Towards Personalized Allele-Specific Antisense Oligonucleotide Therapies for Toxic Gain-of-Function Neurodegenerative Diseases
Abstract
1. Introduction
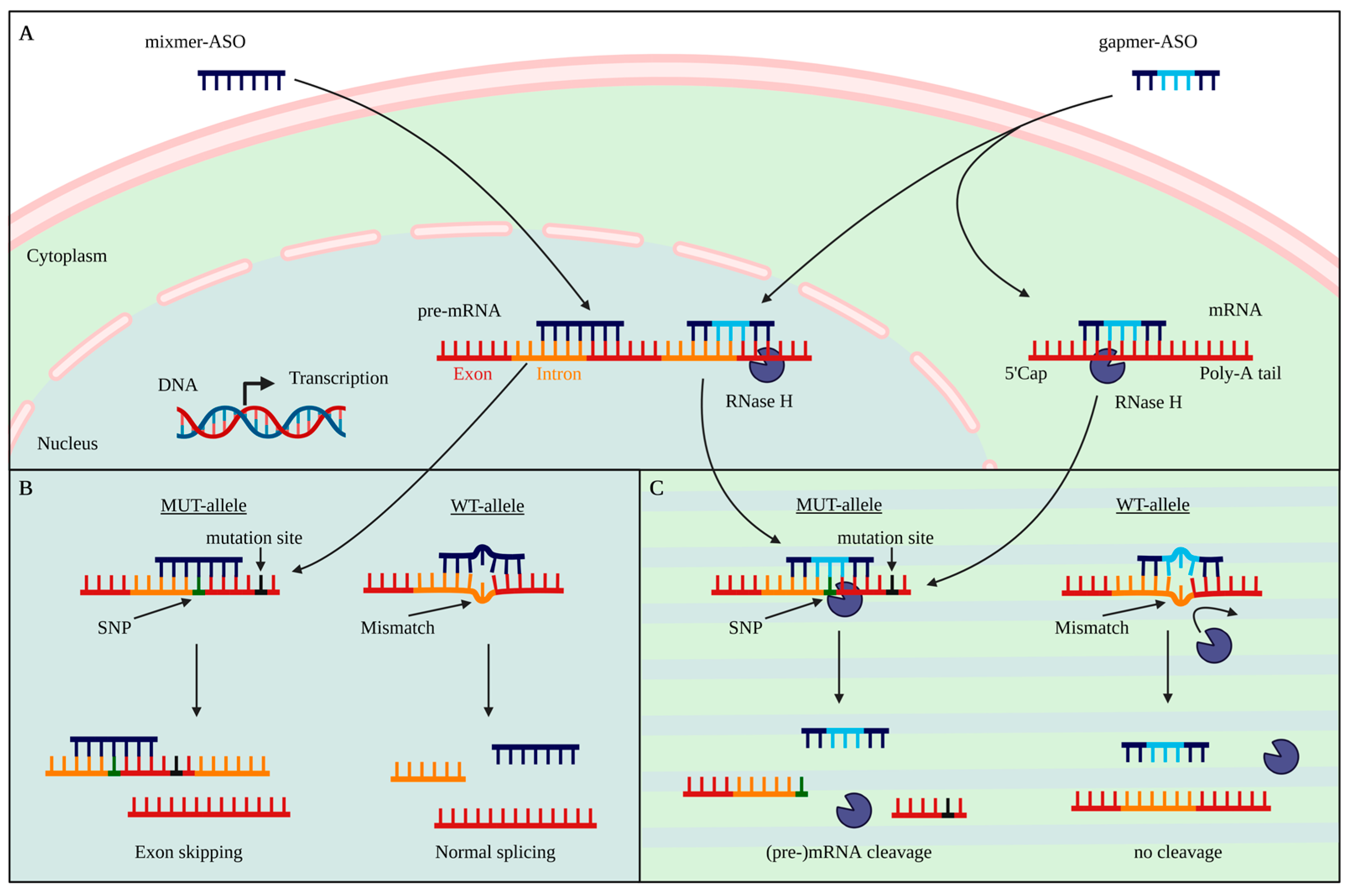
2. Antisense Oligonucleotides
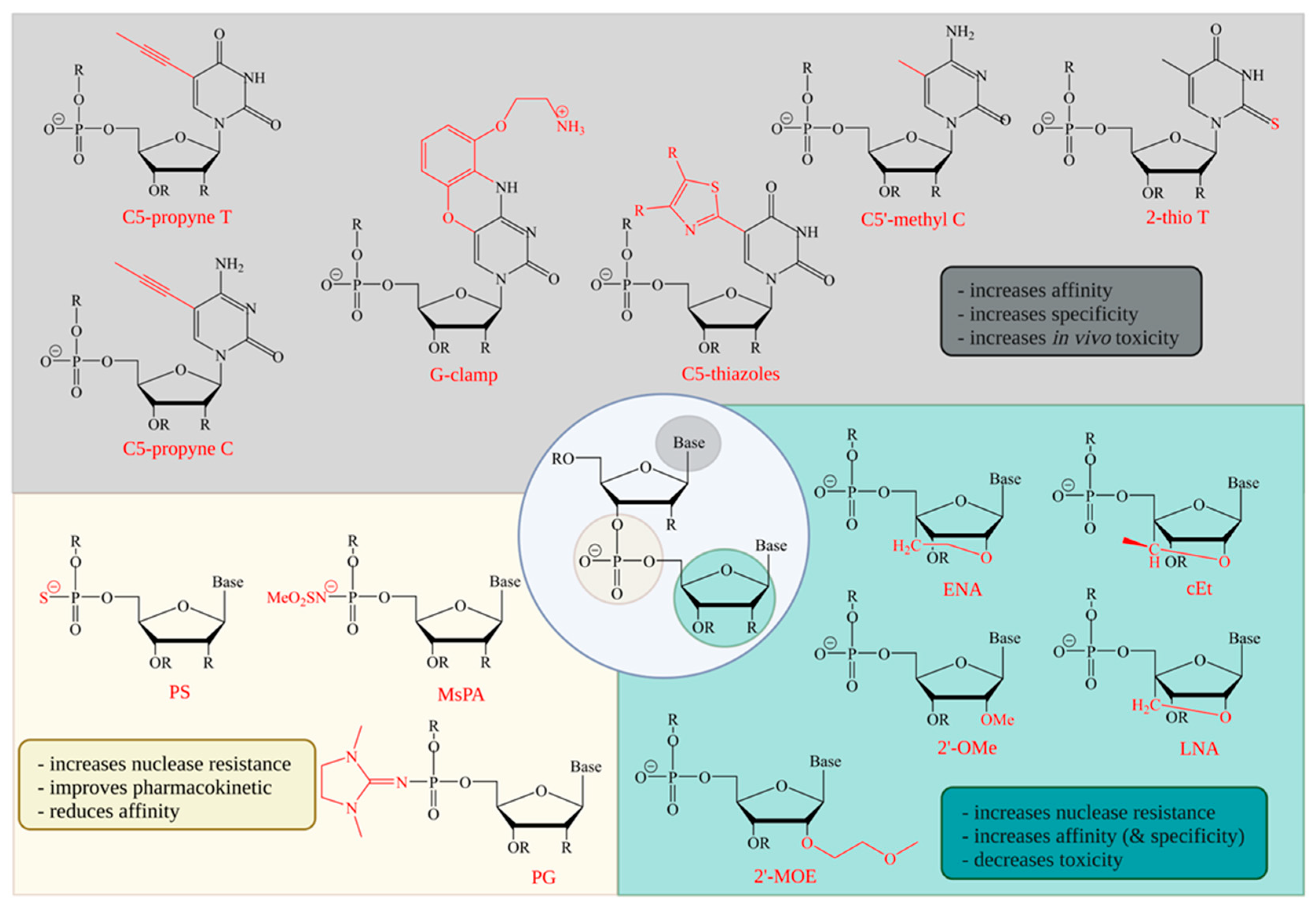
3. Backbone Modifications
4. Sugar Modifications
5. Base Modifications
6. Selective vs. Non-Selective Targeting Strategies
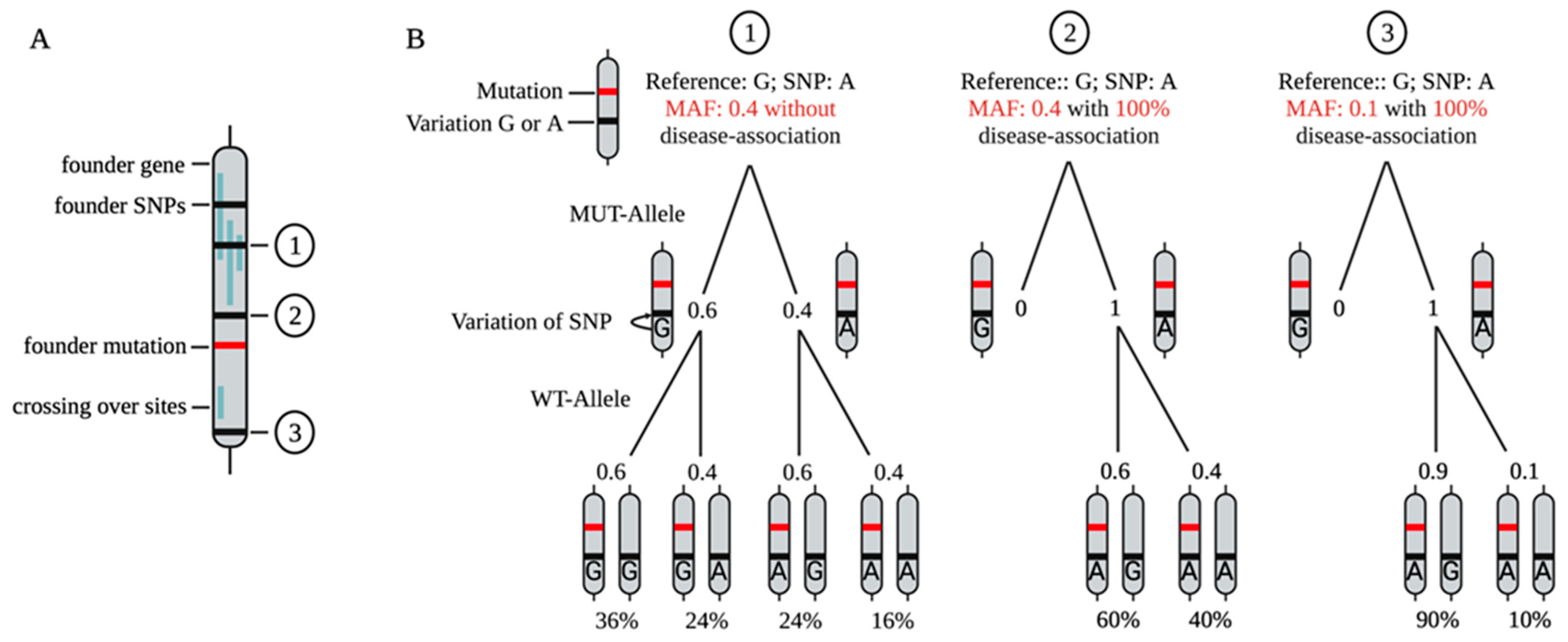
7. SNP-Based Allele-Specific Treatment Strategies
8. Target-Based ASO Design
9. The Challenge of Readout
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Testa, C.M. Antisense Oligonucleotide Therapeutics for Neurodegenerative Disorders. Curr. Geriatr. Rep. 2022, 11, 19–32. [Google Scholar] [CrossRef]
- Rook, M.E.; Southwell, A.L. Antisense Oligonucleotide Therapy: From Design to the Huntington Disease Clinic. BioDrugs 2022, 36, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Baker, B.F.; Crooke, R.M.; Liang, X.-H. Antisense technology: An overview and prospectus. Nat. Rev. Drug Discov. 2021, 20, 427–453. [Google Scholar] [CrossRef] [PubMed]
- Scoles, D.R.; Minikel, E.V.; Pulst, S.M. Antisense oligonucleotides: A primer. Neurol. Genet. 2019, 5, e323. [Google Scholar] [CrossRef] [PubMed]
- Hauser, S.; Helm, J.; Kraft, M.; Korneck, M.; Hübener-Schmid, J.; Schöls, L. Allele-specific targeting of mutant ataxin-3 by antisense oligonucleotides in SCA3-iPSC-derived neurons. Mol. Ther.-Nucleic Acids 2022, 27, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Skotte, N.H.; Southwell, A.L.; Østergaard, M.E.; Carroll, J.B.; Warby, S.C.; Doty, C.N.; Petoukhov, E.; Vaid, K.; Kordasiewicz, H.; Watt, A.T. Allele-specific suppression of mutant huntingtin using antisense oligonucleotides: Providing a therapeutic option for all Huntington disease patients. PLoS ONE 2014, 9, e107434. [Google Scholar] [CrossRef]
