scholar] [CrossRef] [PubMed]
- De Freitas, J.V.B.; Reis, A.V.F.; Silva, A.D.O.; de Sousa, A.C.C.; Martins, J.R.P.; Nogueira, K.A.B.; da Silva Moreira, T.; Petrilli, R.; Eloy, J.O. Monoclonal Antibodies in Nanosystems as a Strategy for Cancer Treatment. In Cancer Nanotechnology; Springer International Publishing: Cham, Switzerland, 2023; pp. 115–176. [Google Scholar]
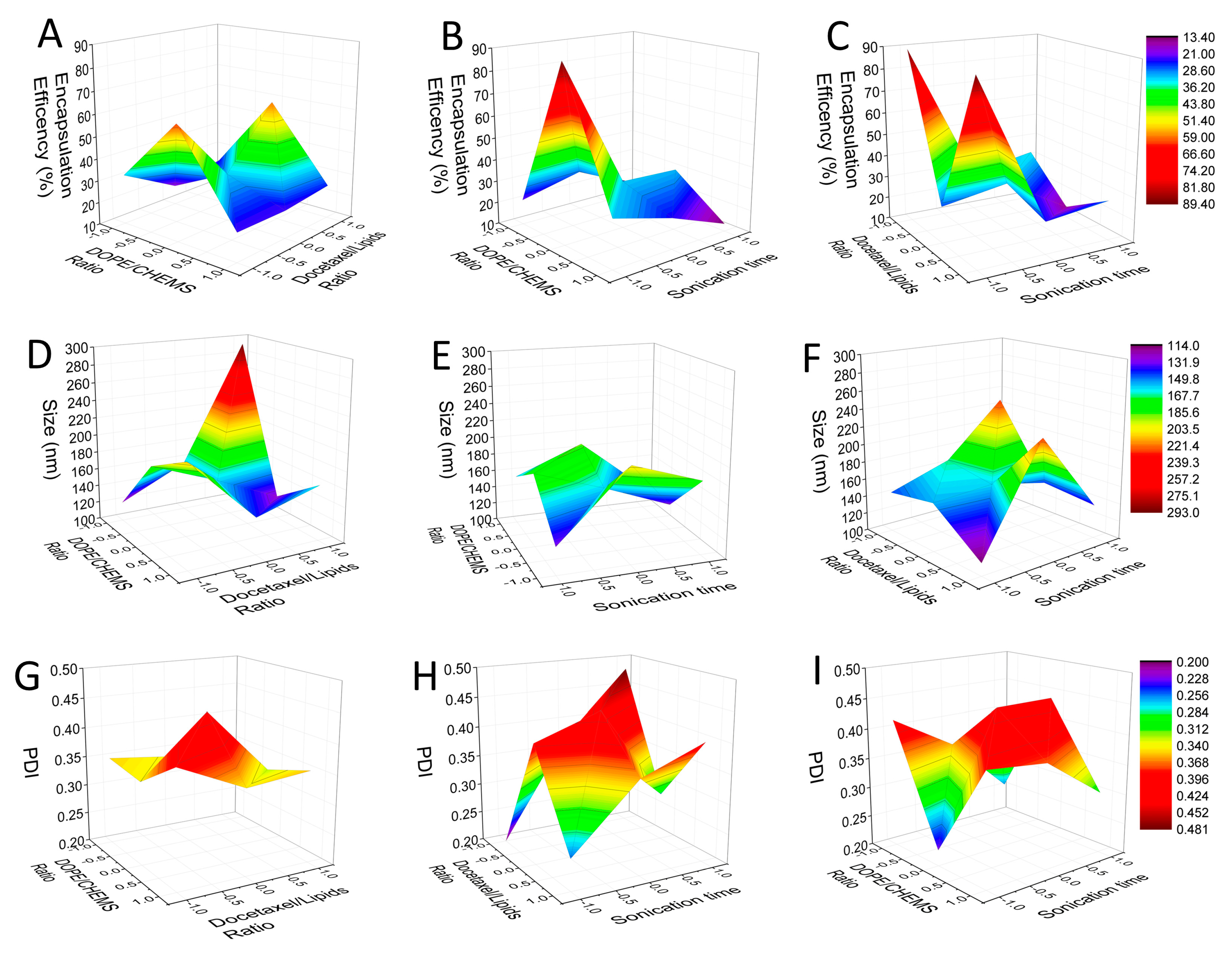
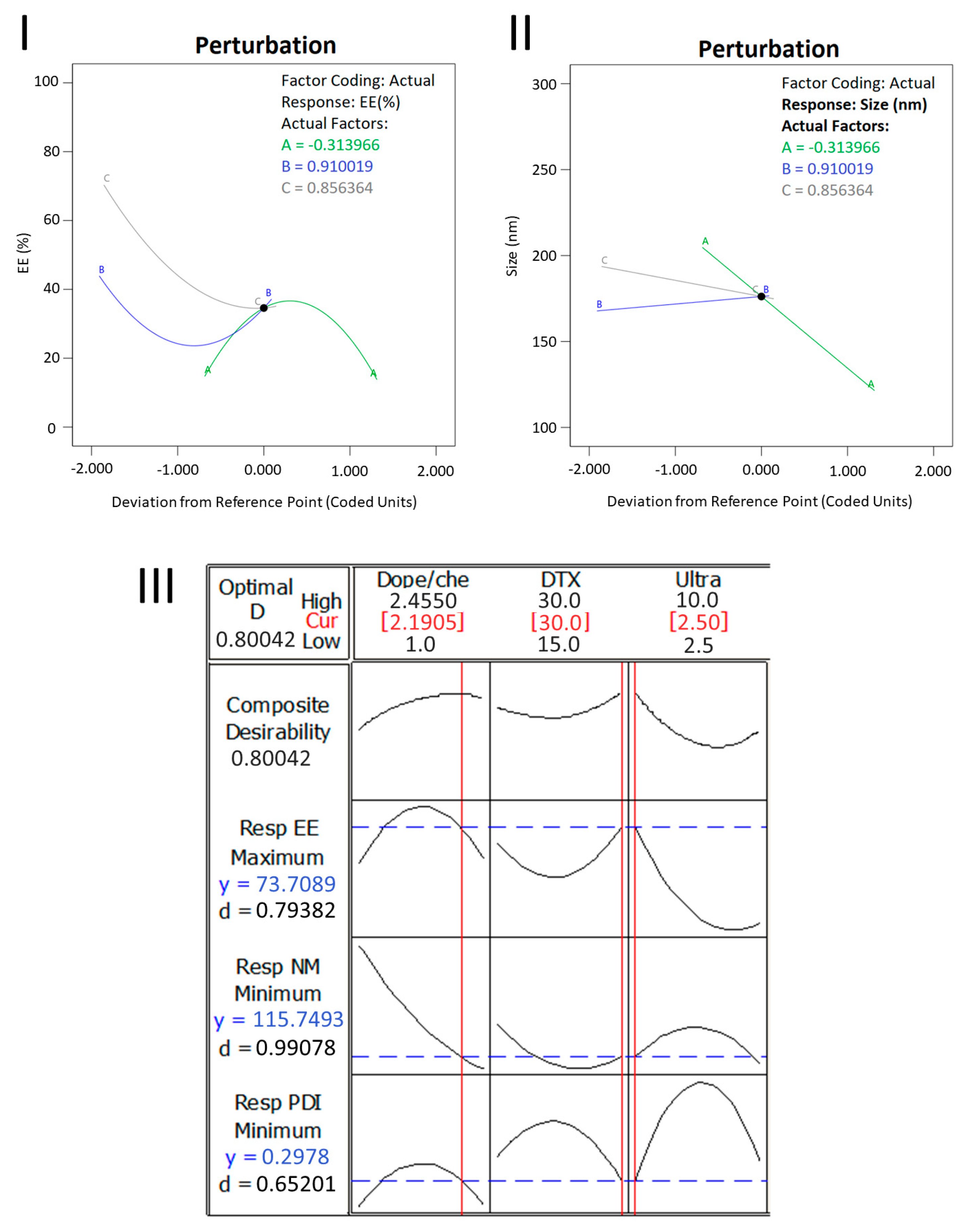


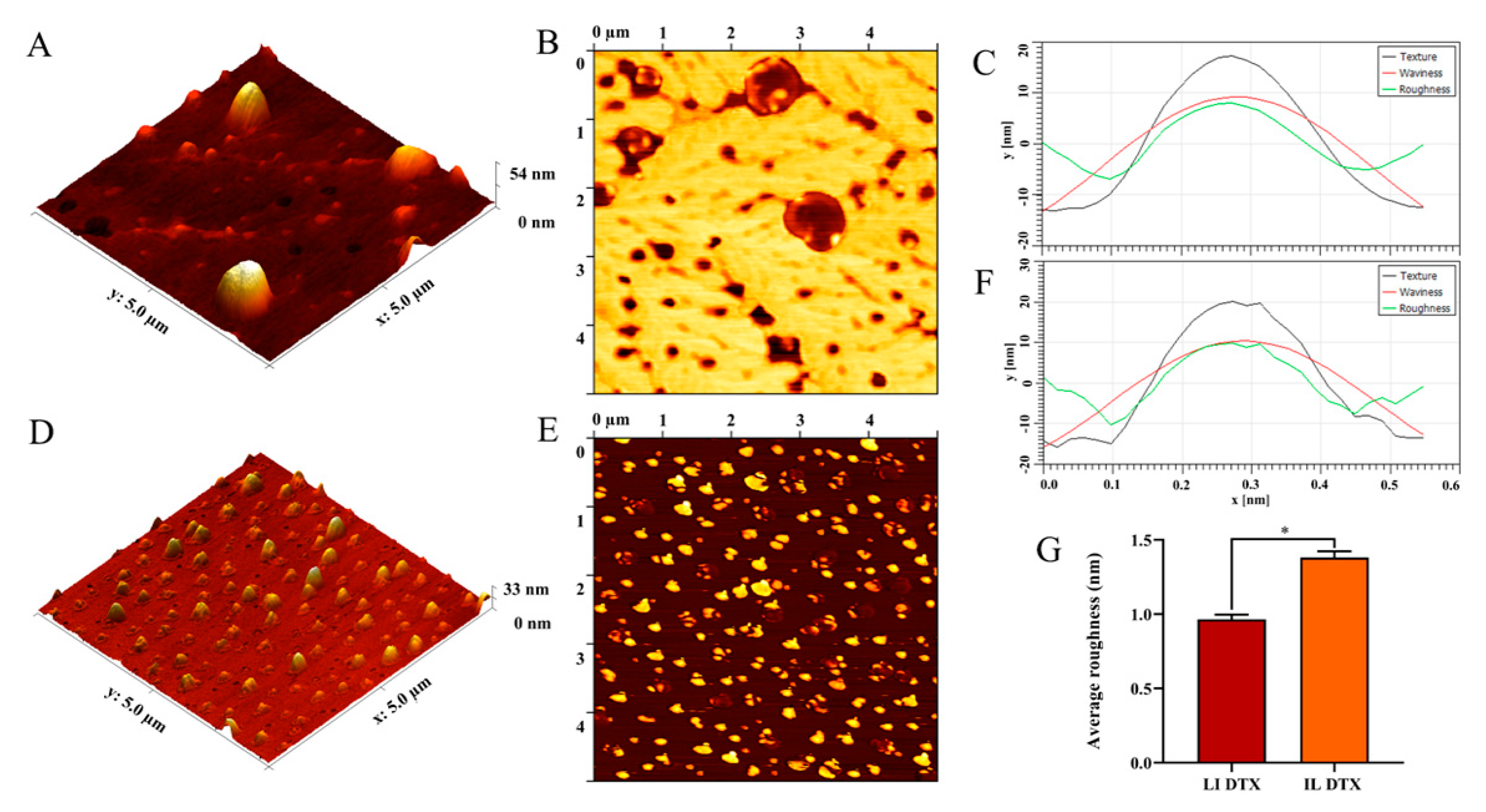
| Independent Variable | Level Used | |||
|---|---|---|---|---|
| Symbol | Low (−1) | Medium (0) | High (+1) | |
| X1: DOPE:CHEMS ratio | DP/CHEM | 1 | 1.5 | 2.5 |
| X2: Docetaxel:lipids ratio | DTX/LP | 1:15 | 1:20 | 1:30 |
| X3: Sonication time | ST | 2.5 min | 5 min | 10 min |
| Fixed Component | DSPE-PEG 0.5 mol | |||
| Response variables | Goal | |||
| Y1: Encapsulation efficiency (%) | EE (%) | Maximum (100%) | ||
| Y2: Particle size (nm) | Size (nm) | Optimum (<200 nm) | ||
| Y3: Polydispersity index | PDI | Minimize | ||
| Formulation Run | X1 (DP/CHEM) | X2 (DTX/LP) | X3 (ST) | Y1 [EE (%)] | Y2 [Size (nm)] | Y3 (PDI) | |||
|---|---|---|---|---|---|---|---|---|---|
| Level | Tested Ratio | Level | Tested Ratio | Level | Time Tested | ||||
| 1 | −1 | 1.0 | −1 | 1:15 | 0 | 5 | 32.4 ± 2.28 | 121.6 ± 2.2 | 0.350 ± 0.019 |
