Bacteriocin-Nanoconjugates (Bac10307-AgNPs) Biosynthesized from Lactobacillus acidophilus-Derived Bacteriocins Exhibit Enhanced and Promising Biological Activities
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Extraction and Purification of Bacteriocin from L. acidophilus
2.3. Analysis of Antibacterial Activity of Partially Purified Bacteriocin-Bac10307
2.3.1. Bacterial Strains and Growth Conditions
2.3.2. Agar Well Diffusion Assay
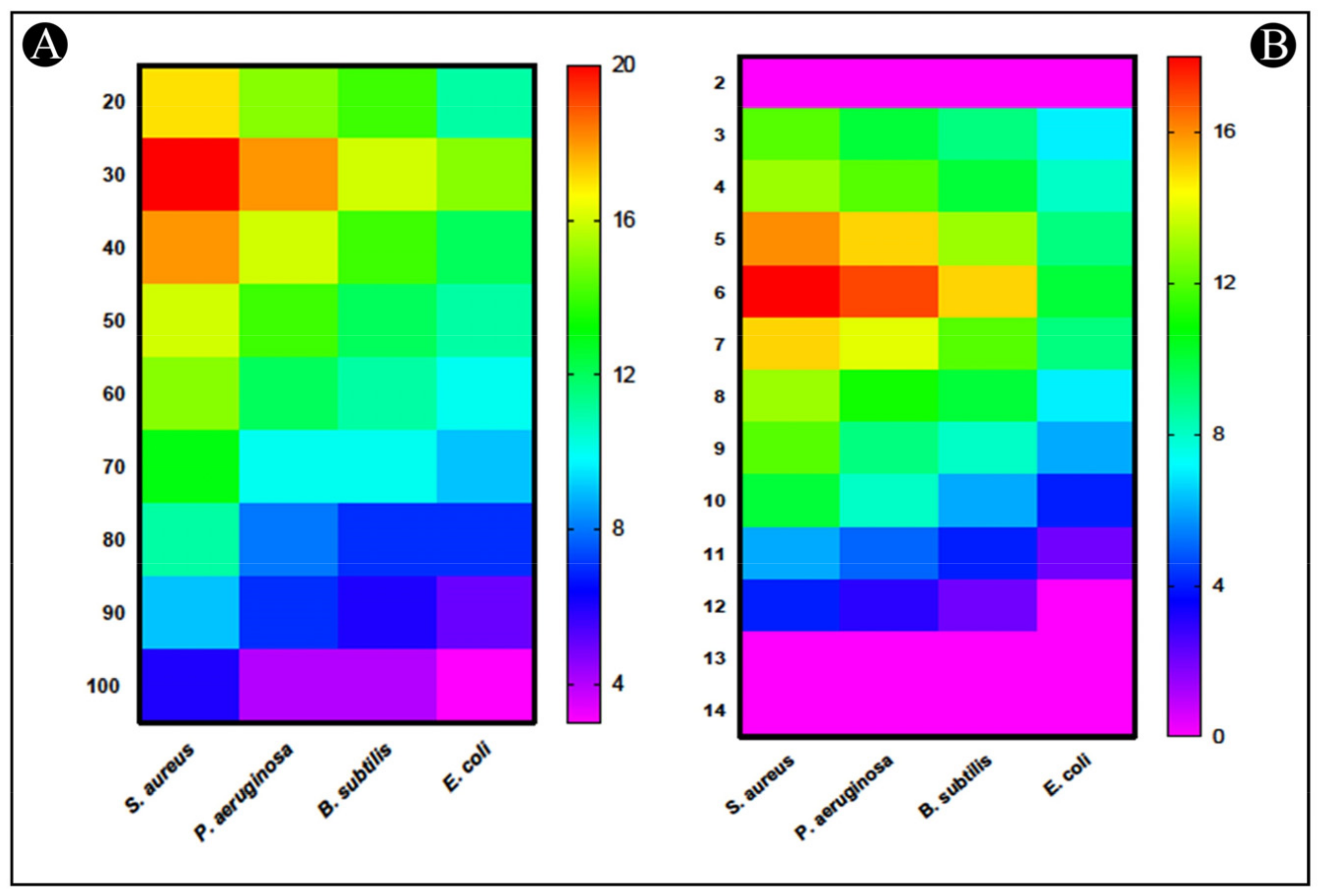
2.4. Assessment of Stability of Bacteriocin-Bac10307
2.4.1. Impact of Temperatures
2.4.2. Impact of pH
2.4.3. Impact of Enzymes
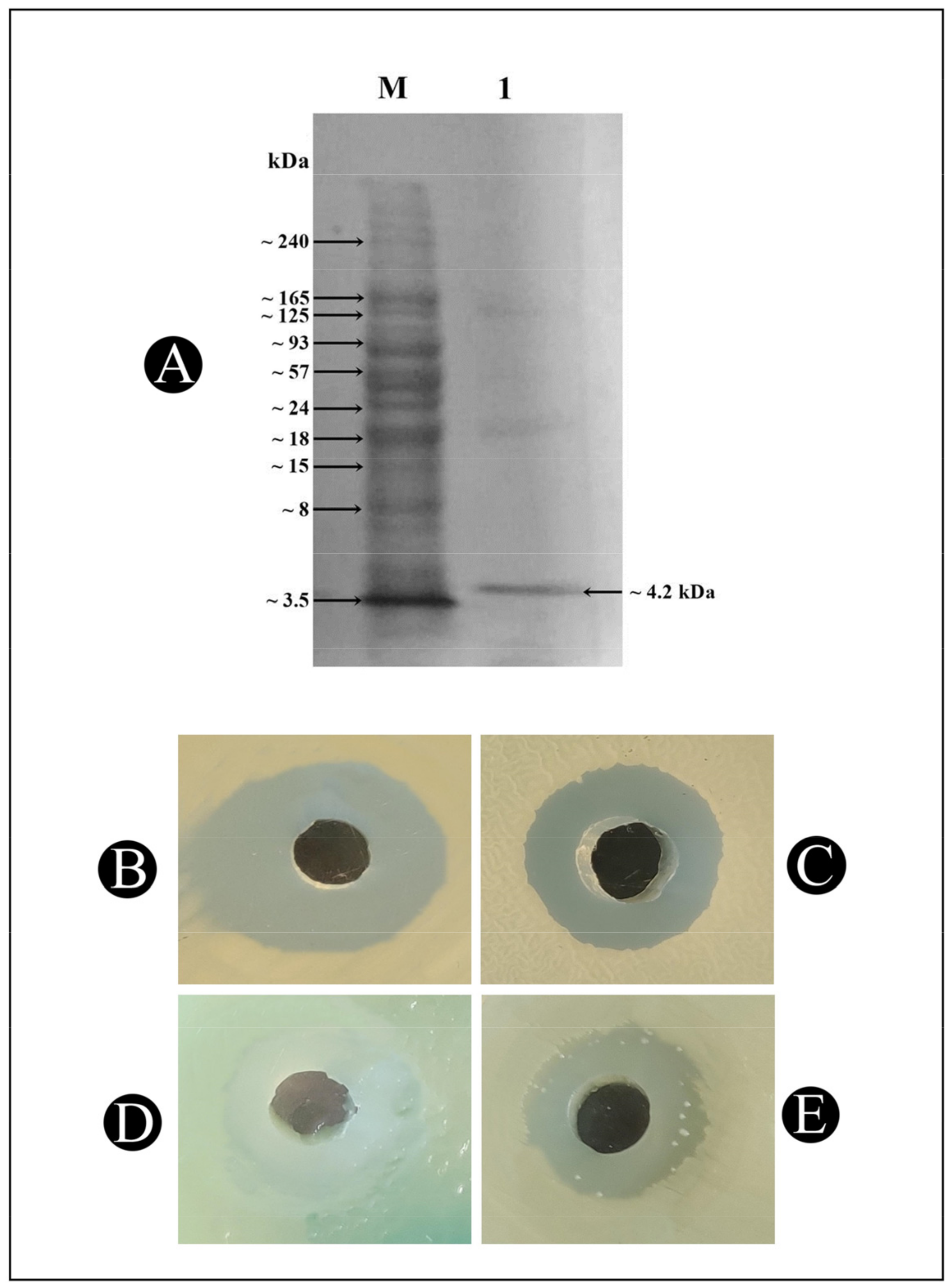
2.5. Molecular Mass Determination of Bacteriocin Bac10307 Using SDS–PAGE
2.6. Synthesis of Bacteriocin-10307-AgNPs
2.7. Characterization of Bac10307-AgNPs
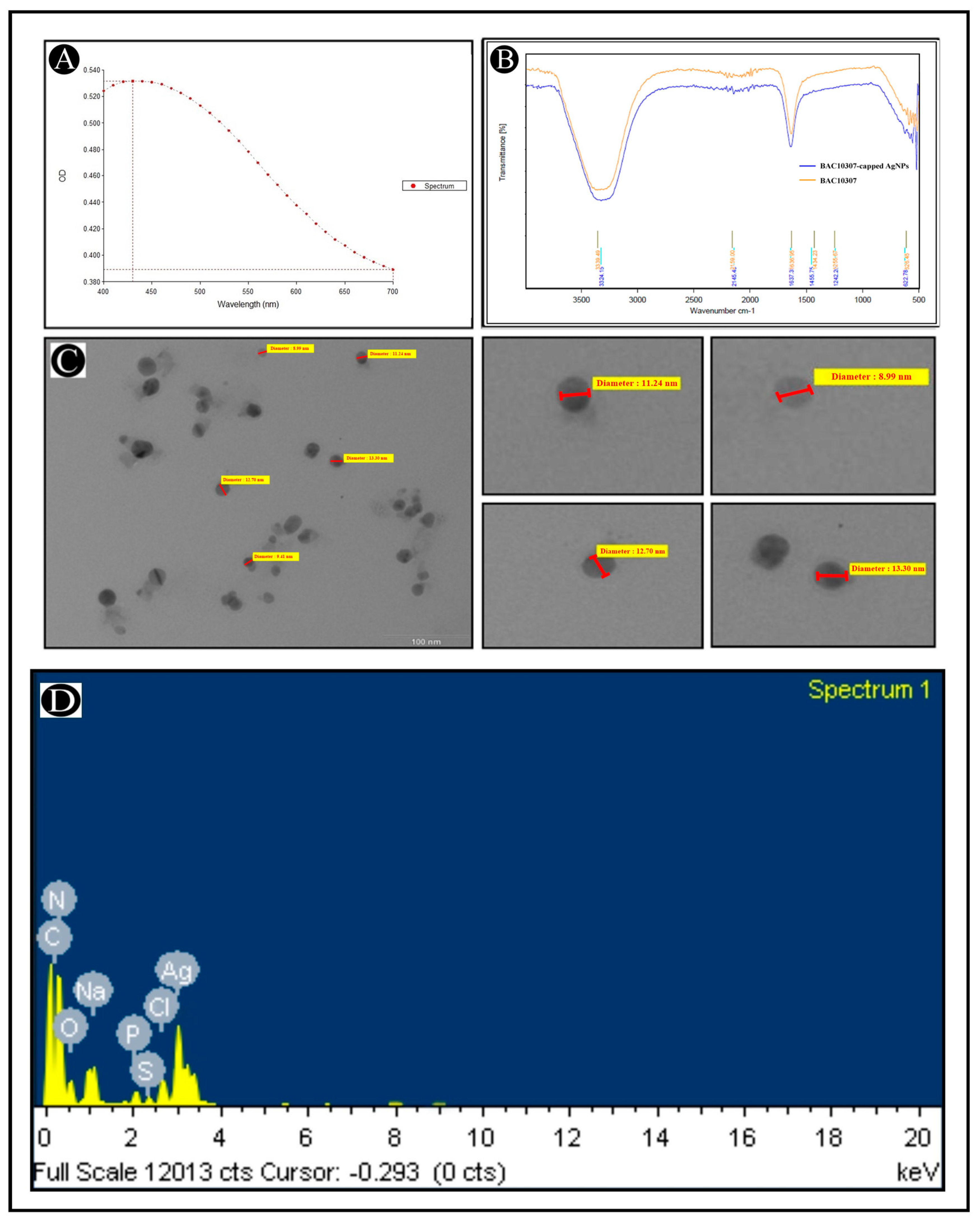
2.7.1. UV–Vis Spectrophotometer
2.7.2. Transmission Electron Microscopy (TEM) and Energy-Dispersive X-Ray (EDX)
2.7.3. Fourier-Transform Infrared (FT-IR) Analysis
2.8. Screening of Antibacterial Activity of Bac10307-AgNPs
2.8.1. Agar Well Diffusion Assay
2.8.2. Assessment of Minimum Inhibitory Concentration (MIC)
2.9. Screening of In Vitro Antioxidant Activity of Bac10307-AgNPs
2.10. Screening of In Vitro Cytotoxicity Assay (MTT Assay) of Bac10307-AgNPs
2.11. Molecular Docking (MD) Assays
2.12. Statistical Analysis
3. Results
3.1. Bacteriocin-Bac10307: Extraction, Purification, and Antibacterial Activity Analysis
3.2. Characterization of Purified Bacteriocin-Bac10307
3.3. Synthesis and Characterization of Bac10307-AgNPs
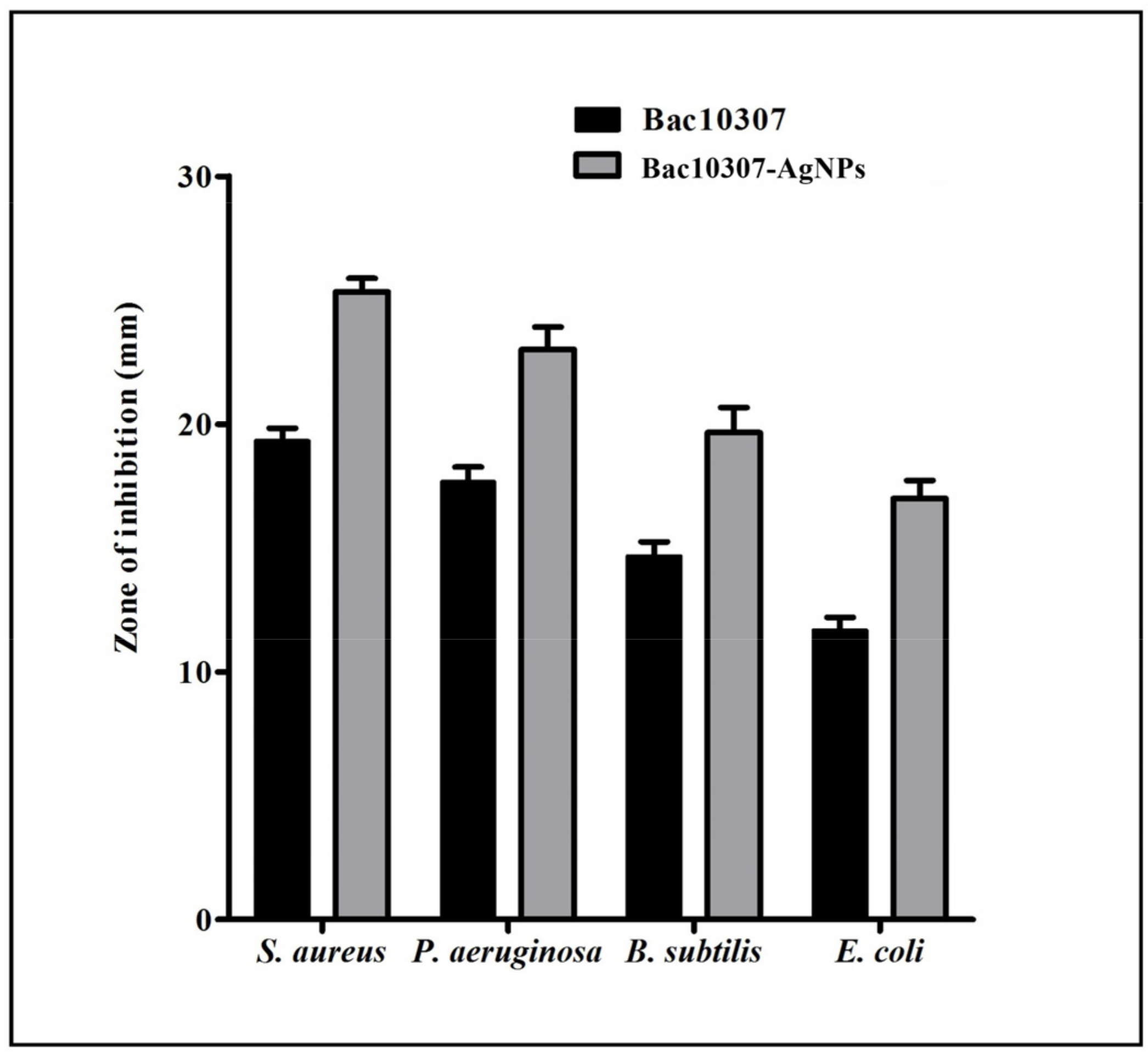
3.4. Antibacterial Activity of Bac10307 and Synthesized Bac10307-AgNPs
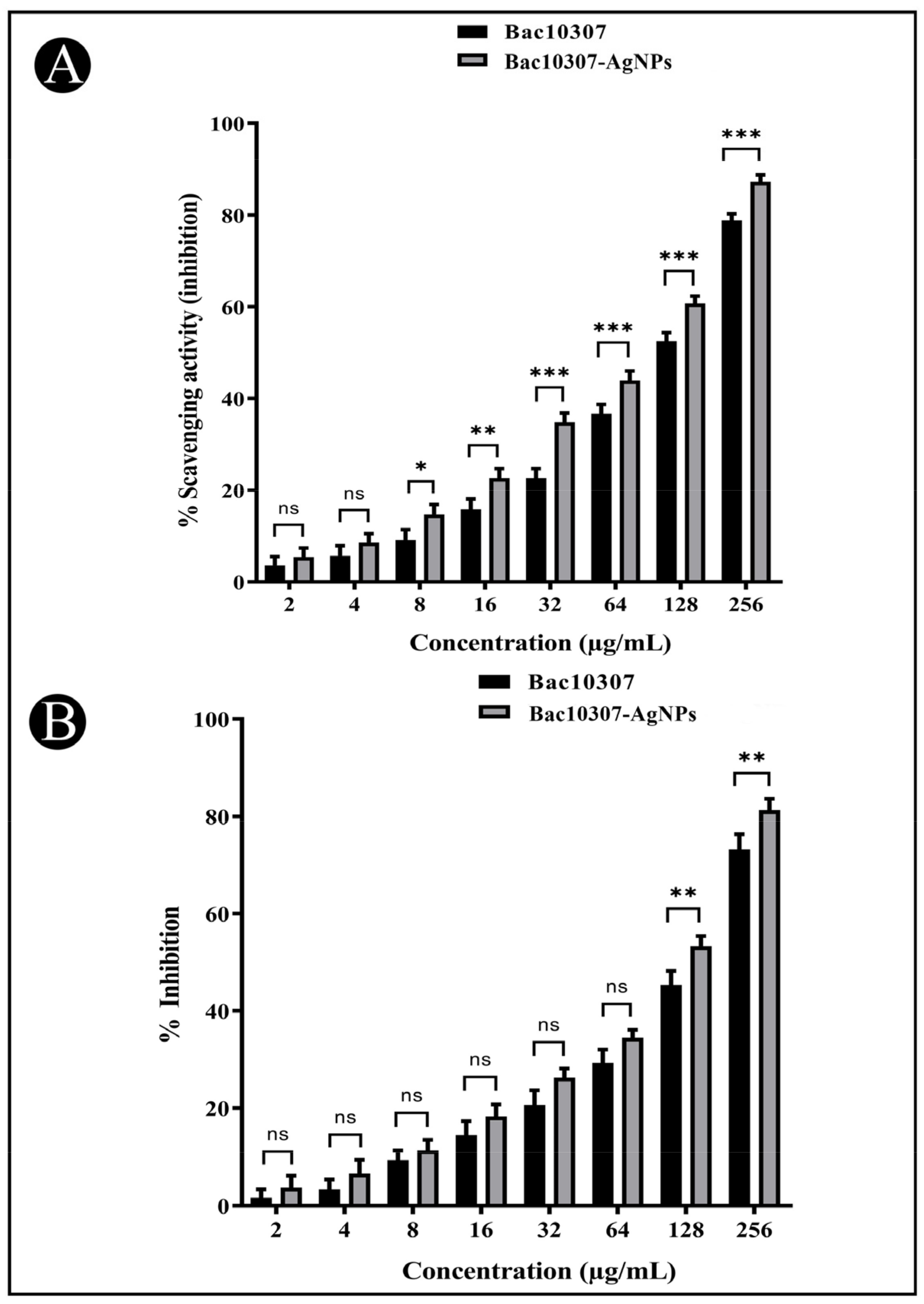
3.5. In Vitro Antioxidant Activity of Synthesized Bac10307-AgNPs
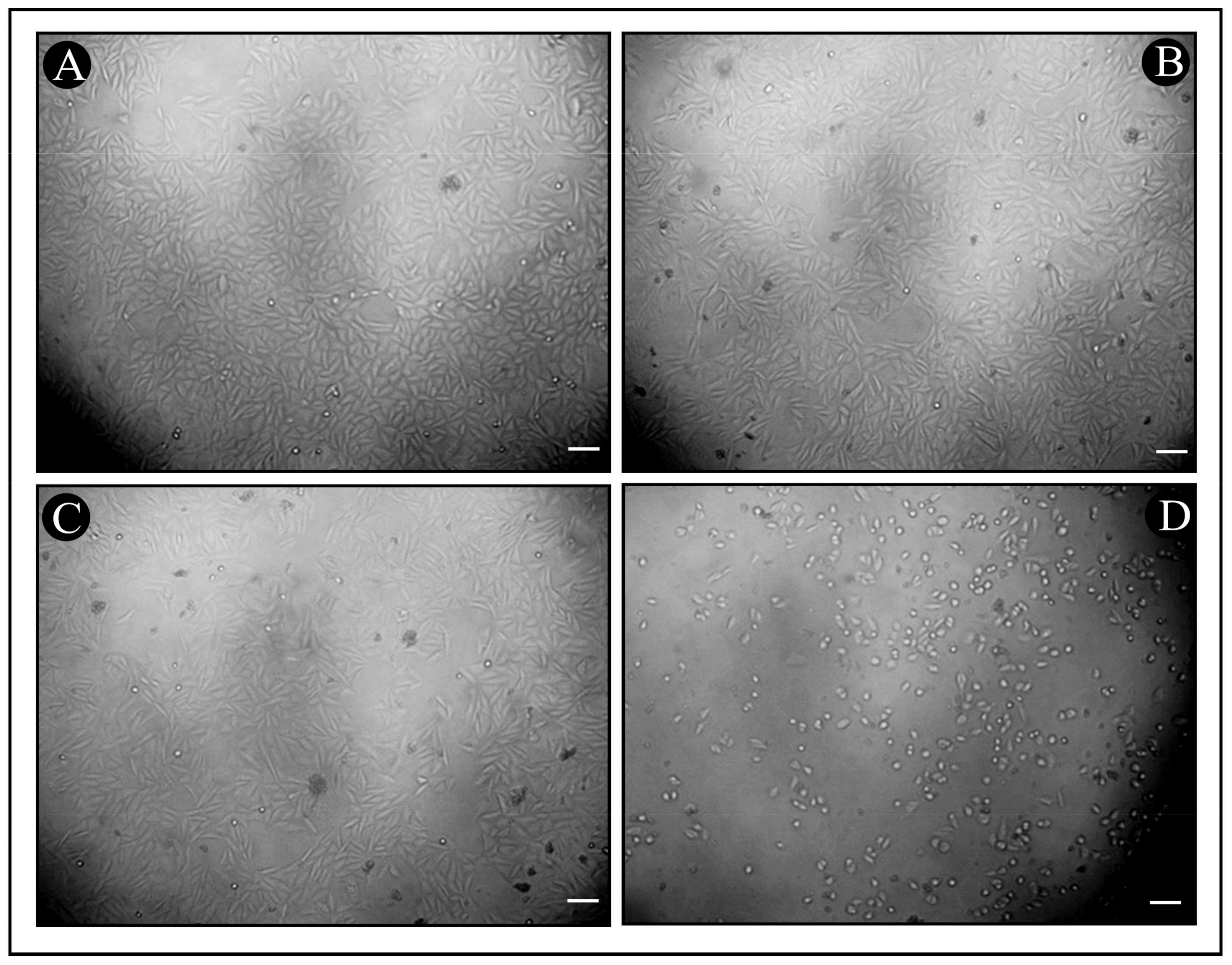
3.6. In Vitro Cytotoxicity of Synthesized Bac10307-AgNPs
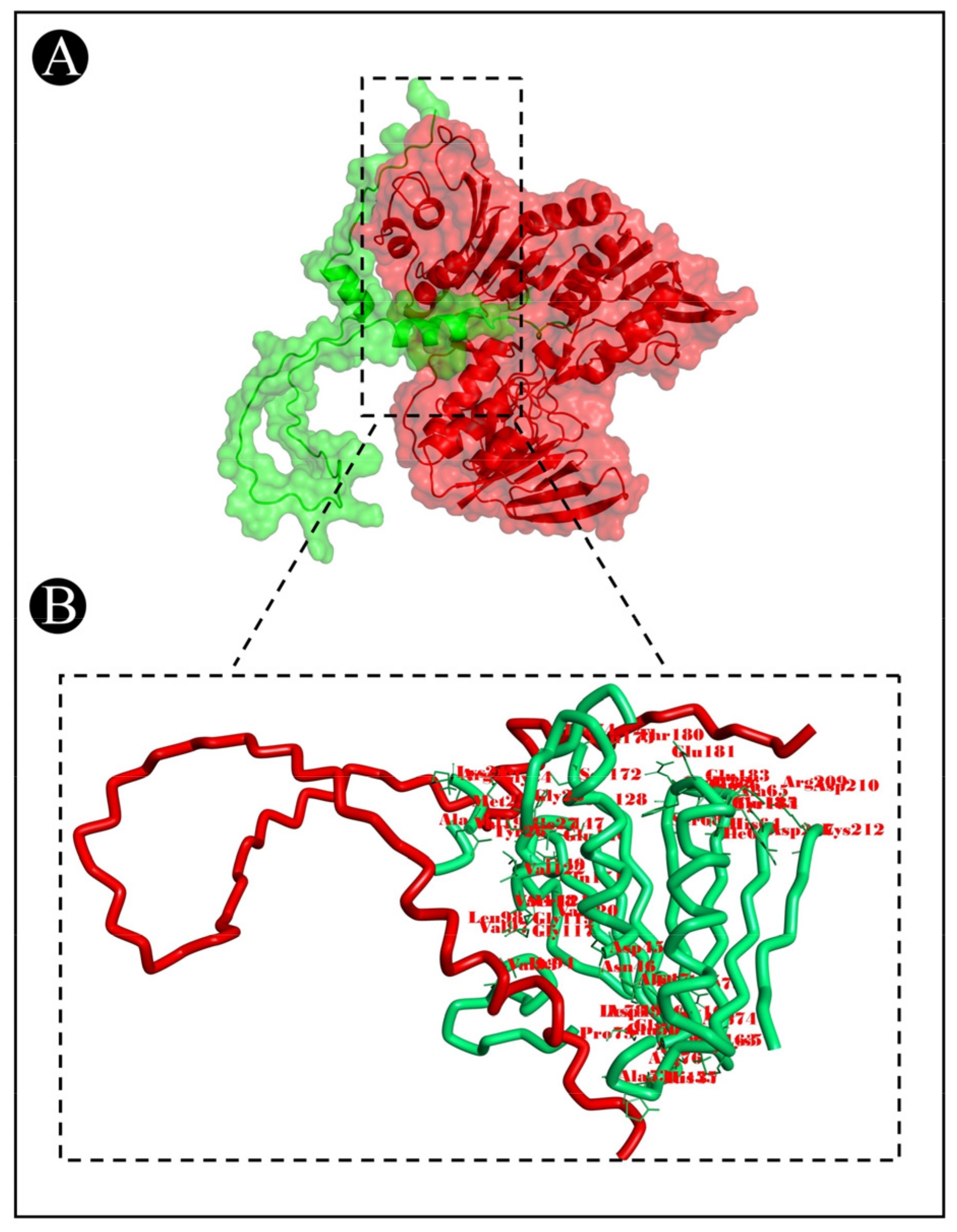
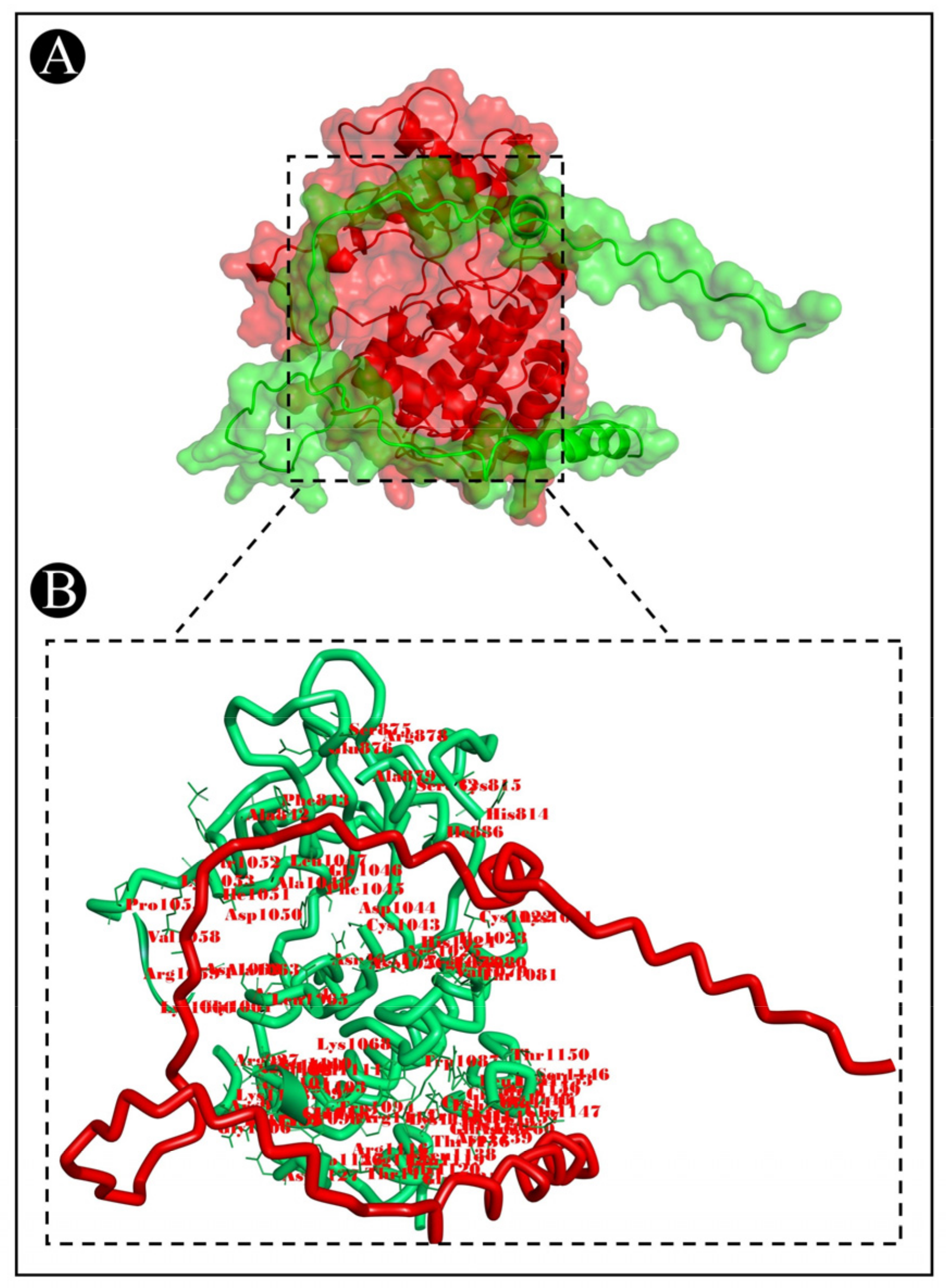
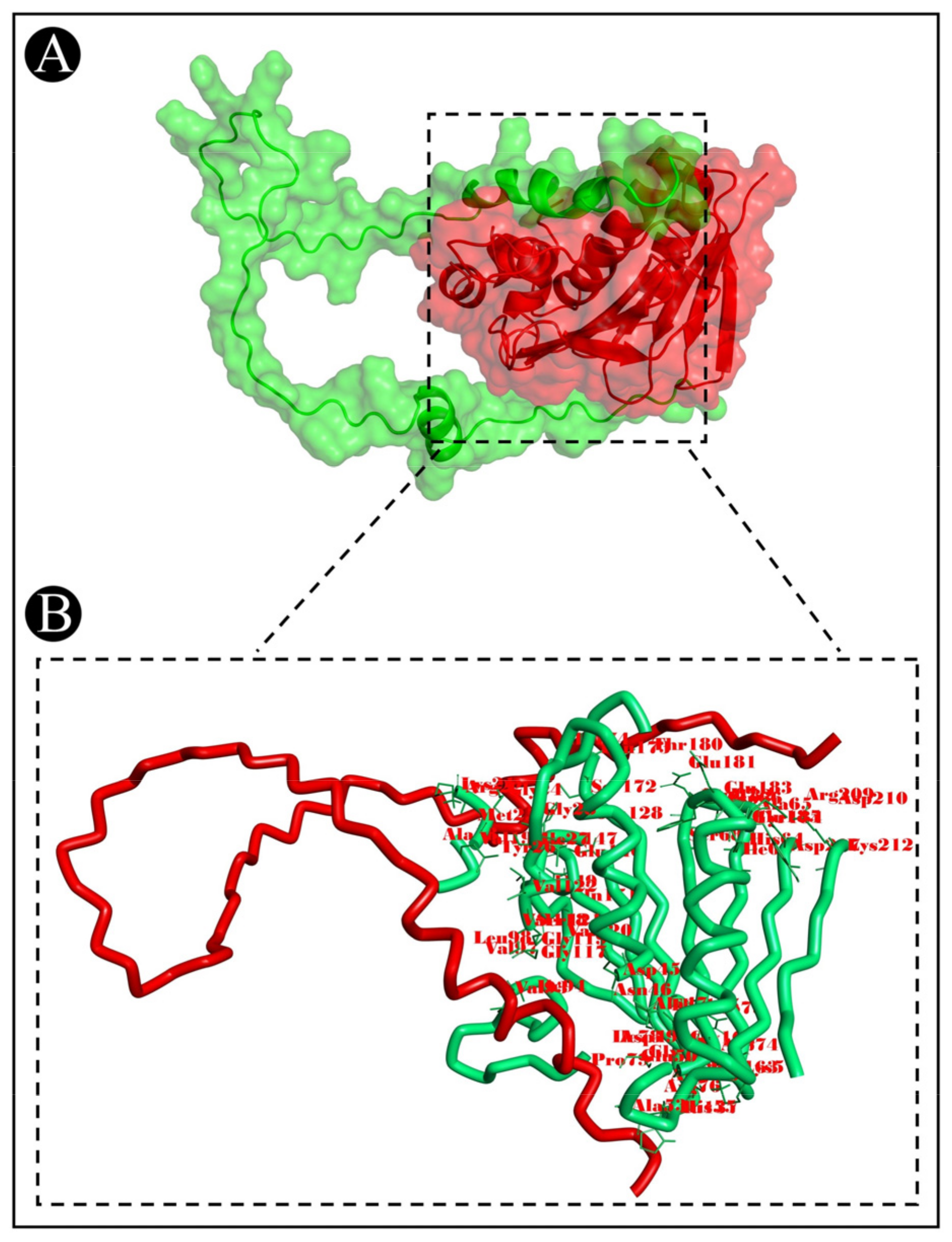
3.7. Molecular Docking Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Irkitova, A.N.; Matsyura, A.V. Ecological and biological characteristics of Lactobacillus acidophilus. Ukr. J. Ecol. 2017, 7, 214–230. [Google Scholar] [CrossRef] [PubMed]
- Villamil, L.; Reyes, C.; Martínez-Silva, M.A. In vivo and in vitro assessment of Lactobacillus acidophilus as probiotic for tilapia (Oreochromis niloticus, Perciformes: Cichlidae) culture improvement. Aquac. Res. 2014, 45, 1116–1125. [Google Scholar] [CrossRef]
- Remes-Troche, J.M.; Coss-Adame, E.; Valdovinos-Díaz, M.; Gómez-Escudero, O.; Icaza-Chávez, M.E.; Chávez-Barrera, J.A.; Zárate-Mondragón, F.; Velasco, J.A.V.-R.; Aceves-Tavares, G.R.; Lira-Pedrín, M.A.; et al. Lactobacillus acidophilus LB: A useful pharmabiotic for the treatment of digestive disorders. Ther. Adv. Gastroenterol. 2020, 13, 1756284820971201. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, C.; Donders, G.G.; Palmeira-De-Oliveira, R.; Queiroz, J.A.; Tomaz, C.; Martinez-De-Oliveira, J. Bacteriocin production of the probiotic Lactobacillus acidophilus KS400. AMB Express 2018, 8, 153. [Google Scholar] [CrossRef]
- Tang, H.W.; Phapugrangkul, P.; Fauzi, H.M.; Tan, J.S. Lactic Acid Bacteria Bacteriocin, an Antimicrobial Peptide Effective Against Multidrug Resistance: A Comprehensive Review. Int. J. Pept. Res. Ther. 2022, 28, 14. [Google Scholar] [CrossRef]
- Silva, S.P.; Ribeiro, S.C.; Teixeira, J.A.; Silva, C.C. Application of an alginate-based edible coating with bacteriocin-producing Lactococcus strains in fresh cheese preservation. LWT 2022, 153, 112486. [Google Scholar] [CrossRef]
- Cotter, P.; Hill, C.; Ross, R. Bacteriocins: Developing innate immunity for food. Nat. Rev. Genet. 2005, 3, 777–788. [Google Scholar] [CrossRef]
- El-Gendy, A.O.; Essam, T.M.; Amin, M.A.; Ahmed, S.H.; Nes, I.F. Clinical Screening for Bacteriocinogenic Enterococcus faecalis Isolated from Intensive Care Unit Inpatient in Egypt. J. Microb. Biochem. Technol. 2013, 4, 161–167. [Google Scholar] [CrossRef]
