Discovering Glioma Tissue through Its Biomarkers’ Detection in Blood by Raman Spectroscopy and Machine Learning
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Raman Spectroscopy
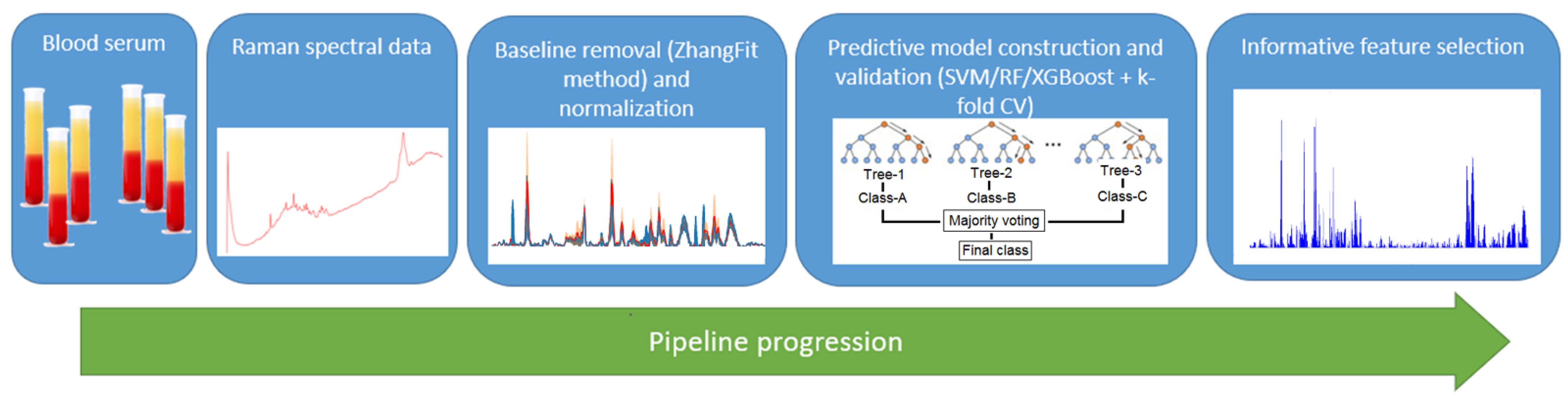
2.3. Machine Learning Pipeline
3. Results
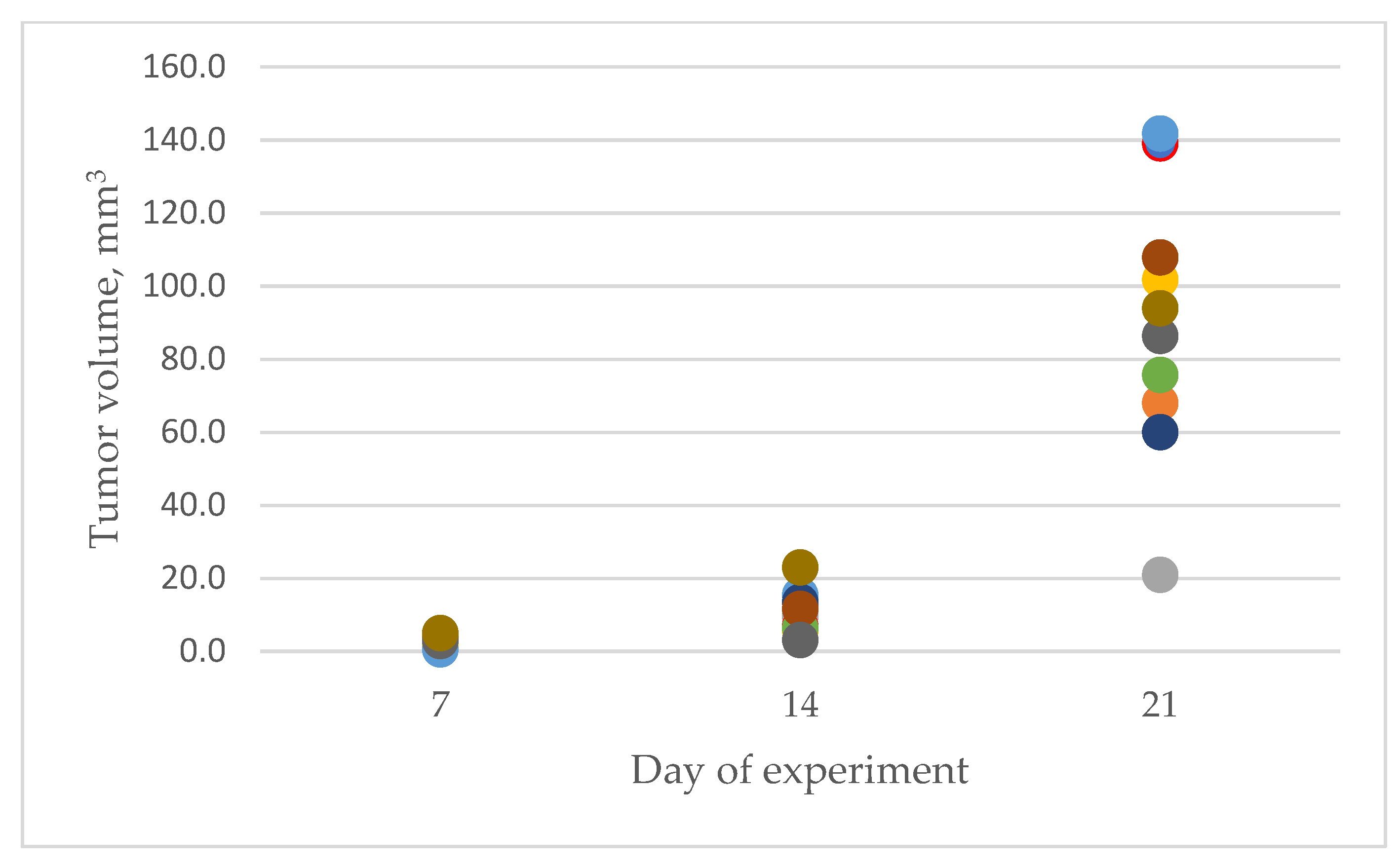
3.1. Characteristics of Experimental Animals
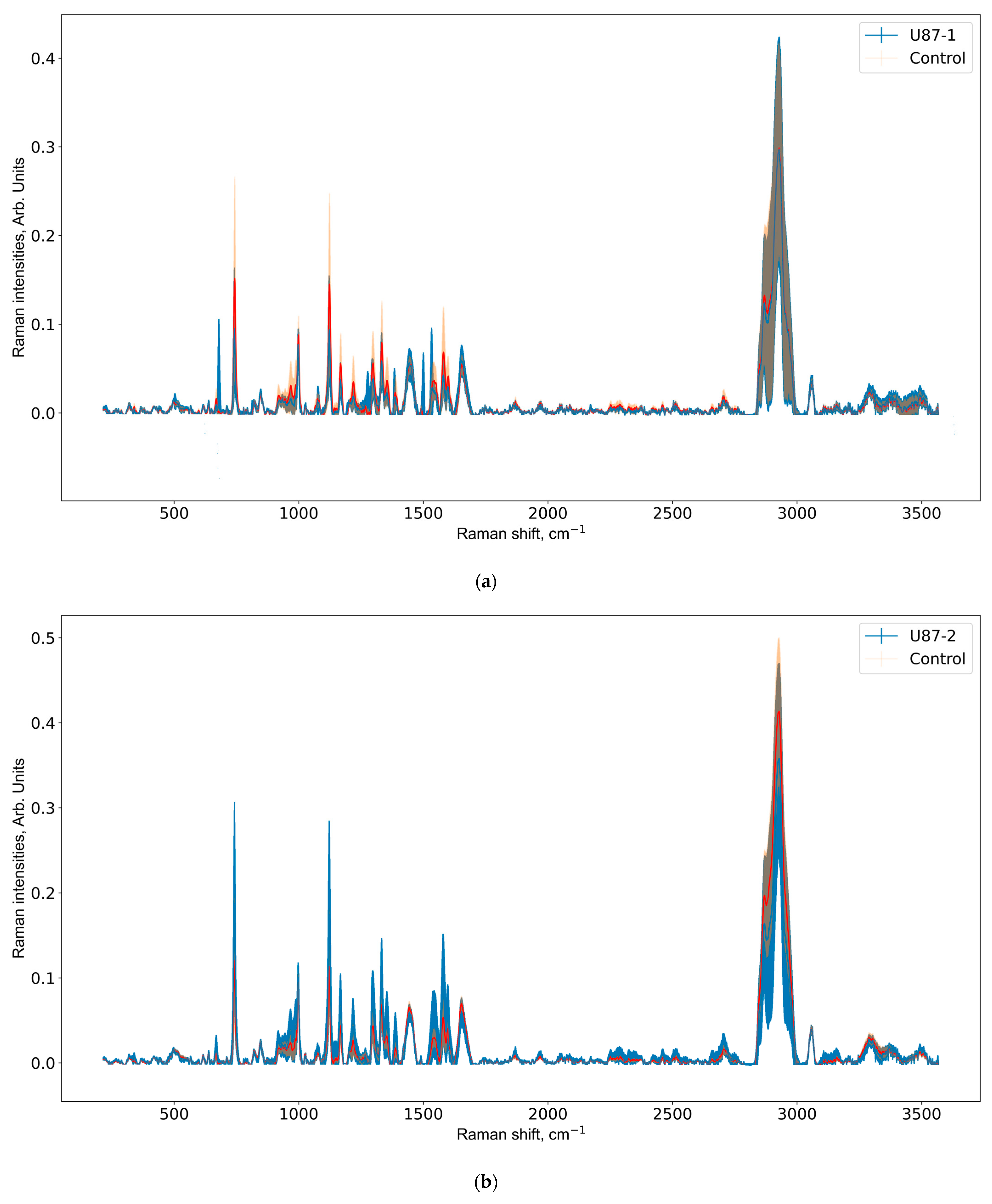
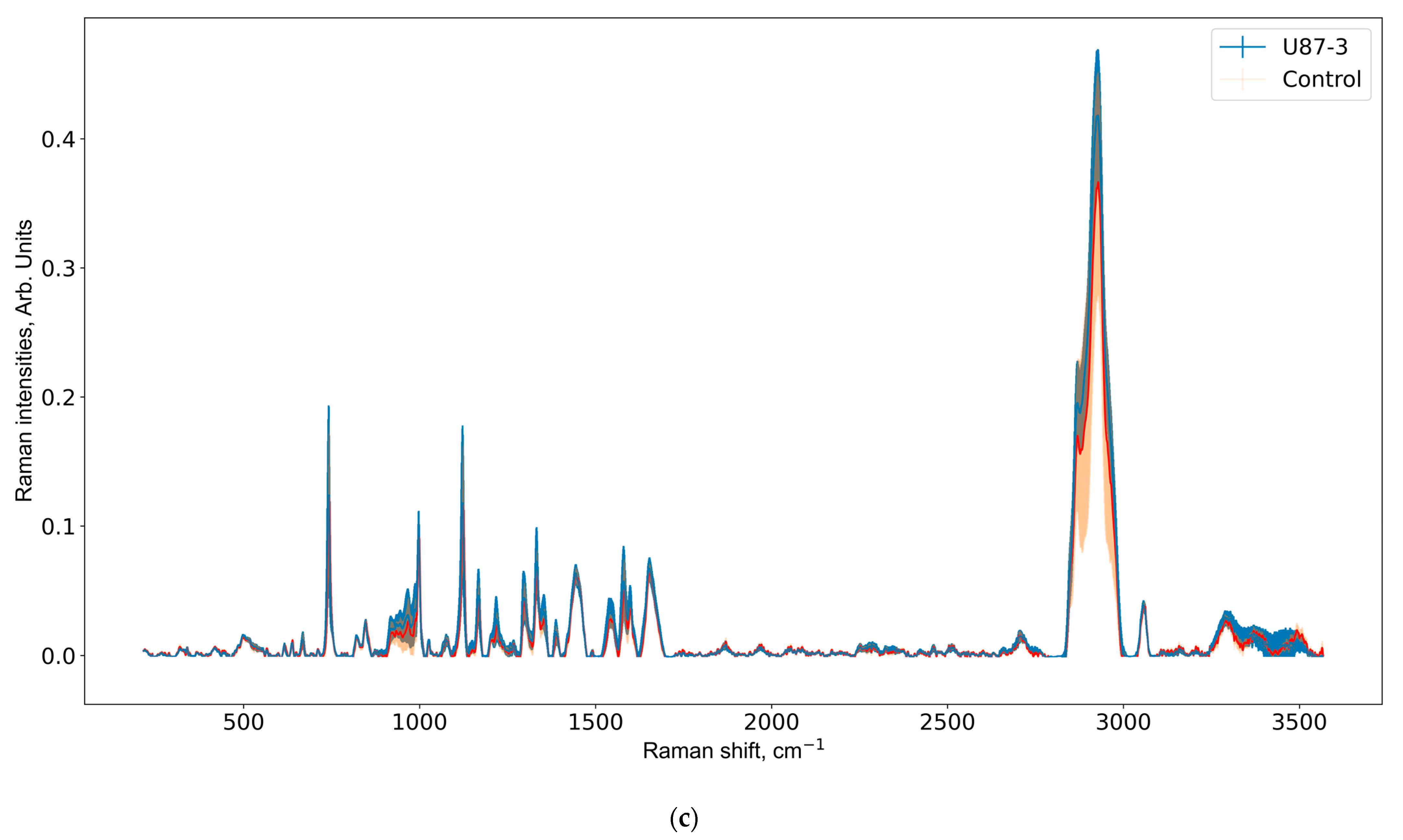
3.2. Raman Spectra Analysis
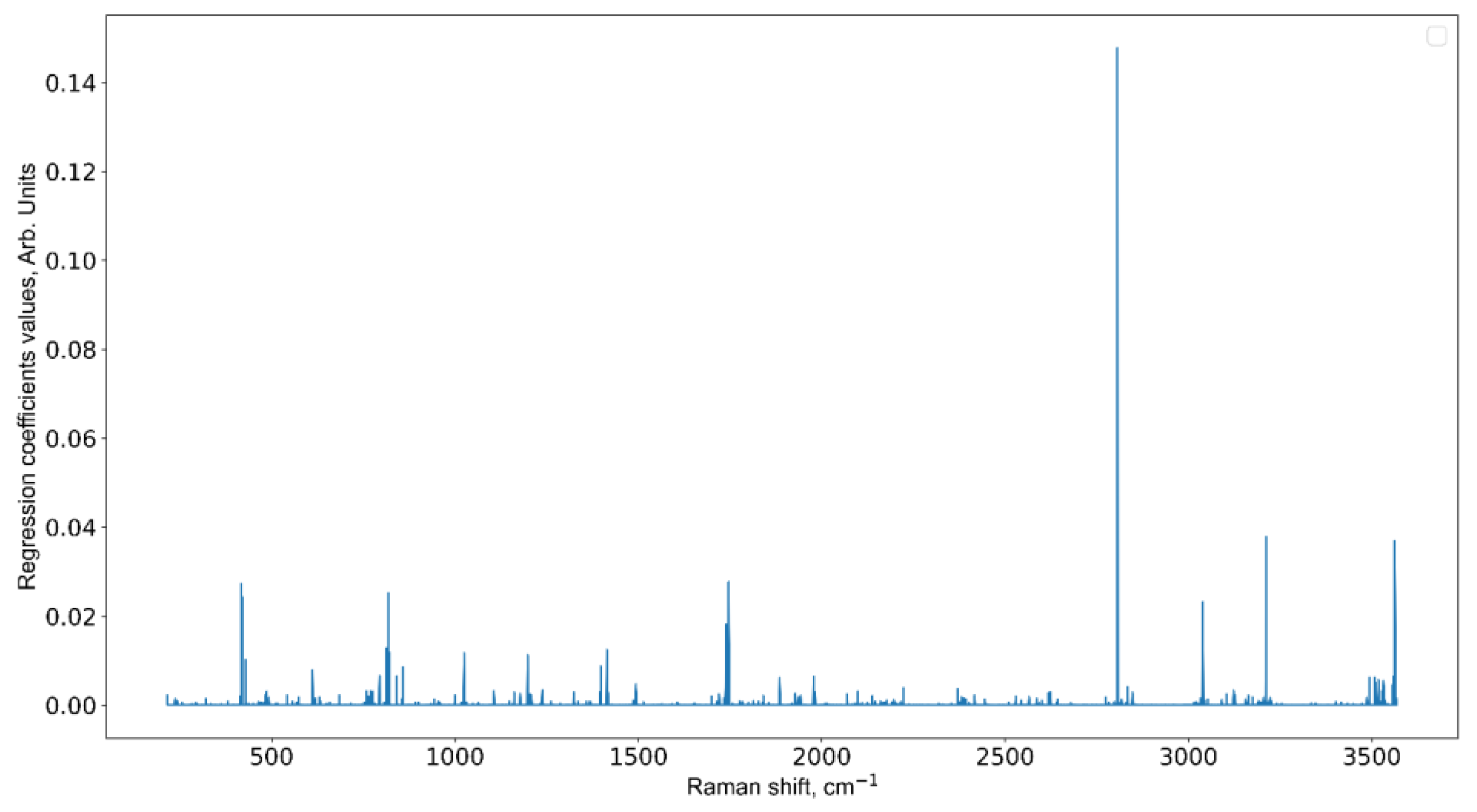
3.3. Informative Feature Selection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Agnihotri, S.; Burrell, K.E.; Wolf, A.; Jalali, S.; Hawkins, C.; Rutka, J.T.; Zadeh, G. Glioblastoma, a brief review of history, molecular genetics, animal models and novel therapeutic strategies. Arch. Immunol. Ther. Exp. 2013, 61, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Wesseling, P.; Capper, D. WHO 2016 classifcation of gliomas. Neuropathol. Appl. Neurobiol. 2018, 44, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro Oncol. 2021, 23 (Suppl. S3), iii1–iii105. [Google Scholar] [CrossRef]
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.M.; Simjee, S.U. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac. J. Cancer Prev. 2017, 18, 3–9. [Google Scholar] [PubMed]
- Torp, S.H.; Solheim, O.; Skjulsvik, A.J. The WHO 2021 Classifcation of Central Nervous System tumours: A practical update on what neurosurgeons need to know—A minireview. Acta Neurochir. 2022, 164, 2453–2464. [Google Scholar] [CrossRef]
- Hammoud, M.A.; Sawaya, R.; Shi, W.; Thall, P.F.; Leeds, N.E. Prognostic significance of preoperative MRI scans in glioblastoma multiforme. J. Neuro-Oncol. 1996, 27, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Wangaryattawanich, P.; Hatami, M.; Wang, J.; Thomas, G.; Flanders, A.; Kirby, J.; Wintermark, M.; Huang, E.S.; Bakhtiari, A.S.; Luedi, M.M.; et al. Multicenter imaging outcomes study of The Cancer Genome Atlas glioblastoma patient cohort: Imaging predictors of overall and progression-free survival. Neuro Oncol. 2015, 17, 1525–1537. [Google Scholar] [CrossRef]
- Bulik, M.; Jancalek, J.; Vanicek, J.; Skoch, A.; Mechl, M. Potential of MR spectroscopy for assessment of glioma grading. Clin. Neurol. Neurosurg. 2013, 115, 146–153. [Google Scholar] [CrossRef]
- Leijenaar, R.T.; Carvalho, S.; Hoebers, F.J.; Aerts, H.J.; van Elmpt, W.J.; Huang, S.H.; Chan, B.; Waldron, J.N.; O’sullivan, B.; Lambin, P. External validation of a prognostic CT-based radiomic signature in oropharyngeal squamous cell carcinoma. Acta Oncol. 2015, 54, 1423–1429. [Google Scholar] [CrossRef]
- Honda, N.; Ishii, K.; Kajimoto, Y.; Kuroiwa, T.; Awazu, K. Determination of optical properties of human brain tumor tissues from 350 to 1000 nm to investigate the cause of false negatives in fluorescence-guided resection with 5-aminolevulinic acid. J. Biomed. Opt. 2018, 23, 075006. [Google Scholar] [CrossRef]
- Vasefi, F.; MacKinnon, N.; Farkas, D.; Kateb, B. Review of the potential of optical technologies for cancer diagnosis in neurosurgery: A step toward intraoperative neurophotonics. Neurophotonics 2016, 4, 011010. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Liu, W.; Zhan, Q.; Li, M.; Zhuang, M.; Liu, Q.H.; Yao, J. Impacts of the murine skull on high-frequency transcranial photoacoustic brain imaging. J. Biophotonics 2019, 12, e201800466. [Google Scholar] [CrossRef] [PubMed]
- Genina, E.A.; Bashkatov, A.N.; Tuchina, D.K.; Dyachenko, P.A.; Navolokin, N.; Shirokov, A.; Khorovodov, A.; Terskov, A.; Klimova, M.; Mamedova, A.; et al. Optical properties of brain tissues at the different stages of glioma development in rats: Pilot study. Biomed. Opt. Express 2019, 10, 5182–5197. [Google Scholar] [CrossRef] [PubMed]
- Cameron, J.M.; Brennan, P.M.; Antoniou, G.; Butler, H.J.; Christie, L.; Conn, J.J.A.; Curran, T.; Gray, E.; Hegarty, M.G.; Jenkinson, M.D.; et al. Clinical validation of a spectroscopic liquid biopsy for earlier detection of brain cancer. Neurooncol. Adv. 2022, 4, vdac024. [Google Scholar] [CrossRef]