- Taylor, J.P.; Hardy, J.; Fischbeck, K.H. Toxic Proteins in Neurodegenerative Disease. Science 2002, 296, 1991–1995. [Google Scholar] [CrossRef]
- Scekic-Zahirovic, J.; Sendscheid, O.; El Oussini, H.; Jambeau, M.; Sun, Y.; Mersmann, S.; Wagner, M.; Dieterlé, S.; Sinniger, J.; Dirrig-Grosch, S. Toxic gain of function from mutant FUS protein is crucial to trigger cell autonomous motor neuron loss. EMBO J. 2016, 35, 1077–1097. [Google Scholar] [CrossRef]
- Yankner, B.A.; Lu, T. Amyloid β-protein toxicity and the pathogenesis of Alzheimer disease. J. Biol. Chem. 2009, 284, 4755–4759. [Google Scholar] [CrossRef]
- Paulson, H.L.; Perez, M.; Trottier, Y.; Trojanowski, J.; Subramony, S.; Das, S.; Vig, P.; Mandel, J.-L.; Fischbeck, K.; Pittman, R. Intranuclear inclusions of expanded polyglutamine protein in spinocerebellar ataxia type 3. Neuron 1997, 19, 333–344. [Google Scholar] [CrossRef]
- Ishikawa, K.; Nagai, Y. Molecular mechanisms and future therapeutics for spinocerebellar ataxia type 31 (SCA31). Neurotherapeutics 2019, 16, 1106–1114. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-C.; Zhang, Y.; Umoh, M.E.; Vaughan, S.W.; Lorenzini, I.; Liu, F.; Sayegh, M.; Donlin-Asp, P.G.; Chen, Y.H.; Duong, D.M. TDP-43 pathology disrupts nuclear pore complexes and nucleocytoplasmic transport in ALS/FTD. Nat. Neurosci. 2018, 21, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Grima, J.C.; Daigle, J.G.; Arbez, N.; Cunningham, K.C.; Zhang, K.; Ochaba, J.; Geater, C.; Morozko, E.; Stocksdale, J.; Glatzer, J.C. Mutant huntingtin disrupts the nuclear pore complex. Neuron 2017, 94, 93–107.e6. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, M.E.; Novelletto, A.; Lin, C.; Tagle, D.; Barnes, G.; Bates, G.; Taylor, S.; Allitto, B.; Altherr, M.; Myers, R. The Huntington’s disease candidate region exhibits many different haplotypes. Nat. Genet. 1992, 1, 99–103. [Google Scholar] [CrossRef] [PubMed]
- Simon-Sanchez, J.; Schulte, C.; Bras, J.M.; Sharma, M.; Gibbs, J.R.; Berg, D.; Paisan-Ruiz, C.; Lichtner, P.; Scholz, S.W.; Hernandez, D.G. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009, 41, 1308–1312. [Google Scholar] [CrossRef]
- Romanul, F.C.; Fowler, H.L.; Radvany, J.; Feldman, R.G.; Feingold, M. Azorean disease of the nervous system. N. Engl. J. Med. 1977, 296, 1505–1508. [Google Scholar] [CrossRef]
- Morita, M.; Al-Chalabi, A.; Andersen, P.; Hosler, B.; Sapp, P.; Englund, E.; Mitchell, J.; Habgood, J.; De Belleroche, J.; Xi, J. A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology 2006, 66, 839–844. [Google Scholar] [CrossRef]
- Chandran, J.S.; Scarrott, J.M.; Shaw, P.J.; Azzouz, M. Gene therapy in the nervous system: Failures and successes. Pers. Med. 2017, 1007, 241–257. [Google Scholar]
- Seidel, K.; Schöls, L.; Nuber, S.; Petrasch-Parwez, E.; Gierga, K.; Wszolek, Z.; Dickson, D.; Gai, W.P.; Bornemann, A.; Riess, O. First appraisal of brain pathology owing to A30P mutant alpha-synuclein. Ann. Neurol. 2010, 67, 684–689. [Google Scholar] [CrossRef]
- Metzger, S.; Rong, J.; Nguyen, H.-P.; Cape, A.; Tomiuk, J.; Soehn, A.S.; Propping, P.; Freudenberg-Hua, Y.; Freudenberg, J.; Tong, L.; et al. Huntingtin-associated protein-1 is a modifier of the age-at-onset of Huntington’s disease. Hum. Mol. Genet. 2008, 17, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Juliano, R.L. Chemical Manipulation of the Endosome Trafficking Machinery: Implications for Oligonucleotide Delivery. Biomedicines 2021, 9, 512. [Google Scholar] [CrossRef]