- Martín, R.; Escobedo, S.; Martín, C.; Crespo, A.; Quiros, L.M.; Suarez, J.E. Chemotherapy, Surface glycosaminoglycans protect eukaryotic cells against membrane-driven peptide bacteriocins. Antimicrob. Agents Chemother. 2015, 59, 677–681. [Google Scholar] [CrossRef]
- Sidhu, P.K.; Nehra, K. Bacteriocin-nanoconjugates as emerging compounds for enhancing antimicrobial activity of bacteriocins. J. King Saud. Univ. 2019, 31, 758–767. [Google Scholar] [CrossRef]
- Ansari, A.; Zohra, R.R.; Tarar, O.M.; Qader, S.A.U.; Aman, A. Screening, purification and characterization of thermostable, protease resistant Bacteriocin active against methicillin resistant Staphylococcus aureus (MRSA). BMC Microbiol. 2018, 18, 192. [Google Scholar] [CrossRef]
- Patil, S.D.; Sharma, R.; Bhattacharyya, T.; Kumar, P.; Gupta, M.; Chaddha, B.S.; Navani, N.K.; Pathania, R. Antibacterial potential of a small peptide from Bacillus sp. RPT-0001 and its capping for green synthesis of silver nanoparticles. J. Microbiol. 2015, 53, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Yang, E.; Fan, L.; Jiang, Y.; Doucette, C.; Fillmore, S. Antimicrobial activity of bacteriocin-producing lactic acid bacteria isolated from cheeses and yogurts. AMB Express 2012, 2, 48. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Siddiqui, A.J.; Hamadou, W.S.; Surti, M.; Awadelkareem, A.M.; Ashraf, S.A.; Alreshidi, M.; Snoussi, M.; Rizvi, S.M.D.; Bardakci, F.; et al. Inhibition of Bacterial Adhesion and Antibiofilm Activities of a Glycolipid Biosurfactant from Lactobacillus rhamnosus with Its Physicochemical and Functional Properties. Antibiotics 2021, 10, 1546. [Google Scholar] [CrossRef]
- Wang, Y.; Shang, N.; Qin, Y.; Zhang, Y.; Zhang, J.; Li, P. The complete genome sequence of Lactobacillus plantarum LPL-1, a novel antibacterial probiotic producing class IIa bacteriocin. J. Biotechnol. 2018, 266, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Elayaraja, S.; Annamalai, N.; Mayavu, P.; Balasubramanian, T. Production, purification and characterization of bacteriocin from Lactobacillus murinus AU06 and its broad antibacterial spectrum. Asian Pac. J. Trop. Biomed. 2014, 4, S305–S311. [Google Scholar] [CrossRef]
- Miao, J.; Guo, H.; Ou, Y.; Liu, G.; Fang, X.; Liao, Z.; Ke, C.; Chen, Y.; Zhao, L.; Cao, Y. Purification and characterization of bacteriocin F1, a novel bacteriocin produced by Lactobacillus paracasei subsp. tolerans FX-6 from Tibetan kefir, a traditional fermented milk from Tibet, China. Food Control 2014, 42, 48–53. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, Y.; Yang, H.; Bu, Y.; Yi, H.; Zhang, L.; Han, X.; Ai, L. Purification and Partial Characterization of Bacteriocin Lac-B23, a Novel Bacteriocin Production by Lactobacillus plantarum J23, Isolated from Chinese Traditional Fermented Milk. Front. Microbiol. 2018, 9, 2165. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.U.; Nayab, H.; Rehman, T.U.; Williamson, M.P.; Haq, K.U.; Shafi, N.; Shafique, F. Characterisation of Bacteriocins Produced by Lactobacillus spp. Isolated from the Traditional Pakistani Yoghurt and Their Antimicrobial Activity against Common Foodborne Pathogens. BioMed Res. Int. 2020, 2020, 8281623. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, P.K.; Nehra, K. Purification and characterization of bacteriocin Bac23 extracted from Lactobacillus plantarum PKLP5 and its interaction with silver nanoparticles for enhanced antimicrobial spectrum against food-borne pathogens. LWT 2021, 139, 110546. [Google Scholar] [CrossRef]
- Adnan, M.; Patel, M.; Reddy, M.N.; Alshammari, E. Formulation, evaluation and bioactive potential of Xylaria primorskensis terpenoid nanoparticles from its major compound xylaranic acid. Sci. Rep. 2018, 8, 1740. [Google Scholar] [CrossRef]