- Diem, M.; Mazur, A.; Lenau, K.; Schubert, J.; Bird, B.; Miljković, M.; Krafft, C.; Popp, J. Molecular pathology via IR and Raman spectral imaging. J. Biophotonics 2013, 6, 855–886. [Google Scholar] [CrossRef]
- Jermyn, M.; Desroches, J.; Mercier, J.; St-Arnaud, K.; Guiot, M.-C.; Leblond, F.; Petrecca, K. Raman spectroscopy detects distant invasive brain cancer cells centimeters beyond MRI capability in humans. Biomed. Opt. Express 2016, 7, 5129–5137. [Google Scholar] [CrossRef]
- Kong, K.; Kendall, C.; Stone, N.; Notingher, I. Raman spectroscopy for medical diagnostics—From in-vitro biofluid assays to in-vivo cancer detection. Adv. Drug Deliv. Rev. 2015, 89, 121–134. [Google Scholar] [CrossRef]
- Zaytsev, K.I.; Dolganova, I.N.; Chernomyrdin, N.V.; Katyba, G.M.; Gavdush, A.A.; Cherkasova, O.P.; Komandin, G.A.; Shchedrina, M.A.; Khodan, A.N.; Ponomarev, D.S.; et al. The progress and perspectives of terahertz technology for diagnosis of neoplasms: A review. J. Opt. 2020, 22, 013001. [Google Scholar] [CrossRef]
- Gavdush, A.A.; Chernomyrdin, N.V.; Malakhov, K.M.; Beshplav, S.T.; Dolganova, I.N.; Kosyrkova, A.V.; Nikitin, P.V.; Musina, G.R.; Katyba, G.M.; Reshetov, I.V.; et al. Terahertz spectroscopy of gelatin-embedded human brain gliomas of different grades: A road toward intraoperative THz diagnosis. J. Biomed. Opt. 2019, 24, 027001. [Google Scholar] [CrossRef]
- Gavdush, A.A.; Chernomyrdin, N.V.; Komandin, G.A.; Dolganova, I.N.; Nikitin, P.V.; Musina, G.R.; Katyba, G.M.; Kucheryavenko, A.S.; Reshetov, I.V.; Potapov, A.A.; et al. Terahertz dielectric spectroscopy of human brain gliomas and intact tissues ex vivo: Double-Debye and double-overdamped-oscillator models of dielectric response. Biomed. Opt. Express 2021, 12, 69–83. [Google Scholar] [CrossRef]
- Bi, J.; Chowdhry, S.; Wu, S.; Zhang, W.; Masui, K.; Mischel, P.S. Altered cellular metabolism in gliomas—An emerging landscape of actionable co-dependency targets. Nat. Rev. Cancer 2020, 20, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Harting, R.; de Vries, R.; Ali, M.; Wurdinger, T.; Best, M.G. Blood-Based Biomarkers for Glioma in the Context of Gliomagenesis: A Systematic Review. Front. Oncol. 2021, 11, 665235. [Google Scholar] [CrossRef] [PubMed]
- Lone, S.N.; Nisar, S.; Masoodi, T.; Singh, M.; Rizwan, A.; Hashem, S.; El-Rifai, W.; Bedognetti, D.; Batra, S.K.; Haris, M.; et al. Liquid biopsy: A step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer 2022, 21, 79. [Google Scholar] [CrossRef] [PubMed]
- Ronvaux, L.; Riva, M.; Coosemans, A.; Herzog, M.; Rommelaere, G.; Donis, N.; D’Hondt, L.; Douxfils, J. Liquid Biopsy in Glioblastoma. Cancers 2022, 14, 3394. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, J.M.; Carter, B.S. Detection of glioblastoma in biofluids. J. Neurosurg. 2018, 129, 334–340. [Google Scholar] [CrossRef]
- Cherkasova, O.; Peng, Y.; Konnikova, M.; Kistenev, Y.; Shi, C.; Vrazhnov, D.; Shevelev, O.; Zavjalov, E.; Kuznetsov, S.; Shkurinov, A. Diagnosis of Glioma Molecular Markers by Terahertz Technologies. Photonics 2021, 8, 22. [Google Scholar] [CrossRef]
- Jelski, W.; Mroczko, B. Molecular and Circulating Biomarkers of Brain Tumors. Int. J. Mol. Sci. 2021, 22, 7039. [Google Scholar] [CrossRef]
- Bonosi, L.; Ferini, G.; Giammalva, G.R.; Benigno, U.E.; Porzio, M.; Giovannini, E.A.; Musso, S.; Gerardi, R.M.; Brunasso, L.; Costanzo, R.; et al. Liquid Biopsy in Diagnosis and Prognosis of High-Grade Gliomas; State-of-the-Art and Literature Review. Life 2022, 12, 407. [Google Scholar] [CrossRef]
- Miyauchi, E.; Furuta, T.; Ohtsuki, S.; Tachikawa, M.; Uchida, Y.; Sabit, H.; Obuchi, W.; Baba, T.; Watanabe, M.; Terasaki, T.; et al. Identification of blood biomarkers in glioblastoma by SWATH mass spectrometry and quantitative targeted absolute proteomics. PLoS ONE 2018, 13, e0193799. [Google Scholar] [CrossRef]
- Lee, J.E.; Jeun, S.S.; Kim, S.H.; Yoo, C.Y.; Baek, H.-M.; Yang, S.H. Metabolic profiling of human gliomas assessed with NMR. J. Clin. Neurosci. 2019, 68, 275–280. [Google Scholar] [CrossRef]