- Crooke, S.T.; Wang, S.; Vickers, T.A.; Shen, W.; Liang, X.-h. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017, 35, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 46–51. [Google Scholar] [CrossRef]
- Wan, W.B.; Seth, P.P. The Medicinal Chemistry of Therapeutic Oligonucleotides. J. Med. Chem. 2016, 59, 9645–9667. [Google Scholar] [CrossRef]
- Seth, P.P.; Siwkowski, A.; Allerson, C.R.; Vasquez, G.; Lee, S.; Prakash, T.P.; Wancewicz, E.V.; Witchell, D.; Swayze, E.E. Short Antisense Oligonucleotides with Novel 2′−4′ Conformationaly Restricted Nucleoside Analogues Show Improved Potency without Increased Toxicity in Animals. J. Med. Chem. 2009, 52, 10–13. [Google Scholar] [CrossRef]
- Kurreck, J.; Wyszko, E.; Gillen, C.; Erdmann, V.A. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002, 30, 1911–1918. [Google Scholar] [CrossRef] [PubMed]
- Lu-Nguyen, N.; Malerba, A.; Herath, S.; Dickson, G.; Popplewell, L. Systemic antisense therapeutics inhibiting DUX4 expression ameliorates FSHD-like pathology in an FSHD mouse model. Hum. Mol. Genet. 2021, 30, 1398–1412. [Google Scholar] [CrossRef]
- Synofzik, M.; van Roon-Mom, W.M.; Marckmann, G.; van Duyvenvoorde, H.A.; Graessner, H.; Schüle, R.; Aartsma-Rus, A. Preparing n-of-1 antisense oligonucleotide treatments for rare neurological diseases in Europe: Genetic, regulatory, and ethical perspectives. Nucleic Acid Ther. 2022, 32, 83–94. [Google Scholar] [CrossRef]
- Crooke, S.T. Molecular mechanisms of antisense oligonucleotides. Nucleic Acid Ther. 2017, 27, 70–77. [Google Scholar] [CrossRef]
- Lim, K.R.Q.; Yokota, T. Invention and early history of gapmers. Gapmers 2020, 2176, 3–19. [Google Scholar]