- Awadelkareem, A.M.; Al-Shammari, E.; Elkhalifa, A.O.; Adnan, M.; Siddiqui, A.J.; Patel, M.; Khan, M.I.; Mehmood, K.; Ashfaq, F.; Badraoui, R. Biosynthesized Silver Nanoparticles from Eruca sativa Miller Leaf Extract Exhibits Antibacterial, Antioxidant, Anti-Quorum-Sensing, Antibiofilm, and Anti-Metastatic Activities. Antibiotics 2022, 11, 853. [Google Scholar] [CrossRef]
- Adnan, M.; Siddiqui, A.J.; Ashraf, S.A.; Snoussi, M.; Badraoui, R.; Alreshidi, M.; Elasbali, A.M.; Al-Soud, W.A.; Alharethi, S.H.; Sachidanandan, M. Polyhydroxybutyrate (PHB)-Based Biodegradable Polymer from Agromyces indicus: Enhanced Production, Characterization, and Optimization. Polymers 2022, 14, 3982. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E.W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef]
- Awadelkareem, A.M.; Al-Shammari, E.; Elkhalifa, A.E.O.; Adnan, M.; Siddiqui, A.J.; Snoussi, M.; Khan, M.I.; Azad, Z.R.A.A.; Patel, M.; Ashraf, S.A. Phytochemical and In Silico ADME/Tox Analysis of Eruca sativa Extract with Antioxidant, Antibacterial and Anticancer Potential against Caco-2 and HCT-116 Colorectal Carcinoma Cell Lines. Molecules 2022, 27, 1409. [Google Scholar] [CrossRef]
- Reddy, M.N.; Adnan, M.; Alreshidi, M.M.; Saeed, M.; Patel, M. Evaluation of Anticancer, Antibacterial and Antioxidant Properties of a Medicinally Treasured Fern Tectaria coadunata with its Phytoconstituents Analysis by HR-LCMS. Anti-Cancer Agents Med. Chem. 2020, 20, 1845–1856. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Beglov, D.; Bohnuud, T.; Mottarella, S.E.; Xia, B.; Hall, D.R.; Vajda, S. How good is automated protein docking? Proteins Struct. Funct. Bioinform. 2013, 81, 2159–2166. [Google Scholar] [CrossRef]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Desta, I.T.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and Its Limits in Rigid Body Protein-Protein Docking. Structure 2020, 28, 1071–1081.e3. [Google Scholar] [CrossRef]
- Lagos, R.; Tello, M.; Mercado, G.; García, V.; Monasterio, O. Antibacterial and antitumorigenic properties of microcin E492, a pore-forming bacteriocin. Curr. Pharm. Biotechnol. 2009, 10, 74–85. [Google Scholar] [CrossRef]
- Guzmán-Rodríguez, J.J.; Ochoa-Zarzosa, A.; López-Gómez, R.; López-Meza, J.E. Plant Antimicrobial Peptides as Potential Anticancer Agents. BioMed Res. Int. 2015, 2015, 735087. [Google Scholar] [CrossRef]
- Deslouches, B.; Steckbeck, J.D.; Craigo, J.K.; Doi, Y.; Burns, J.L.; Montelaro, R.C. Engineered Cationic Antimicrobial Peptides To Overcome Multidrug Resistance by ESKAPE Pathogens. Antimicrob. Agents Chemother. 2015, 59, 1329–1333. [Google Scholar] [CrossRef]
- Agrawal, S.; Acharya, D.; Adholeya, A.; Barrow, C.; Deshmukh, S.K. Nonribosomal Peptides from Marine Microbes and Their Antimicrobial and Anticancer Potential. Front. Pharmacol. 2017, 8, 828. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Chen, D.; Wang, L.; Lin, C.; Ma, C.; Xi, X.; Chen, T.; Shaw, C.; Zhou, M. Dermaseptin-PH: A Novel Peptide with Antimicrobial and Anticancer Activities from the Skin Secretion of the South American Orange-Legged Leaf Frog, Pithecopus (Phyllomedusa) hypochondrialis. Molecules 2017, 22, 1805. [Google Scholar] [CrossRef] [PubMed]
- Delves-Brougthon, J. Nisin and its uses as a food preservative. Food Technol. 1990, 44, 100–117. [Google Scholar]
- Bradshaw, J.P. Cationic antimicrobial peptides. BioDrugs 2003, 17, 233–240. [Google Scholar] [CrossRef]
- Schilliager, U.; Geisen, R.; Holzapfel, W.H. Potential of antagonistic microganisms and bacteriocins for the biological preservation of food. Trends Food Sci Tech. 1997, 7, 158–164. [Google Scholar] [CrossRef]
- Davidson, P.M.; Sofos, J.N.; Branen, A.L. Antimicrobials in Food; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Riley, M.A.; Wertz, J.E. Bacteriocins: Evolution, Ecology, and Application. Annu. Rev. Microbiol. 2002, 56, 117–137. [Google Scholar] [CrossRef]