| 2 | 1 | 2.5 | −1 | 1:15 | 0 | 5 | 22.88 ± 1.12 | 213.3 ± 7.3 | 0.394 ± 0.032 |
| 3 | −1 | 1.0 | 1 | 1:30 | 0 | 5 | 23.88 ± 4.89 | 292.9 ± 21.9 | 0.361 ± 0.044 |
| 4 | 1 | 2.5 | 1 | 1:30 | 0 | 5 | 25.15 ± 6.00 | 163.1 ± 0.8 | 0.355 ± 0.004 |
| 5 | −1 | 1.0 | 0 | 1:20 | −1 | 2.5 | 21.42 ± 2.08 | 182.5 ± 1.8 | 0.416 ± 0.005 |
| 6 | 1 | 2.5 | 0 | 1:20 | −1 | 2.5 | 31.73 ± 3.21 | 126.8 ± 3.1 | 0.386 ± 0.037 |
| 7 | −1 | 1.0 | 0 | 1:20 | 1 | 10 | 16.36 ± 3.70 | 130.9 ± 4.3 | 0.259 ± 0.001 |
| 8 | 1 | 2.5 | 0 | 1:20 | 1 | 10 | 13.45 ± 0.57 | 156 ± 0.4 | 0.303 ± 0.018 |
| 9 | 0 | 1.5 | −1 | 1:15 | −1 | 2.5 | 89.36 ± 8.77 | 145.1 ± 0.7 | 0.200 ± 0.004 |
| 10 | 0 | 1.5 | 1 | 1:30 | −1 | 2.5 | 88.97 ± 13.78 | 114.1 ± 0.6 | 0.253 ± 0.009 |
| 11 | 0 | 1.5 | −1 | 1:15 | 1 | 10 | 33.26 ± 9.17 | 228.8 ± 6.2 | 0.481 ± 0.047 |
| 12 | 0 | 1.5 | 1 | 1:30 | 1 | 10 | 28.01 ± 6.43 | 137.7 ± 1.2 | 0.400 ± 0.022 |
| 13 | 0 | 1.5 | 0 | 1:20 | 0 | 5 | 32.72 ± 6.85 | 144.0 ± 6.4 | 0.480 ± 0.013 |
| 14 | 0 | 1.5 | 0 | 1:20 | 0 | 5 | 43.17 ± 5.24 | 193.9 ± 27.2 | 0.453 ± 0.043 |
| 15 | 0 | 1.5 | 0 | 1:20 | 0 | 5 | 19.31 ± 0.74 | 137.1 ± 1.9 | 0.387 ± 0.198 |
| Y1 [EE (%)] | Y2 [Size (nm)] | Y3 (PDI) | ||||
|---|---|---|---|---|---|---|