- Lima, W.; Wu, H.; Crooke, S.T. The RNase H mechanism. Antisense Drug Technol. Princ. Strateg. Appl. 2007, 2, 47–74. [Google Scholar]
- Liang, X.-H.; Sun, H.; Nichols, J.G.; Crooke, S.T. RNase H1-dependent antisense oligonucleotides are robustly active in directing RNA cleavage in both the cytoplasm and the nucleus. Mol. Ther. 2017, 25, 2075–2092. [Google Scholar] [CrossRef]
- Freier, S.M.; Altmann, K.H. The ups and downs of nucleic acid duplex stability: Structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997, 25, 4429–4443. [Google Scholar] [CrossRef]
- Migawa, M.T.; Shen, W.; Wan, W.B.; Vasquez, G.; Oestergaard, M.E.; Low, A.; De Hoyos, C.L.; Gupta, R.; Murray, S.; Tanowitz, M.; et al. Site-specific replacement of phosphorothioate with alkyl phosphonate linkages enhances the therapeutic profile of gapmer ASOs by modulating interactions with cellular proteins. Nucleic Acids Res. 2019, 47, 5465–5479. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; De Hoyos, C.L.; Migawa, M.T.; Vickers, T.A.; Sun, H.; Low, A.; Bell, T.A., 3rd; Rahdar, M.; Mukhopadhyay, S.; Hart, C.E.; et al. Chemical modification of PS-ASO therapeutics reduces cellular protein-binding and improves the therapeutic index. Nat. Biotechnol. 2019, 37, 640–650. [Google Scholar] [CrossRef]
- Østergaard, M.E.; De Hoyos, C.L.; Wan, W.B.; Shen, W.; Low, A.; Berdeja, A.; Vasquez, G.; Murray, S.; Migawa, M.T.; Liang, X.-h.; et al. Understanding the effect of controlling phosphorothioate chirality in the DNA gap on the potency and safety of gapmer antisense oligonucleotides. Nucleic Acids Res. 2020, 48, 1691–1700. [Google Scholar] [CrossRef]