- Saraniya, A.; Jeevaratnam, K. Optimization of nutritional and non-nutritional factors involved for production of antimicrobial compounds from Lactobacillus pentosus SJ65 using response surface methodology. Braz. J. Microbiol. 2014, 45, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Salmaso, S.; Elvassore, N.; Bertucco, A.; Lante, A.; Caliceti, P. Nisin-loaded poly-l-lactide nano-particles produced by CO2 anti-solvent precipitation for sustained antimicrobial activity. Int. J. Pharm. 2004, 287, 163–173. [Google Scholar] [CrossRef]
- De Giani, A.; Bovio, F.; Forcella, M.; Fusi, P.; Sello, G.; Di Gennaro, P. Identification of a bacteriocin-like compound from Lactobacillus plantarum with antimicrobial activity and effects on normal and cancerogenic human intestinal cells. AMB Express 2019, 9, 88. [Google Scholar] [CrossRef] [PubMed]
- Danilova, T.A.; Adzhieva, A.A.; Danilina, G.A.; Polyakov, N.B.; Soloviev, A.I.; Zhukhovitsky, V.G. Antimicrobial Activity of Supernatant of Lactobacillus plantarum against Pathogenic Microorganisms. Bull. Exp. Biol. Med. 2019, 167, 751–754. [Google Scholar] [CrossRef]
- Abo-Amer, A.E. Characterization of a bacteriocin-like inhibitory substance produced by Lactobacillus plantarum isolated from Egyptian home-made yogurt. Sci. Asia. 2007, 33, 313–319. [Google Scholar] [CrossRef]
- Djadouni, F.; Kihal, M. Characterization and determination of the factors affecting anti-listerial bacteriocins from Lactobacillus plantarum and Pediococcus pentosaceus isolated from dairy milk products. Afr. J. Food Sci. 2013, 7, 35–44. [Google Scholar]
- Moghaddam, M.Z.; Sattari, M.; Mobarez, A.M.; Doctorzadeh, F. Inhibitory effect of yogurt Lactobacilli bacteriocins on growth and verotoxins production of enterohemorrhgic Escherichia coli O157: H7. Pak. J. Biol. Sci. 2006, 9, 2112–2116. [Google Scholar] [CrossRef]
- Milioni, C.; Martínez, B.; Degl’Innocenti, S.; Turchi, B.; Fratini, F.; Cerri, D.; Fischetti, R. A novel bacteriocin produced by Lactobacillus plantarum LpU4 as a valuable candidate for biopreservation in artisanal raw milk cheese. Dairy Sci. Technol. 2015, 95, 479–494. [Google Scholar] [CrossRef]
- Song, J.Y.; Kim, B.S. Rapid biological synthesis of silver nanoparticles using plant leaf extracts. Bioproc. Biosyst. Eng. 2009, 32, 79–84. [Google Scholar] [CrossRef]
- Hata, T.; Tanaka, R.; Ohmomo, S. Isolation and characterization of plantaricin ASM1: A new bacteriocin produced by Lactobacillus plantarum A-1. Int. J. Food Microbiol. 2010, 137, 94–99. [Google Scholar] [CrossRef]
- Atrih, A.; Rekhif, N.; Moir, A.; Lebrihi, A.; Lefebvre, G. Mode of action, purification and amino acid sequence of plantaricin C19, an anti-Listeria bacteriocin produced by Lactobacillus plantarum C19. Int. J. Food Microbiol. 2001, 68, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Zhao, H.; Zhang, C.; Yu, J.; Lu, Z. Purification and Characterization of Plantaricin 163, a Novel Bacteriocin Produced by Lactobacillus plantarum 163 Isolated from Traditional Chinese Fermented Vegetables. J. Agric. Food Chem. 2013, 61, 11676–11682. [Google Scholar] [CrossRef]
- Chen, Y.-S.; Wang, Y.-C.; Chow, Y.-S.; Yanagida, F.; Liao, C.-C.; Chiu, C.-M. Purification and characterization of plantaricin Y, a novel bacteriocin produced by Lactobacillus plantarum 510. Arch. Microbiol. 2014, 196, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qin, Y.; Zhang, Y.; Wu, R.; Li, P. Antibacterial mechanism of plantaricin LPL-1, a novel class IIa bacteriocin against Listeria monocytogenes. Food Control 2019, 97, 87–93. [Google Scholar] [CrossRef]
- Mills, S.; Ross, R.; Hill, C. Bacteriocins and bacteriophage; a narrow-minded approach to food and gut microbiology. FEMS Microbiol. Rev. 2017, 41, S129–S153. [Google Scholar] [CrossRef] [PubMed]
- Lazzari, S.; Moscatelli, D.; Codari, F.; Salmona, M.; Morbidelli, M.; Diomede, L. Colloidal stability of polymeric nanoparticles in biological fluids. J. Nanoparticle Res. 2012, 14, 920. [Google Scholar] [CrossRef]