- Baranovičová, E.; Galanda, T.; Galanda, M.; Hatok, J.; Kolarovszki, B.; Richterová, R.; Račay, P. Metabolomic profiling of blood plasma in patients with primary brain tumours: Basal plasma metabolites correlated with tumour grade and plasma biomarker analysis predicts feasibility of the successful statistical discrimination from healthy subjects—A preliminary study. IUBMB Life 2019, 71, 1994–2002. [Google Scholar] [CrossRef] [PubMed]
- Mörén, L.; Bergenheim, A.T.; Ghasimi, S.; Brännström, T.; Johansson, M.; Antti, H. Metabolomic screening of tumour tissue and serum in glioma patients reveals diagnostic and prognostic information. Metabolites 2015, 5, 502–520. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Heimberger, A.B.; Lu, Z.; Wu, X.; Hodges, T.R.; Song, R.; Shen, J. Metabolomics profiling in plasma samples from glioma patients correlates with tumor phenotypes. Oncotarget 2016, 7, 20486–20495. [Google Scholar] [CrossRef] [PubMed]
- Strain, S.K.; Groves, M.D.; Olino, K.L.; Emmett, M.R. Measurement of 2-hydroxyglutarate enantiomers in serum by chiral gas chromatography-tandem mass spectrometry and its application as a biomarker for IDH mutant gliomas. Clin. Mass Spectrom. 2020, 15, 16–24. [Google Scholar] [CrossRef]
- Peng, Y.; Shi, C.; Kistenev, Y.V.; Vrazhnov, D.A.; Cherkasova, O.P. Indirect Imaging. In Advances in Brain Imaging Techniques; Mazumder, N., Gangadharan, G., Kistenev, Y.V., Eds.; Springer: Singapore, 2022. [Google Scholar] [CrossRef]
- Gray, E.; Butler, H.J.; Board, R.; Brennan, P.M.; Chalmers, A.J.; Dawson, T.; Goodden, J.; Hamilton, W.; Hegarty, M.G.; James, A.; et al. Health economic evaluation of a serum-based blood test for brain tumour diagnosis: Exploration of two clinical scenarios. BMJ Open 2018, 8, e017593. [Google Scholar] [CrossRef]
- Gray, E.; Cameron, J.M.; Butler, H.J.; Jenkinson, M.D.; Hegarty, M.G.; Palmer, D.S.; Brennan, P.M.; Baker, M.J. Early economic evaluation to guide the development of a spectroscopic liquid biopsy for the detection of brain cancer. Int. J. Technol. Assess Health Care 2021, 37, E41. [Google Scholar] [CrossRef]
- Cameron, J.M.; Butler, H.J.; Smith, B.R.; Hegarty, M.G.; Jenkinson, M.D.; Syed, K.; Brennan, P.M.; Ashton, K.; Dawson, T.; Palmer, D.S.; et al. Developing infrared spectroscopic detection for stratifying brain tumour patients: Glioblastoma multiforme vs. lymphoma. Analyst 2019, 144, 6736–6750. [Google Scholar] [CrossRef]
- Butler, H.J.; Brennan, P.M.; Cameron, J.M.; Finlayson, D.; Hegarty, M.G.; Jenkinson, M.D.; Palmer, D.S.; Smith, B.R.; Baker, M.J. Development of high-throughput ATR-FTIR technology for rapid triage of brain cancer. Nat. Commun. 2019, 10, 4501. [Google Scholar] [CrossRef]
- Theakstone, A.G.; Brennan, P.M.; Jenkinson, M.D.; Mills, S.J.; Syed, K.; Rinaldi, C.; Xu, Y.; Goodacre, R.; Butler, H.J.; Palmer, D.S.; et al. Rapid Spectroscopic Liquid Biopsy for the Universal Detection of Brain Tumours. Cancers 2021, 13, 3851. [Google Scholar] [CrossRef]
- Brennan, P.M.; Butler, H.J.; Christie, L.; Hegarty, M.G.; Jenkinson, M.D.; Keerie, C.; Norrie, J.; O’Brien, R.; Palmer, D.S.; Smith, B.R.; et al. Early diagnosis of brain tumours using a novel spectroscopic liquid biopsy. Brain Commun. 2021, 3, fcab056. [Google Scholar] [CrossRef]
- Wartewing, S. IR and Raman Spectroscopy: Fundamental Processing; Wiley-VCH Verlag GbmH & Co.: Weinheim, Germany, 2003. [Google Scholar]
- Depciuch, J.; Barnaś, E.; Skręt-Magierło, J.; Skręt, A.; Kaznowska, E.; Łach, K.; Jakubczyk, P.; Cebulski, J. Spectroscopic evaluation of carcinogenesis in endometrial cancer. Sci. Rep. 2021, 11, 9079. [Google Scholar] [CrossRef]
- Auner, G.W.; Koya, S.K.; Huang, C.; Broadbent, B.; Trexler, M.; Auner, Z.; Elias, A.; Mehne, K.C.; Brusatori, M.A. Applications of Raman spectroscopy in cancer diagnosis. Cancer Metastasis Rev. 2018, 37, 691–717. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Han, Y.; Sun, B.; Zhang, W.; Liu, S.; Liu, J.; Lv, H.; Zhang, G.; Kang, X. Label-free serum detection based on Raman spectroscopy for the diagnosis and classification of glioma. J. Raman Spectrosc. 2020, 51, 1977–1985. [Google Scholar] [CrossRef]