| Coeff. | p-Value | Coeff. | p-Value | Coeff. | p-Value | |
| Intercept | 139.426 | 0.345 | 108.439 | 0.822 | −0.545 | 0.580 |
| X1 | 155.460 | 0.094 | 2.211 | 0.993 | 0.331 | 0.553 |
| X2 | −15.354 | 0.138 | −5.294 | 0.866 | 0.039 | 0.545 |
| X3 | −19.119 | 0.178 | 41.911 | 0.362 | 0.089 | 0.336 |
| X12 | −47.378 | 0.051 | 36.928 | 0.587 | −0.076 | 0.581 |
| X22 | 0.346 | 0.109 | 0.480 | 0.466 | −0.001 | 0.549 |
| X32 | 1.388 | 0.109 | −2.482 | 0.355 | −0.006 | 0.284 |
| X1X2 | 0.304 | 0.843 | −7.727 | 0.182 | −0.002 | 0.847 |
| X1X3 | 0.289 | 0.925 | 4.016 | 0.704 | −0.003 | 0.882 |
| X2X3 | −0.159 | 0.597 | −0.684 | 0.509 | 0.000 | 0.917 |
| R2 | 81.81(%) | 51.42(%) | 30.66(%) | |||
| Regression equation of the fitted model | ||||||
| Immunoliposome with 2 mg Antibody | |||
| Blank Immunoliposome * | DTX Immunoliposome * | ||
| Size (nm) | 107.4 ± 2.05 | 106.8 ± 4.45 | |
| PDI | 0.224 ± 0.013 | 0.224 ± 0.013 | |
| Zeta (mV) | −16.7 ± 1.42 | -17.1 ± 1.50 | |
| EC% | 9.84% | 10.68% | |
| EE% | - | 85.84 ± 3.70 | |
| Immunoliposome with 3 mg Antibody | |||
| Blank immunoliposome * | DTX immunoliposome * | DTX immunoliposome at 37 °C | |
| Size (nm) | 111.07 ± 0.51 | 111.47 ± 0.49 | 156.77 ± 1.67 |
| PDI | 0.197 ± 0.01 | 0.245 ± 0.01 | 0.245 ± 0.00 |
| Zeta (mV) | −17.77 ± 1.20 | −17.77 ± 1.20 | −17.67 ± 1.35 |