- Niaz, T.; Shabbir, S.; Noor, T.; Imran, M. Antimicrobial and antibiofilm potential of bacteriocin loaded nano-vesicles functionalized with rhamnolipids against foodborne pathogens. LWT 2019, 116, 108583. [Google Scholar] [CrossRef]
- Kumar, C.G.; Mamidyala, S.K. Extracellular synthesis of silver nanoparticles using culture supernatant of Pseudomonas aeruginosa. Colloids Surf. B Biointerfaces 2011, 84, 462–466. [Google Scholar] [CrossRef]
- Saravana, K.; Annalakshmi, A. Enhancing the antimicrobial activity of nisin by encapsulating on silver nanoparticle synthesized by Bacillus sp. Int. J. Pharma. Biol. Arch. 2012, 3, 406–410. [Google Scholar]
- Gomaa, E.Z. Synergistic Antibacterial Efficiency of Bacteriocin and Silver Nanoparticles Produced by Probiotic Lactobacillus paracasei Against Multidrug Resistant Bacteria. Int. J. Pept. Res. Ther. 2019, 25, 1113–1125. [Google Scholar] [CrossRef]
- Thirumurugan, A.; Ramachandran, S.; Gowri, A.S. Combined effect of bacteriocin with gold nanoparticles against food spoiling bacteria-an approach for food packaging material preparation. Int. Food Res. J. 2013, 20, 1909–1912. [Google Scholar]
- Singh, A.K.; Bai, X.; Amalaradjou, M.A.R.; Bhunia, A.K. Antilisterial and Antibiofilm Activities of Pediocin and LAP Functionalized Gold Nanoparticles. Front. Sustain. Food Syst. 2018, 2, 74. [Google Scholar] [CrossRef]
- Morales-Avila, E.; Ferro-Flores, G.; Ocampo-García, B.E.; López-Téllez, G.; López-Ortega, J.; Rogel-Ayala, D.G.; Sánchez-Padilla, D. Antibacterial Efficacy of Gold and Silver Nanoparticles Functionalized with the Ubiquicidin (29–41) Antimicrobial Peptide. J. Nanomater. 2017, 2017, 5831959. [Google Scholar] [CrossRef]
- Elkhalifa, A.E.O.; Al-Shammari, E.; Alam, M.J.; Alcantara, J.C.; Khan, M.A.; Eltoum, N.E.; Ashraf, S.A. Okra-derived dietary Carotenoid lutein against Breast Cancer, with an Approach towards Developing a Nutraceutical Product: A Meta-analysis Study. J. Pharm. Res. Int. 2021, 33, 135–142. [Google Scholar] [CrossRef]
- Cao, W.; Chen, H.-D.; Yu, Y.-W.; Li, N.; Chen, W.-Q. Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020. Chin. Med. J. 2021, 134, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-C.; Lin, H.-H.; Hsu, C.-H.; Wang, C.-J.; Chiang, T.-A.; Chen, J.-H. Inhibitory effects of andrographolide on migration and invasion in human non-small cell lung cancer A549 cells via down-regulation of PI3K/Akt signaling pathway. Eur. J. Pharmacol. 2010, 632, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Loza-Mejía, M.A.; Salazar, J.R.; Sánchez-Tejeda, J.F. In Silico Studies on Compounds Derived from Calceolaria: Phenylethanoid Glycosides as Potential Multitarget Inhibitors for the Development of Pesticides. Biomolecules 2018, 8, 121. [Google Scholar] [CrossRef]
- Lee, K.; Kim, D. In-Silico Molecular Binding Prediction for Human Drug Targets Using Deep Neural Multi-Task Learning. Genes 2019, 10, 906. [Google Scholar] [CrossRef]
- Bharathi, A.; Roopan, S.M.; Vasavi, C.S.; Munusami, P.; Gayathri, G.A.; Gayathri, M. In silico molecular docking and in vitro antidiabetic studies of dihydropyrimido [4,5-a] acridin-2-amines. BioMed Res. Int. 2014, 2014, 971569. [Google Scholar] [CrossRef]
- Comeau, S.R.; Gatchell, D.W.; Vajda, S.; Camacho, C.J. ClusPro: A fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004, 32, W96–W99. [Google Scholar] [CrossRef]
- Xue, L.C.; Dobbs, D.; Honavar, V. HomPPI: A class of sequence homology based protein-protein interface prediction methods. BMC Bioinform. 2011, 12, 244. [Google Scholar] [CrossRef]
- Hao, T.; Peng, W.; Wang, Q.; Wang, B.; Sun, J. Reconstruction and Application of Protein–Protein Interaction Network. Int. J. Mol. Sci. 2016, 17, 907. [Google Scholar] [CrossRef]