- Atkins, C.G.; Buckley, K.; Blades, M.W.; Turner, R.F.B. Raman Spectroscopy of Blood and Blood Components. Appl. Spectrosc. 2017, 71, 767–793. [Google Scholar] [CrossRef] [PubMed]
- Pichardo-Molina, J.L.; Frausto-Reyes, C.; Barbosa-García, O.; Huerta-Franco, R.; González-Trujillo, J.L.; Ramírez-Alvarado, C.A.; Gutiérrez-Juárez, G.; Medina-Gutiérrez, C. Raman spectroscopy and multivariate analysis of serum samples from breast cancer patients. Lasers Med. Sci. 2007, 22, 229–236. [Google Scholar] [CrossRef]
- Taleb, I.; Thiéfin, G.; Gobinet, C.; Untereiner, V.; Bernard-Chabert, B.; Heurgué, A.; Truntzer, C.; Hillon, P.; Manfait, M.; Ducoroy, P.; et al. Diagnosis of hepatocellular carcinoma in cirrhotic patients: A proof-of-concept study using serum micro-Raman spectroscopy. Analyst 2013, 138, 4006–4014. [Google Scholar] [CrossRef]
- Zavjalov, E.L.; Razumov, I.A.; Gerlinskaya, L.A.; Romashchenko, A.V. In vivo MRI Visualization of U87 Glioblastoma Development Dynamics in the Model of Orthotopic Xenotransplantation to the SCID Mouse. Russ. J. Genet. Appl. Res. 2016, 6, 448–453. [Google Scholar] [CrossRef]
- Vrazhnov, D.; Knyazkova, A.; Konnikova, M.; Shevelev, O.; Razumov, I.; Zavjalov, E.; Kistenev, Y.; Shkurinov, A.; Cherkasova, O. Analysis of Mouse Blood Serum in the Dynamics of U87 Glioblastoma by Terahertz Spectroscopy and Machine Learning. Appl. Sci. 2022, 12, 10533. [Google Scholar] [CrossRef]
- Mankova, A.A.; Cherkasova, O.P.; Lazareva, E.N.; Bucharskaya, A.B.; Dyachenko, P.A.; Kistenev, Y.V.; Vrazhnov, D.A.; Skiba, V.E.; Tuchin, V.V.; Shkurinov, A.P. Study of Blood Serum in Rats with Transplanted Cholangiocarcinoma Using Raman Spectroscopy. Opt. Spectrosc. 2020, 128, 964–971. [Google Scholar] [CrossRef]
- Cherkasova, O.; Mankova, A.; Vrazhnov, D. Raman spectroscopy of blood plasma for cancer diagnosis. In XV International Conference on Pulsed Lasers and Laser Applications; SPIE: Bellingham, WA, USA, 2021; Volume 12086, pp. 344–350. [Google Scholar]
- Zhang, Z.M.; Chen, S.; Liang, Y.Z. Baseline correction using adaptive iteratively reweighted penalized least squares. Analyst 2010, 135, 1138–1146. [Google Scholar] [CrossRef]
- Karmenyan, A.V.; Kistenev, Y.V.; Perevedentseva, E.V.; Krivokharchenko, A.S.; Sarmiento, M.N.; Barus, E.L.; Cheng, C.-L.; Vrazhnov, D.A. Machine learning methods for the in-vitro analysis of preimplantation embryo Raman micro-spectroscopy. In Proceedings of the Fourth International Conference on Terahertz and Microwave Radiation: Generation, Detection, and Applications, Tomsk, Russia, 24–26 August 2020; Volume 11582, pp. 115820W1–115820W5. [Google Scholar] [CrossRef]
- Jolliffe, I.T. A note on the use of principal components in regression. J. R. Stat. Soc. C Appl. Stat. 1982, 31, 300–303. [Google Scholar] [CrossRef]
- Blake, N.; Gaifulina, R.; Griffin, L.D.; Bell, I.M.; Thomas, G.M.H. Machine Learning of Raman Spectroscopy Data for Classifying Cancers: A Review of the Recent Literature. Diagnostics 2022, 12, 1491. [Google Scholar] [CrossRef] [PubMed]
- Schölkopf, B.; Smola, A.J.; Bach, F. Learning with Kernels: Support Vector Machines, Regularization, Optimization, and Beyond; MIT Press: Cambridge, MA, USA, 2002. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K. Xgboost: Extreme gradient boosting. R Package Version 0.4-2 2015, 1, 1–4. [Google Scholar]
- Rodriguez, J.D.; Perez, A.; Lozano, J.A. Sensitivity analysis of k-fold cross validation in prediction error estimation. IEEE Trans. Pattern Anal. Mach. Intell. 2009, 32, 569–575. [Google Scholar] [CrossRef] [PubMed]
- Haenssle, H.A.; Fink, C.; Schneiderbauer, R.; Toberer, F.; Buhl, T.; Blum, A.; Zalaudek, I. Man against machine: Diagnostic performance of a deep learning convolutional neural network for dermoscopic melanoma recognition in comparison to 58 dermatologists. Ann. Oncol. 2018, 29, 1836–1842. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://xgboost.readthedocs.io/en/latest/R-package/discoverYourData.html#special-note-what-about-random-forests (accessed on 1 January 2022).