| EC% | 11.65% | 6.91% | 14.06% |
| EE% | - | 75.8 ± 15.30 | 86.0 ± 14.29 |
| PC3 | ||
| Formulation | IC50 (nM) | Confidence interval |
| DTX solution | 55.77 ± 9.21 | 38.76 to 81.80 |
| DTX liposome | 65.74 ± 14.61 | 41.17 to 110.3 |
| DTX immunoliposome | 152.1 ± 25.43 | 106.2 to 225.5 |
| Cetuximab | - | - |
| DU145 | ||
| Formulation | IC50 (nM) | Confidence interval |
| DTX solution | 33.55 ± 7.20 | 20.91 to 55.33 |
| DTX liposome | 28.28 ± 4.60 | 19.80 to 40.99 |
| DTX immunoliposome | 12.60 ± 2.50 | 8.128 to 19.91 |
| Cetuximab | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moreira, T.d.S.; Silva, A.D.O.; Vasconcelos, B.R.F.; Santos, E.d.S.; de Sousa, A.C.C.; de Freitas, J.V.B.; de Oliveira, Y.S.; Vidal, L.M.T.; Ribeiro, F.d.O.S.; de Araújo, A.R.; et al. DOPE/CHEMS-Based EGFR-Targeted Immunoliposomes for Docetaxel Delivery: Formulation Development, Physicochemical Characterization and Biological Evaluation on Prostate Cancer Cells. Pharmaceutics 2023, 15, 915. https://doi.org/10.3390/pharmaceutics15030915
Moreira TdS, Silva ADO, Vasconcelos BRF, Santos EdS, de Sousa ACC, de Freitas JVB, de Oliveira YS, Vidal LMT, Ribeiro FdOS, de Araújo AR, et al. DOPE/CHEMS-Based EGFR-Targeted Immunoliposomes for Docetaxel Delivery: Formulation Development, Physicochemical Characterization and Biological Evaluation on Prostate Cancer Cells. Pharmaceutics. 2023; 15(3):915. https://doi.org/10.3390/pharmaceutics15030915