- Alfonso, E.E.; Deng, Z.; Boaretto, D.; Hood, B.L.; Vasile, S.; Smith, L.H.; Chambers, J.W.; Chapagain, P.; Leng, F. Novel and Structurally Diversified Bacterial DNA Gyrase Inhibitors Discovered through a Fluorescence-Based High-Throughput Screening Assay. ACS Pharmacol. Transl. Sci. 2022, 5, 932–944. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.-A.; Arnold, R.S.; Lassegue, B.; Shi, J.; Xu, X.X.; Sorescu, D.; Chung, A.B.; Griendling, K.K.; Lambeth, J.D. Cell transformation by the superoxide-generating oxidase Mox1. Nat. Cell Biol. 1999, 401, 79–82. [Google Scholar] [CrossRef]
- Dupuy, C.; Ohayon, R.; Valent, A.; Noël-Hudson, M.-S.; Dème, D.; Virion, A. Purification of a Novel Flavoprotein Involved in the Thyroid NADPH Oxidase. J. Biol. Chem. 1999, 274, 37265–37269. [Google Scholar] [CrossRef]
- Bedard, K.; Krause, K.-H. The NOX Family of ROS-Generating NADPH Oxidases: Physiology and Pathophysiology. Physiol. Rev. 2007, 87, 245–313. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.-M.; Zhou, H.; Hong, J.-S. NADPH oxidases: Novel therapeutic targets for neurodegenerative diseases. Trends Pharmacol. Sci. 2012, 33, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Gordon, E.; Claesson-Welsh, L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 2016, 17, 611–625. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, Y.; Leung, L.-H.; Liu, L.; Yang, F.; Yao, X. Efficacy and safety of angiogenesis inhibitors in advanced gastric cancer: A systematic review and meta-analysis. J. Hematol. Oncol. 2016, 9, 111. [Google Scholar] [CrossRef]
| Test Organisms | Zone of Inhibition (mm) | MIC (μg/mL) Bac10307-AgNPs | MBC (μg/mL) Bac10307-AgNPs | ||
|---|---|---|---|---|---|
| Bac10307 | AgNPs | Bac10307- AgNPs | |||
| S. aureus | 19.66 | 11.66 | 25.33 | 8 | 16 |
| P. aeruginosa | 17.33 | 10.33 | 23.00 | 8 | 16 |
| B. subtilis | 14.66 | 8.33 | 19.66 | 16 | 32 |
| E. coli | 11.66 | 7.66 | 17.00 | 64 | 128 |
| Proteins | Receptor-Ligand | Interection Type | Distance |
|---|---|---|---|
| 2CDU-Acidocin A | A:LYS78:HZ2–A:ASP282:OD2 | Salt Bridge;Attractive Charge | 1.76041 |
| A:LYS78:HZ3–A:ASP282:OD1 | Salt Bridge;Attractive Charge | 1.75112 | |
| A:CYS133:HN–A:HIS81:NE2 | Conventional Hydrogen Bond | 2.43145 | |
| A:ILE160:HN–A:PHE80:O | Conventional Hydrogen Bond | 2.31372 | |
| A:GLY244:HN–A:HIS81:O | Conventional Hydrogen Bond | 1.88916 | |
| A:ALA300:HN–A:PHE76:O | Conventional Hydrogen Bond | 2.81082 | |
| A:THR301:HN–A:GLY74:O | Conventional Hydrogen Bond | 2.55908 | |
| A:ARG305:HH11–A:TRP71:O | Conventional Hydrogen Bond | 1.81072 | |
| A:ARG305:HH21–A:TRP71:O | Conventional Hydrogen Bond | 1.84157 | |
| A:ARG431:HH12–A:SER3:O | Conventional Hydrogen Bond | 2.70789 | |
| A:PHE433:HN–A:ILE5:O | Conventional Hydrogen Bond | 2.04607 | |
| A:SER3:HN–A:GLU366:OE1 | Conventional Hydrogen Bond | 2.29836 | |
| A:SER7:HG–A:PRO432:O | Conventional Hydrogen Bond | 1.84543 | |
| A:GLN9:HE22–A:ASP422:OD1 | Conventional Hydrogen Bond | 2.12445 | |
| A:THR79:HN–A:PRO298:O | Conventional Hydrogen Bond | 2.73643 | |
| A:THR79:HG1–A:PRO298:O | Conventional Hydrogen Bond | 1.85808 | |
| A:SER41:CB–A:GLY77:O | Conventional Hydrogen Bond | 3.70135 | |
| A:ARG431:CD–A:SER3:O | Carbon Hydrogen Bond | 3.15632 | |
| A:HIS81:CE1–A:LEU241:O | Carbon Hydrogen Bond | 3.13598 | |
| A:LYS187:NZ–A:PHE80 | Pi-Cation | 3.51826 | |
| A:ARG305:NH1–A:TRP71 | Pi-Cation | 4.77402 | |
| A:SER7:CA–A:PHE433 | Pi-Sigma | 3.67277 | |
| A:ILE243:C,O;GLY244:N–A:PHE80 | Amide-Pi Stacked | 3.93853 | |
| A:ALA300–A:LYS78 | Alkyl | 4.9093 | |
| A:ILE438–A:ILE5 | Alkyl | 4.68217 | |
| A:LYS10–A:MET420 | Alkyl | 4.61562 | |
| A:PHE367–A:ILE2 | Pi-Alkyl | 5.07639 | |
| A:TRP71–A:VAL304 | Pi-Alkyl | 5.08256 | |
| A:TRP71–A:ARG305 | Pi-Alkyl | 4.70863 | |
| A:TRP71–A:ARG305 | Pi-Alkyl | 4.06273 | |
| A:TRP71–A:ARG308 | Pi-Alkyl | 5.17719 | |
| A:PHE76–A:LEU330 | Pi-Alkyl | 4.80412 | |
| A:HIS81–A:PRO117 | Pi-Alkyl | 4.38714 | |
| A:HIS81–A:LEU132 | Pi-Alkyl | 5.41817 | |
| 6f86-Acidocin A | A:MET1:N–A:ASP210:OD2 | Attractive Charge | 5.4936 |