- Short, K.W.; Carpenter, S.; Freyer, J.P.; Mourant, J.R. Raman Spectroscopy Detects Biochemical Changes Due to Proliferation in Mammalian Cell Cultures. Biophys. J. 2005, 88, 4274–4288. [Google Scholar] [CrossRef]
- Sciortino, T.; Secoli, R.; d’Amico, E.; Moccia, S.; Conti Nibali, M.; Gay, L.; Rossi, M.; Pecco, N.; Castellano, A.; De Momi, E.; et al. Raman Spectroscopy and Machine Learning for IDH Genotyping of Unprocessed Glioma Biopsies. Cancers 2021, 13, 4196. [Google Scholar] [CrossRef]
- Riva, M.; Sciortino, T.; Secoli, R.; D’Amico, E.; Moccia, S.; Fernandes, B.; Conti Nibali, M.; Gay, L.; Rossi, M.; De Momi, E.; et al. Glioma biopsies Classification Using Raman Spectroscopy and Machine Learning Models on Fresh Tissue Samples. Cancers 2021, 13, 1073. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, C.H.; Wu, B.; Yu, X.; Cheng, G.; Zhu, K.; Wang, K.; Zhang, C.; Zhao, M.; Zong, R.; et al. Optical biopsy identification and grading of gliomas using label-free visible resonance Raman spectroscopy. J. Biomed. Opt. 2019, 24, 095001. [Google Scholar] [CrossRef]
- Kowalska, A.A.; Berus, S.; Szleszkowski, L.; Kamińska, A.; Kmiecik, A.; Ratajczak-Wielgomas, K.; Jurek, T.; Zadka, L. Brain tumour homogenates analysed by surface-enhanced Raman spectroscopy: Discrimination among healthy and cancer cells. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2018, 231, 117769. [Google Scholar] [CrossRef] [PubMed]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Kalkanis, S.N.; Kast, R.E.; Rosenblum, M.R.; Mikkelsen, T.; Yurgelevic, S.M.; Nelson, K.M.; Raghunathan, A.; Poisson, L.M.; Auner, G.W. Raman spectroscopy to distinguish grey matter, necrosis, and glioblastoma multiforme in frozen tissue sections. J. Neurooncol. 2014, 116, 477–485. [Google Scholar] [CrossRef] [PubMed]
- Uckermann, O.; Galli, R.; Tamosaityte, S.; Leipnitz, E.; Geiger, K.D.; Schackert, G.; Koch, E.; Steiner, G.; Kirsch, M. Label-Free Delineation of Brain Tumors by Coherent Anti-Stokes Raman Scattering Microscopy in an Orthotopic Mouse Model and Human Glioblastoma. PLoS ONE 2014, 9, e107115. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q. The Efects of Dissolved Hydrophobic and Hydrophilic Groups on Water Structure. J. Solut. Chem. 2020, 49, 1473–1484. [Google Scholar] [CrossRef]