Chicago/Turabian StyleMoreira, Thais da Silva, Alan Denis Olivindo Silva, Bianca Rodrigues Farias Vasconcelos, Elias da Silva Santos, Ana Carolina Cruz de Sousa, João Vito Barroso de Freitas, Yara Santiago de Oliveira, Laura Maria Teodorio Vidal, Fábio de Oliveira Silva Ribeiro, Alyne Rodrigues de Araújo, and et al. 2023. "DOPE/CHEMS-Based EGFR-Targeted Immunoliposomes for Docetaxel Delivery: Formulation Development, Physicochemical Characterization and Biological Evaluation on Prostate Cancer Cells" Pharmaceutics 15, no. 3: 915. https://doi.org/10.3390/pharmaceutics15030915
APA StyleMoreira, T. d. S., Silva, A. D. O., Vasconcelos, B. R. F., Santos, E. d. S., de Sousa, A. C. C., de Freitas, J. V. B., de Oliveira, Y. S., Vidal, L. M. T., Ribeiro, F. d. O. S., de Araújo, A. R., Vieira Neto, J. d. B., Pessoa, C. d. Ó., Petrilli, R., & Eloy, J. O. (2023). DOPE/CHEMS-Based EGFR-Targeted Immunoliposomes for Docetaxel Delivery: Formulation Development, Physicochemical Characterization and Biological Evaluation on Prostate Cancer Cells. Pharmaceutics, 15(3), 915. https://doi.org/10.3390/pharmaceutics15030915