| A:ARG22:HH11–A:LEU59:O | Conventional Hydrogen Bond | 1.77015 | |
| A:ARG22:HH21–A:SER58:OG | Conventional Hydrogen Bond | 1.72382 | |
| A:TYR26:HH–A:ASP64:OD2 | Conventional Hydrogen Bond | 1.97086 | |
| A:ASN46:HD22–A:ALA72:O | Conventional Hydrogen Bond | 1.98784 | |
| A:ASN46:HD22–A:THR73:O | Conventional Hydrogen Bond | 2.63274 | |
| A:ARG76:HH12–A:GLY77:O | Conventional Hydrogen Bond | 1.8161 | |
| A:ARG136:HH11–A:PHE76:O | Conventional Hydrogen Bond | 2.53417 | |
| A:ARG136:HH11–A:LYS78:O | Conventional Hydrogen Bond | 2.17681 | |
| A:ARG136:HH21–A:LYS78:O | Conventional Hydrogen Bond | 1.72262 | |
| A:THR180:HG1–A:SER7:OG | Conventional Hydrogen Bond | 1.87972 | |
| A:SER3:HN–A:ARG209:O | Conventional Hydrogen Bond | 2.36379 | |
| A:SER3:HG–A:ARG209:O | Conventional Hydrogen Bond | 1.86756 | |
| A:GLN9:HE21–A:GLU174:OE2 | Conventional Hydrogen Bond | 2.01259 | |
| A:LYS10:HN–A:GLN128:OE1 | Conventional Hydrogen Bond | 2.17849 | |
| A:ALA18:HN–A:ASP14:O | Conventional Hydrogen Bond | 1.93332 | |
| A:ALA18:HN–A:LYS15:O | Conventional Hydrogen Bond | 2.65481 | |
| A:SER21:HN–A:ALA18:O | Conventional Hydrogen Bond | 2.35624 | |
| A:GLY23:HN–A:ALA18:O | Conventional Hydrogen Bond | 1.89654 | |
| A:LYS24:HZ3–A:TYR26:OH | Conventional Hydrogen Bond | 1.63416 | |
| A:TYR26:HN–A:LYS24:O | Conventional Hydrogen Bond | 2.39531 | |
| A:LYS62:HZ1–A:TYR26:O | Conventional Hydrogen Bond | 2.61388 | |
| A:LYS62:HZ2–A:TYR26:O | Conventional Hydrogen Bond | 2.68718 | |
| A:LYS62:HZ3–A:MET25:O | Conventional Hydrogen Bond | 1.66846 | |
| A:LEU68:HN–A:LEU98:O | Conventional Hydrogen Bond | 2.64427 | |
| A:THR73:HG1–A:ASP49:OD1 | Conventional Hydrogen Bond | 1.98276 | |
| A:THR79:HG1–A:ARG76:O | Conventional Hydrogen Bond | 1.81635 | |
| A:ARG76:CD–A:GLY77:O | Carbon Hydrogen Bond | 3.35609 | |
| A:MET4:CA–A:GLU181:OE2 | Carbon Hydrogen Bond | 3.26235 | |
| A:TRP71:CD1–A:VAL97:O | Carbon Hydrogen Bond | 3.75115 | |
| A:GLU50:OE1–A:PHE76 | Pi-Anion | 3.4452 | |
| A:ALA18–A:ILE61 | Alkyl | 4.4781 | |
| A:VAL118–A:LEU65 | Alkyl | 4.41069 | |
| A:LYS62–A:MET25 | Alkyl | 5.37497 | |
| A:ALA72–A:VAL120 | Alkyl | 5.07682 | |
| A:ALA75–A:PRO79 | Alkyl | 5.06784 | |
| A:ALA75–A:ILE94 | Alkyl | 4.19361 | |
| A:TYR26–A:LYS62 | Pi-Alkyl | 4.48685 | |
| A:TYR26–A:LYS24 | Pi-Alkyl | 4.99044 | |
| A:TRP71–A:VAL97 | Pi-Alkyl | 4.62974 | |
| A:TRP71–A:VAL97 | Pi-Alkyl | 5.0319 | |
| A:PHE76–A:ILE78 | Pi-Alkyl | 5.21439 | |
| A:PHE80–A:ARG76 | Pi-Alkyl | 4.69087 | |
| 2OH4-Acidocin A | A:ARG1078:HH11–A:ASP14:OD1 | Salt Bridge;Attractive Charge | 1.77815 |
| A:ARG1122:HH22–A:ASP64:OD2 | Salt Bridge;Attractive Charge | 1.82447 | |
| A:LYS24:HZ1–A:ASP1044:OD1 | Salt Bridge;Attractive Charge | 2.0004 | |
| A:LYS24:HZ3–A:ASP1044:OD1 | Salt Bridge;Attractive Charge | 2.03938 | |
| A:ARG55:HH21–A:GLU1112:OE2 | Salt Bridge;Attractive Charge | 1.89818 | |
| A:LYS56:HZ1–A:GLU1111:OE1 | Salt Bridge;Attractive Charge | 1.69962 | |
| A:LYS56:HZ3–A:GLU1111:OE2 | Salt Bridge;Attractive Charge | 1.80997 | |
| A:LYS78:HZ3–A:GLU1156:OE2 | Salt Bridge;Attractive Charge | 1.71364 | |
| A:ARG1078:NH2–A:ASP14:OD2 | Attractive Charge | 2.75179 | |
| A:ARG1122:NH1–A:ASP64:OD1 | Attractive Charge | 4.71658 | |
| A:ARG55:NH1–A:ASP1110:OD2 | Attractive Charge | 5.55384 | |
| A:ARG55:NH1–A:GLU1112:OE1 | Attractive Charge | 2.72487 | |
| A:ARG878:HH11–A:TYR27:OH | Conventional Hydrogen Bond | 1.773 | |
| A:ARG927:HH11–A:SER38:O | Conventional Hydrogen Bond | 3.0835 | |
| A:ARG927:HH21–A:SER38:O | Conventional Hydrogen Bond | 1.74776 | |
| A:ILE1023:HN–A:SER21:O | Conventional Hydrogen Bond | 1.98064 | |
| A:ARG1025:HH11–A:SER21:O | Conventional Hydrogen Bond | 1.79986 | |
| A:ARG1025:HH21–A:SER21:OG | Conventional Hydrogen Bond | 1.96334 | |
| A:ARG1025:HH21–A:GLY22:O | Conventional Hydrogen Bond | 2.07187 | |
| A:LYS1060:HZ2–A:LYS36:O | Conventional Hydrogen Bond | 1.63582 | |
| A:GLY1061:HN–A:THR35:O | Conventional Hydrogen Bond | 2.45869 | |