- Cassanas, G.; Morssli, M.; Fabrègue, E.; Bardet, L. Vibrational spectra of lactic acid and lactates. J. Raman Spectrosc. 1991, 22, 409–413. [Google Scholar] [CrossRef]
- Kopec, M.; Błaszczyk, M.; Radek, M.; Abramczyk, N. Raman imaging and statistical methods for analysis various type of human brain tumors and breast cancers. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2021, 262, 120091. [Google Scholar] [CrossRef]
- Kouri, M.A.; Spyratou, E.; Karnachoriti, M.; Kalatzis, D.; Danias, N.; Arkadopoulos, N.; Seimenis, I.; Raptis, Y.S.; Kontos, A.G.; Efstathopoulos, E.P. Raman Spectroscopy: A Personalized Decision-Making Tool on Clinicians’ Hands for In Situ Cancer Diagnosis and Surgery Guidance. Cancers 2022, 14, 1144. [Google Scholar] [CrossRef]
- Ghita, A.; Hubbard, T.; Matousek, P.; Stone, N. Noninvasive Detection of Differential Water Content Inside Biological Samples Using Deep Raman Spectroscopy. Anal. Chem. 2020, 92, 9449–9453. [Google Scholar] [CrossRef]
- Miyai, M.; Tomita, H.; Soeda, A.; Yano, H.; Iwama, T.; Hara, A. Current trends in mouse models of glioblastoma. J. Neurooncol. 2017, 135, 423–432. [Google Scholar] [CrossRef]
- Davis, A.E. Mechanisms of traumatic brain injury: Biomechanical, structural and cellular considerations. Crit. Care Nurs. Q. 2000, 23, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Cernak, I. Animal models of head trauma. NeuroRx 2005, 2, 410–422. [Google Scholar] [CrossRef] [PubMed]
- Cherkasova, O.; Konnikova, M.; Heinz, T.; Nazarov, M.; Kuryanova, A.; Aksenova, N.; Mankova, A.; Vrazhnov, D.; Kistenev, Y.; Peng, Y.; et al. Terahertz and infrared absorption spectra of mouse blood plasma in glioma development. In Proceedings of the 46th International Conference on Infrared, Millimeter and Terahertz Waves (IRMMW-THz 2021), Chengdu, China, 29 August–3 September 2021; pp. 1–2. [Google Scholar]
- Wang, Y.; Wang, G.; Xu, D.; Jiang, B.; Ge, M.; Wu, L.; Yang, C.; Mu, N.; Wang, S.; Chang, C.; et al. Terahertz spectroscopic diagnosis of early blast-induced traumatic brain injury in rats. Biomed. Opt. Express 2020, 11, 4085–4098. [Google Scholar] [CrossRef] [PubMed]
- Borisov, A.V.; Syrkina, A.G.; Kuzmin, D.A.; Ryabov, V.V.; Boyko, A.A.; Zaharova, O.; Zasedatel, V.S.; Kistenev, Y.V. Application of machine learning and laser optical-acoustic spectroscopy to study the profile of exhaled air volatile markers of acute myocardial infarction. J. Breath Res. 2021, 15, 027104. [Google Scholar] [CrossRef] [PubMed]