| A:ALA1063:HN–A:THR35:OG1 | Conventional Hydrogen Bond | 2.01064 | |
| A:LYS1108:HZ1–A:ILE49:O | Conventional Hydrogen Bond | 2.21003 | |
| A:LYS1108:HZ3–A:GLY51:O | Conventional Hydrogen Bond | 1.71447 | |
| A:ARG1115:HH12–A:LYS56:O | Conventional Hydrogen Bond | 1.81567 | |
| A:ARG1116:HH11–A:GLN57:OE1 | Conventional Hydrogen Bond | 1.89055 | |
| A:ARG1116:HH21–A:GLN57:OE1 | Conventional Hydrogen Bond | 1.9005 | |
| A:ARG1116:HH22–A:SER58:O | Conventional Hydrogen Bond | 2.01369 | |
| A:ARG1122:HE–A:ASP64:OD1 | Conventional Hydrogen Bond | 2.45807 | |
| A:GLY1143:HN–A:ASP64:OD2 | Conventional Hydrogen Bond | 2.04712 | |
| A:LYS24:HZ1–A:HIS1024:NE2 | Conventional Hydrogen Bond | 2.30731 | |
| A:LYS24:HZ2–A:HIS1024:O | Conventional Hydrogen Bond | 1.64274 | |
| A:HIS33:HN–A:ASP1050:O | Conventional Hydrogen Bond | 2.89181 | |
| A:THR35:HG1–A:ARG1059:O | Conventional Hydrogen Bond | 2.54504 | |
| A:LYS36:HZ1–A:VAL1107:O | Conventional Hydrogen Bond | 1.69429 | |
| A:ARG55:HN–A:GLU1112:OE1 | Conventional Hydrogen Bond | 2.65589 | |
| A:LYS56:HN–A:GLU1112:OE1 | Conventional Hydrogen Bond | 2.47201 | |
| A:GLN57:HN–A:GLU1112:OE1 | Conventional Hydrogen Bond | 1.99761 | |
| A:SER58:HG–A:GLU1119:OE2 | Conventional Hydrogen Bond | 1.94121 | |
| A:LEU59:HN–A:GLU1119:OE1 | Conventional Hydrogen Bond | 2.04219 | |
| A:ILE61:HN–A:GLU1119:O | Conventional Hydrogen Bond | 1.97082 | |
| A:GLN63:HE21–A:GLY1120:O | Conventional Hydrogen Bond | 2.03591 | |
| A:LEU68:HN–A:ASP1139:OD2 | Conventional Hydrogen Bond | 2.48216 | |
| A:LYS78:HZ1–A:GLN1163:OE1 | Conventional Hydrogen Bond | 1.98774 | |
| A:LYS78:HZ2–A:ASN1160:OD1 | Conventional Hydrogen Bond | 1.69053 | |
| A:SER58:CA–A:GLU1119:OE1 | Carbon Hydrogen Bond | 2.9455 | |
| A:LYS24:NZ–A:HIS1024 | Pi-Cation | 3.6145 | |
| A:ASP64:OD2–A:HIS1142 | Pi-Anion | 4.79084 | |
| A:VAL1058:CG1–A:HIS33 | Pi-Sigma | 3.82332 | |
| A:LYS1021–A:ILE20 | Alkyl | 4.85838 | |
| A:ALA1101–A:LEU39 | Alkyl | 4.05593 | |
| A:PRO1149–A:LEU68 | Alkyl | 5.33682 | |
| A:HIS1142–A:LEU65 | Pi-Alkyl | 5.30386 | |
| A:TYR27–A:ALA879 | Pi-Alkyl | 4.63121 | |
| A:TRP40–A:PRO1105 | Pi-Alkyl | 4.91592 | |
| A:TRP71–A:PRO1149 | Pi-Alkyl | 4.42062 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siddiqui, A.J.; Patel, M.; Adnan, M.; Jahan, S.; Saxena, J.; Alshahrani, M.M.; Abdelgadir, A.; Bardakci, F.; Sachidanandan, M.; Badraoui, R.; et al. Bacteriocin-Nanoconjugates (Bac10307-AgNPs) Biosynthesized from Lactobacillus acidophilus-Derived Bacteriocins Exhibit Enhanced and Promising Biological Activities. Pharmaceutics 2023, 15, 403. https://doi.org/10.3390/pharmaceutics15020403
Siddiqui AJ, Patel M, Adnan M, Jahan S, Saxena J, Alshahrani MM, Abdelgadir A, Bardakci F, Sachidanandan M, Badraoui R, et al. Bacteriocin-Nanoconjugates (Bac10307-AgNPs) Biosynthesized from Lactobacillus acidophilus-Derived Bacteriocins Exhibit Enhanced and Promising Biological Activities. Pharmaceutics. 2023; 15(2):403. https://doi.org/10.3390/pharmaceutics15020403
Chicago/Turabian StyleSiddiqui, Arif Jamal, Mitesh Patel, Mohd Adnan, Sadaf Jahan, Juhi Saxena, Mohammed Merae Alshahrani, Abdelmushin Abdelgadir, Fevzi Bardakci, Manojkumar Sachidanandan, Riadh Badraoui, and et al. 2023. "Bacteriocin-Nanoconjugates (Bac10307-AgNPs) Biosynthesized from Lactobacillus acidophilus-Derived Bacteriocins Exhibit Enhanced and Promising Biological Activities" Pharmaceutics 15, no. 2: 403. https://doi.org/10.3390/pharmaceutics15020403
APA StyleSiddiqui, A. J., Patel, M., Adnan, M., Jahan, S., Saxena, J., Alshahrani, M. M., Abdelgadir, A., Bardakci, F., Sachidanandan, M., Badraoui, R., Snoussi, M., & Ouhtit, A. (2023). Bacteriocin-Nanoconjugates (Bac10307-AgNPs) Biosynthesized from Lactobacillus acidophilus-Derived Bacteriocins Exhibit Enhanced and Promising Biological Activities. Pharmaceutics, 15(2), 403. https://doi.org/10.3390/pharmaceutics15020403