- De Gelder, J.; De Gussem, K.; Vandenabeele, P.; Moens, L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Kyne, M.; Wang, C.; Yu, X.-Y. Data mining in Raman imaging in a cellular biological system. Comput. Struct. Biotechnol. J. 2020, 18, 2920–2930. [Google Scholar] [CrossRef]
- Byrne, H.J.; Bonnier, F.; McIntyre, J.; Parachalil, D.R. Quantitative analysis of human blood serum using vibrational spectroscopy. Clin. Spectrosc. 2020, 2, 100004. [Google Scholar] [CrossRef]
- Butler, H.J.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef]
- Kaushik, R.; Rani, C.; Neeshu, K.; Tanwar, M.; Pathak, D.K.; Chaudhary, A.; Siraj, F.; Jha, H.C.; Kumar, R. Brain Tumour Detection and Grading Using Raman Scattering: Analogy from Semiconductors for Solving Biological Problem. Adv. Mater. Process Technol. 2020, 8, 703–714. [Google Scholar] [CrossRef]
- Smolyanskaya, O.A.; Chernomyrdin, N.V.; Konovko, A.A.; Zaytsev, K.I.; Ozheredov, I.A.; Cherkasova, O.P.; Nazarov, M.M.; Guillet, J.P.; Kozlov, S.A.; Kistenev, Y.V.; et al. Terahertz biophotonics as a tool for studies of dielectric and spectral properties of biological tissues and liquids. Prog. Quantum Electron. 2018, 62, 1–77. [Google Scholar] [CrossRef]
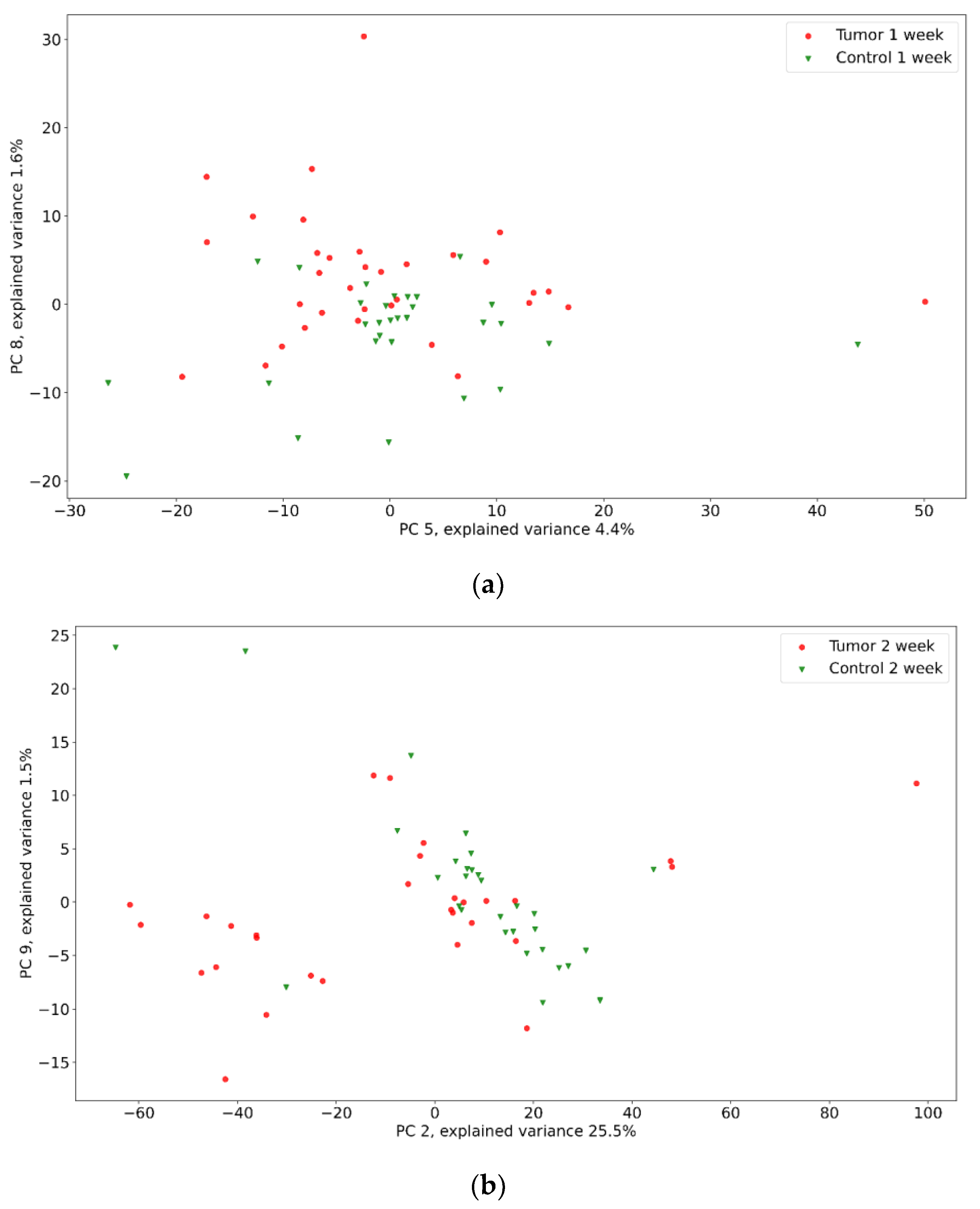
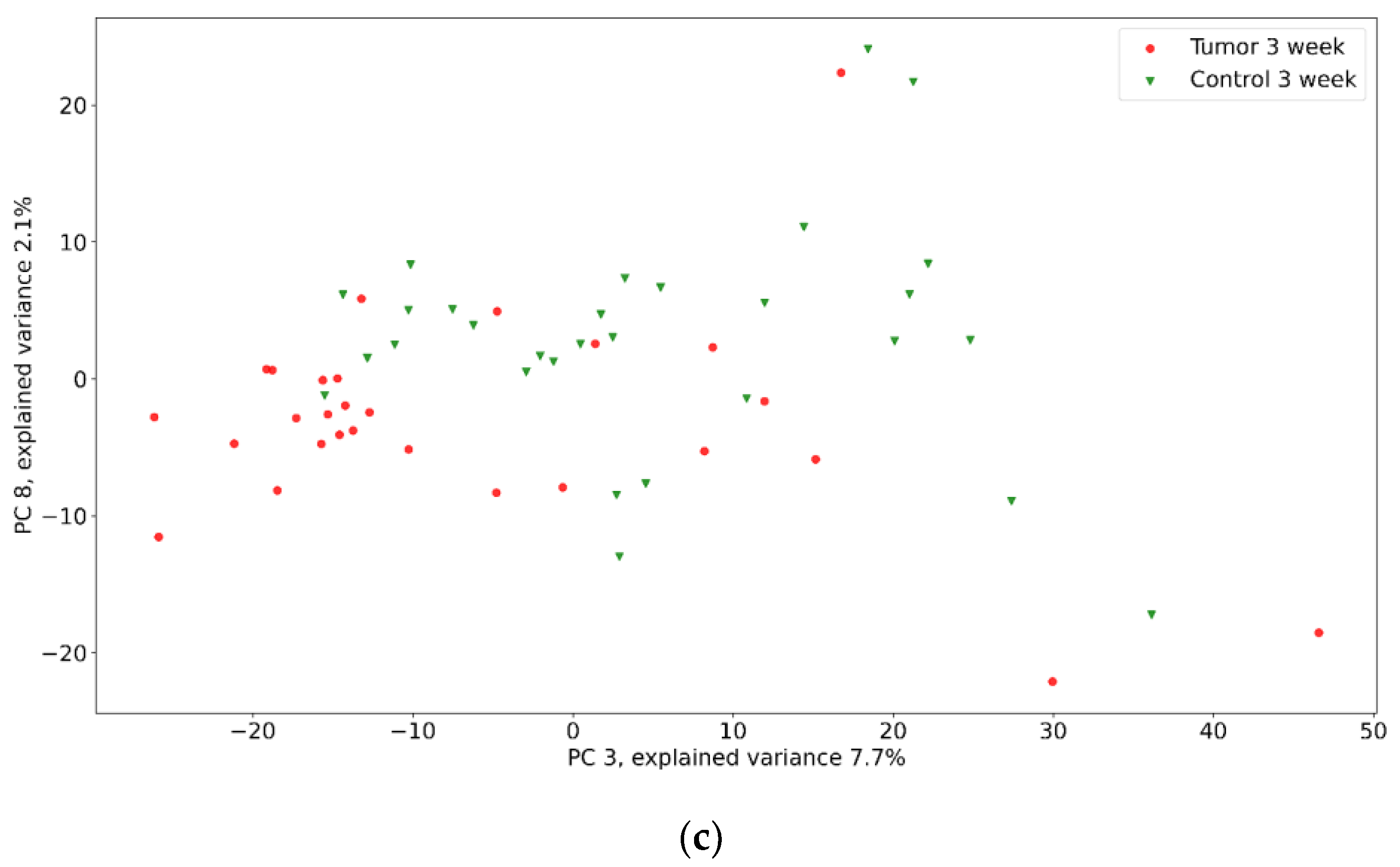
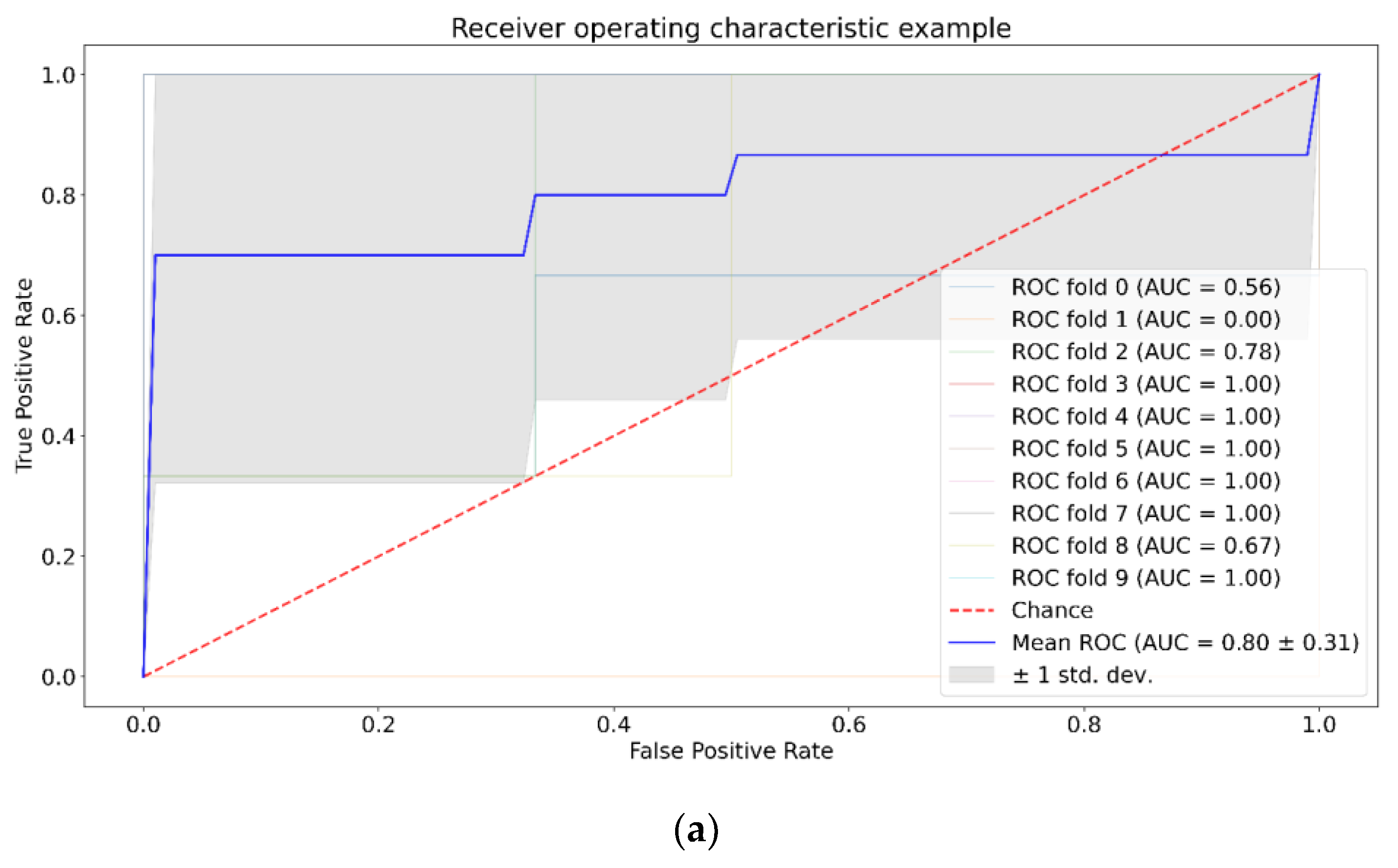
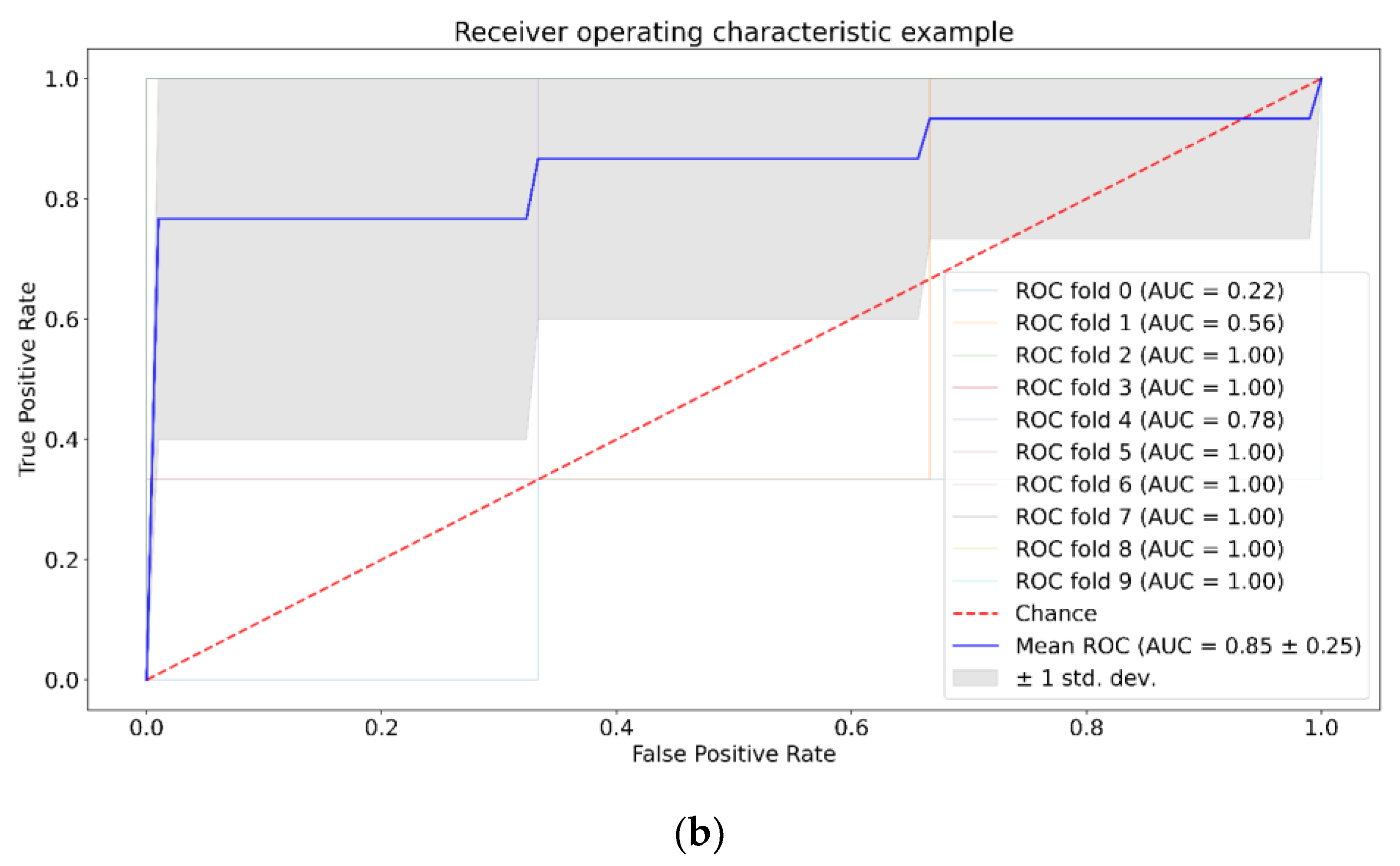
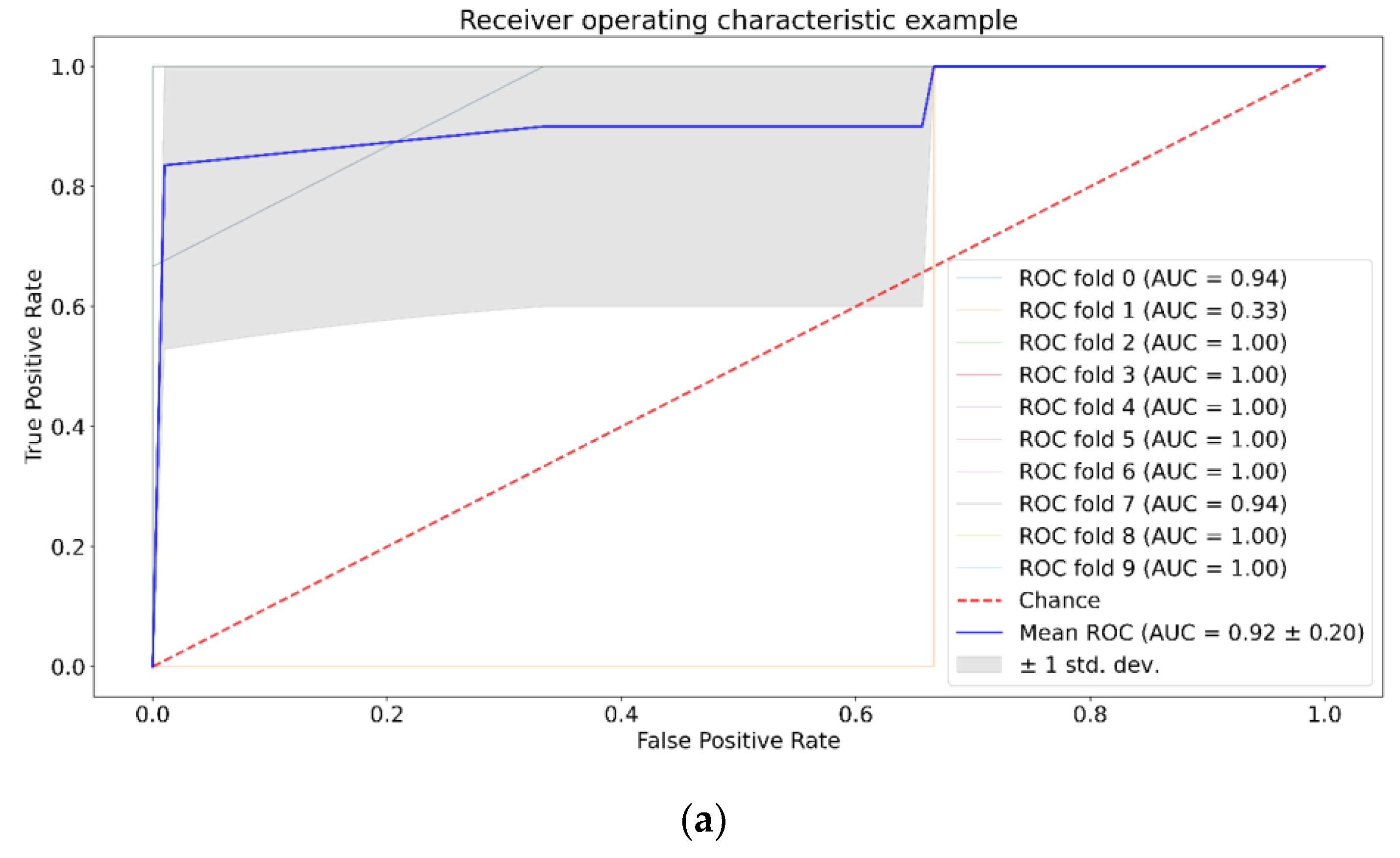
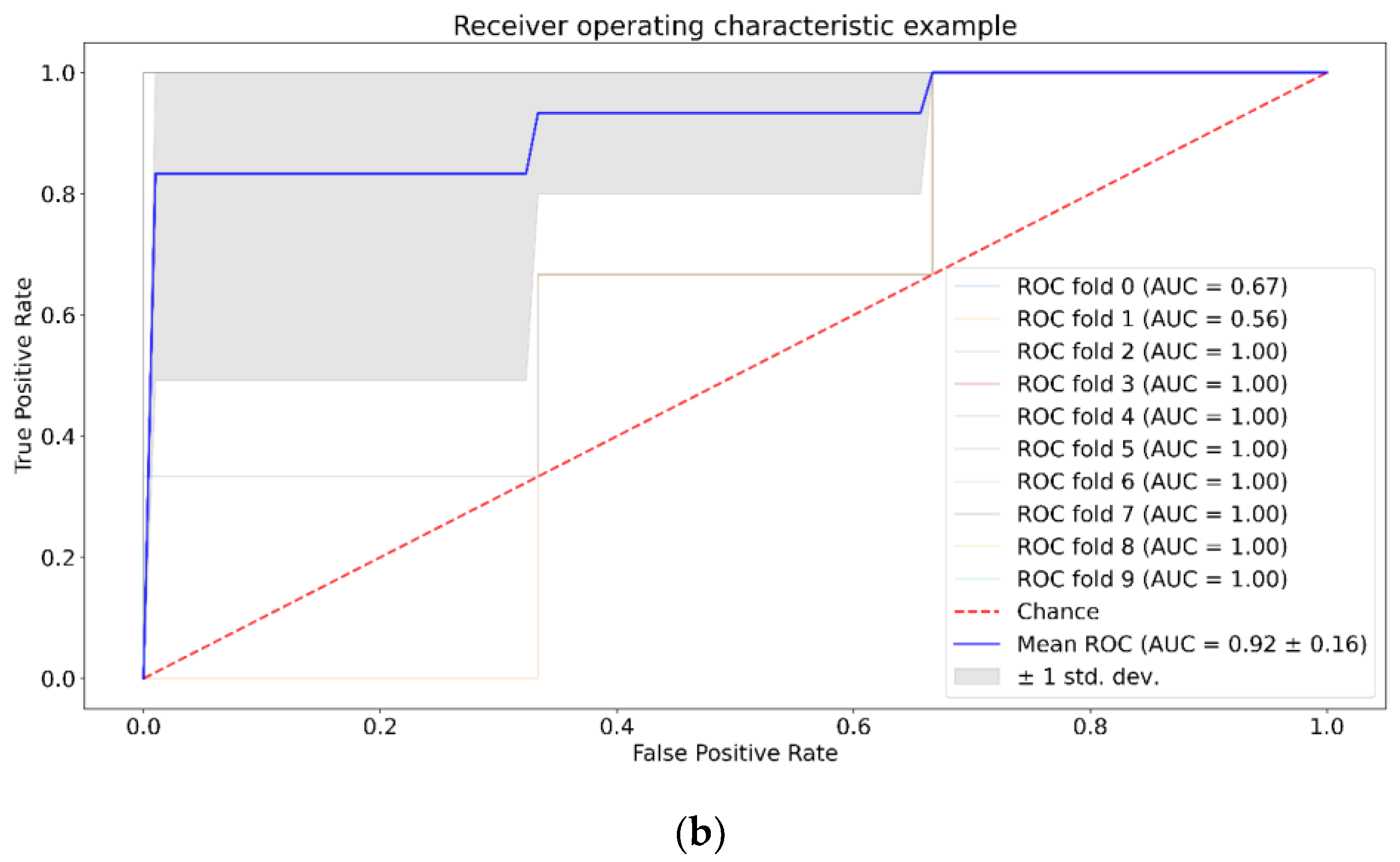
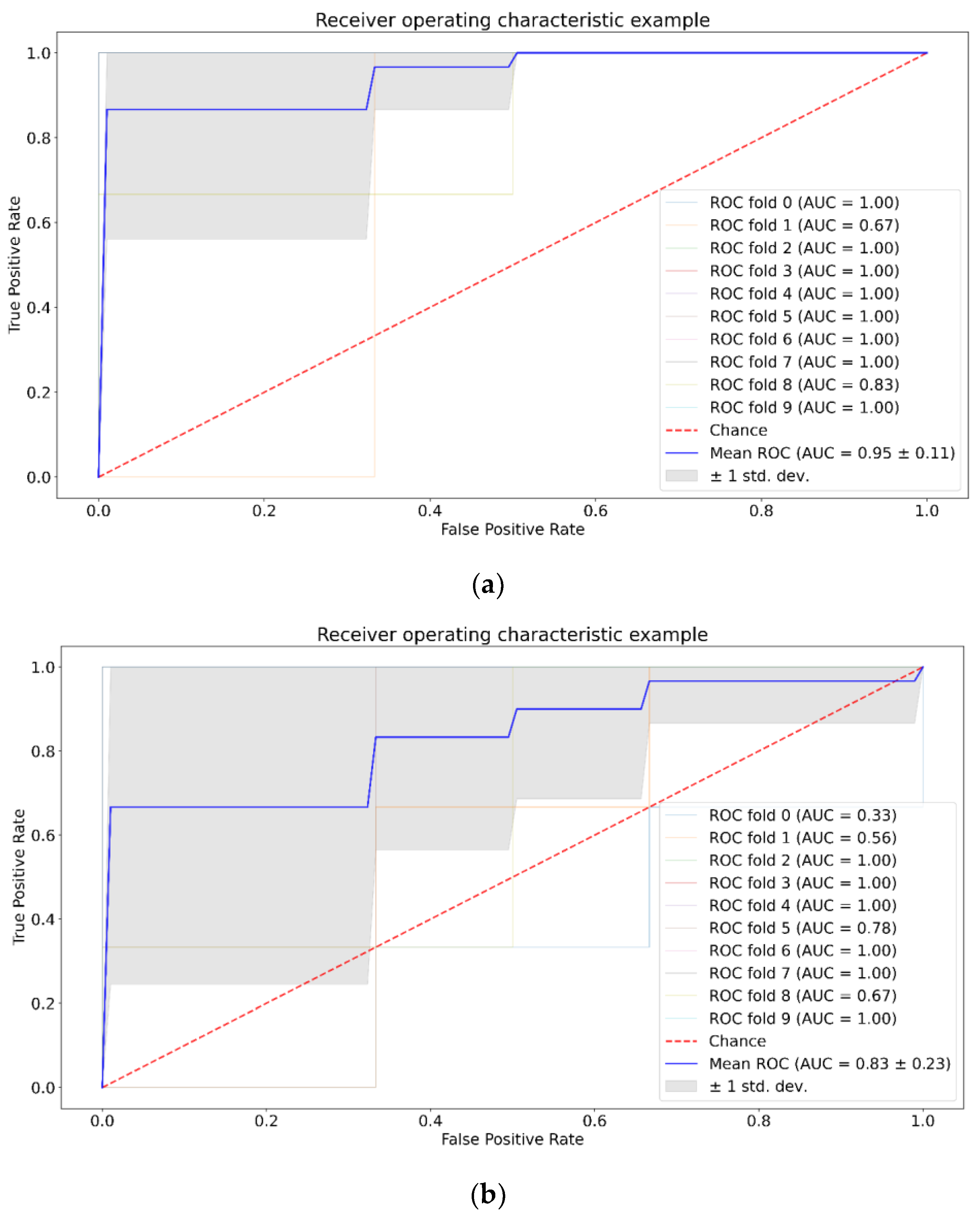
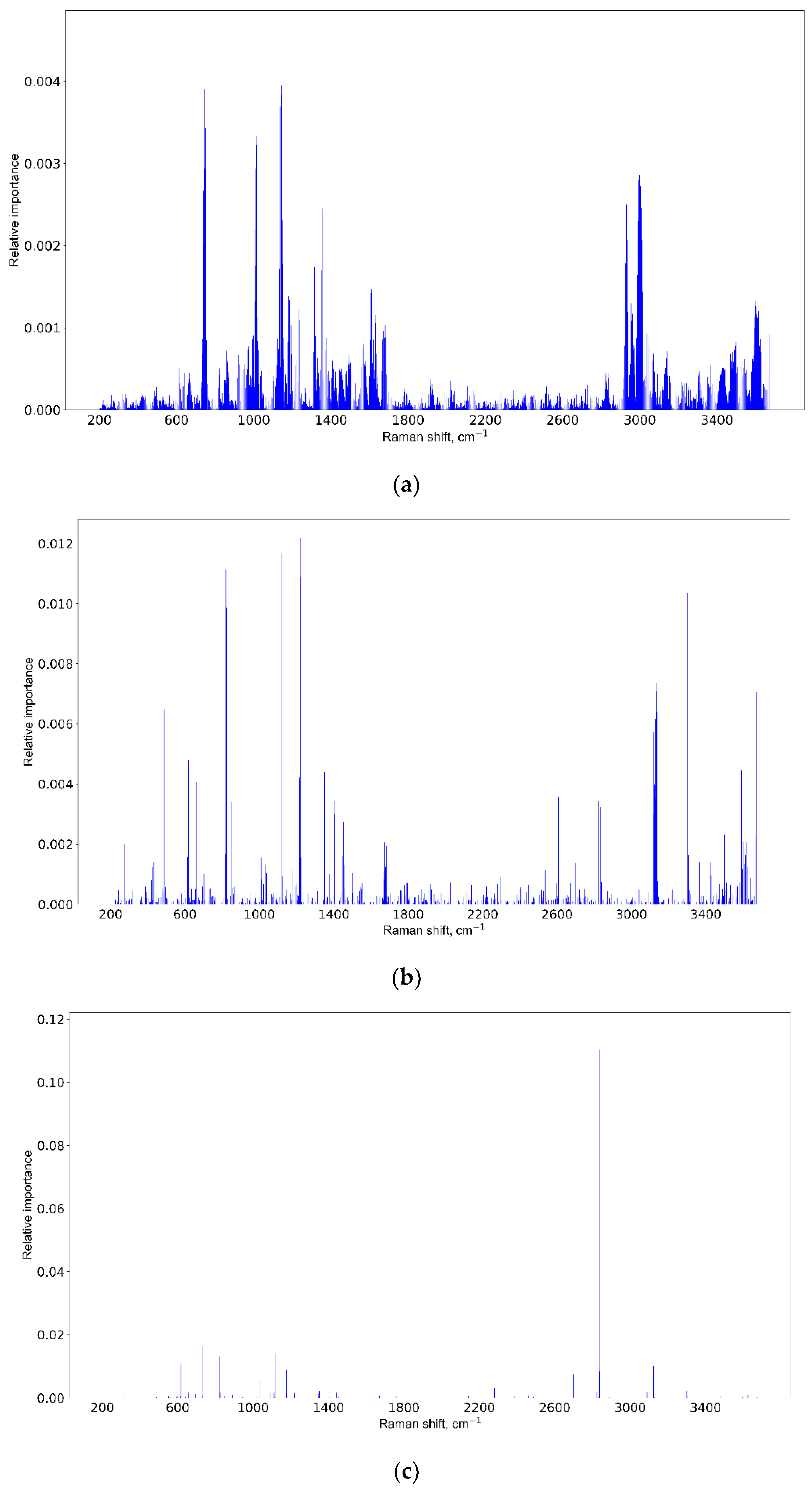
| Positive (Model Prediction) | Negative (Model Prediction) | |
|---|---|---|
| Positive (expert label) | True positives (TP) | False negative (FN) |
| Negative (expert label) | False positive (FP) | True negative (TN) |
| SVM no PCA | SVM with PCA | RF no PCA | PF with PCA | XGBoost no PCA | XGBoost with PCA | |
|---|---|---|---|---|---|---|
| U87-1 | 0.72 ± 0.38 | 0.91 ± 0.17 | 0.81 ± 0.29 | 0.78 ± 0.21 | 0.92 ± 0.13 | 0.87 ± 0.19 |
| U87-2 | 0.80 ± 0.28 | 0.68 ± 0.32 | 0.73 ± 0.22 | 0.78 ± 0.29 | 0.82 ± 0.0.18 | 0.72 ± 0.27 |
| U87-3 | 0.80 ± 0.31 | 0.85 ± 0.25 | 0.92 ± 0.20 | 0.92 ± 0.16 | 0.95 ± 0.11 | 0.83 ± 023 |
| Raman Peaks (cm−1) | Assignments | |||
|---|---|---|---|---|
| This Paper Blood Serum | Glioma Brain tissues | Modes | Molecules | References |
| 417 420 | 391 | Protein, lipids Cholesterol/cholesterol ester | [63] | |
| 419 | [64] | |||
| 431 | [65] | |||
| 818 | 754 | CC, CN2, CH2 rock | Nucleic acids, DNA, Trp, | [66] [67] [65] [65] [68] |
| Nucleoproteins, Cyt | ||||
| 800 | C-C stretching | L-Tryptophane | ||
| 817 | Collagen | |||
| 826 | Tyr, proline | |||
| 825–827 | O-P-O stretch | DNA | ||
| 1746 | 1736 | C=O | Lipids | [66] |
| 1737 | Cholesterol | [69] | ||
| 1739 | Cholesterol ester | [69] | ||
| 1740 | Protein | [63] | ||
| 2807 | 2850 | CH2 | Lipids, fatty acids | [44,66,70] |
| 3039 | 3040 | -OH CH3 CH3 | Water | [71] |
| 2930 | Proteins and DNA | [44] | ||
| 2985 | Lactate | [72] | ||
| 2932 | Lipoprotein | [66] | ||
| 2939 | Lactate, lipoprotein | [66] | ||
| 3039 | 3064 | NH3 | Nucleic acids/ protein, Trp | [66] |
| 3212 | 3174 | NH, CH3 | Amid B, L-glutamine | [66] |
| 3164 | str (NH), str (OH) | Amide, water | [73] | |
| >3200 | str (OH) | Water | [74] | |
| 3563 | 3350–3550 | OH | Water | [72,75] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vrazhnov, D.; Mankova, A.; Stupak, E.; Kistenev, Y.; Shkurinov, A.; Cherkasova, O. Discovering Glioma Tissue through Its Biomarkers’ Detection in Blood by Raman Spectroscopy and Machine Learning. Pharmaceutics 2023, 15, 203. https://doi.org/10.3390/pharmaceutics15010203
Vrazhnov D, Mankova A, Stupak E, Kistenev Y, Shkurinov A, Cherkasova O. Discovering Glioma Tissue through Its Biomarkers’ Detection in Blood by Raman Spectroscopy and Machine Learning. Pharmaceutics. 2023; 15(1):203. https://doi.org/10.3390/pharmaceutics15010203
Chicago/Turabian StyleVrazhnov, Denis, Anna Mankova, Evgeny Stupak, Yury Kistenev, Alexander Shkurinov, and Olga Cherkasova. 2023. "Discovering Glioma Tissue through Its Biomarkers’ Detection in Blood by Raman Spectroscopy and Machine Learning" Pharmaceutics 15, no. 1: 203. https://doi.org/10.3390/pharmaceutics15010203
APA StyleVrazhnov, D., Mankova, A., Stupak, E., Kistenev, Y., Shkurinov, A., & Cherkasova, O. (2023). Discovering Glioma Tissue through Its Biomarkers’ Detection in Blood by Raman Spectroscopy and Machine Learning. Pharmaceutics, 15(1), 203. https://doi.org/10.3390/pharmaceutics15010203