- Stein, D.; Foster, E.; Huang, S.B.; Weller, D.; Summerton, J. A specificity comparison of four antisense types: Morpholino, 2’-O-methyl RNA, DNA, and phosphorothioate DNA. Antisense Nucleic Acid Drug Dev. 1997, 7, 151–157. [Google Scholar] [CrossRef]
- Miroshnichenko, S.K.; Patutina, O.A.; Burakova, E.A.; Chelobanov, B.P.; Fokina, A.A.; Vlassov, V.V.; Altman, S.; Zenkova, M.A.; Stetsenko, D.A. Mesyl phosphoramidate antisense oligonucleotides as an alternative to phosphorothioates with improved biochemical and biological properties. Proc. Natl. Acad. Sci. USA 2019, 116, 1229–1234. [Google Scholar] [CrossRef]
- Anderson, B.A.; Freestone, G.C.; Low, A.; De-Hoyos, C.L.; Iii, W.J.D.; Østergaard, M.E.; Migawa, M.T.; Fazio, M.; Wan, W.B.; Berdeja, A. Towards next generation antisense oligonucleotides: Mesylphosphoramidate modification improves therapeutic index and duration of effect of gapmer antisense oligonucleotides. Nucleic Acids Res. 2021, 49, 9026–9041. [Google Scholar] [CrossRef]
- Lomzov, A.A.; Kupryushkin, M.S.; Shernyukov, A.V.; Nekrasov, M.D.; Dovydenko, I.S.; Stetsenko, D.A.; Pyshnyi, D.V. Diastereomers of a mono-substituted phosphoryl guanidine trideoxyribonucleotide: Isolation and properties. Biochem. Biophys. Res. Commun. 2019, 513, 807–811. [Google Scholar] [CrossRef] [PubMed]
- Kupryushkin, M.S.; Filatov, A.V.; Mironova, N.L.; Patutina, O.A.; Chernikov, I.V.; Chernolovskaya, E.L.; Zenkova, M.A.; Pyshnyi, D.V.; Stetsenko, D.A.; Altman, S. Antisense oligonucleotide gapmers containing phosphoryl guanidine groups reverse MDR1-mediated multiple drug resistance of tumor cells. Mol. Ther.-Nucleic Acids 2022, 27, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Dyudeeva, E.; Kupryushkin, M.; Lomzov, A.; Pyshnaya, B.; Pyshnyi, D. Physicochemical properties of the phosphoryl guanidine oligodeoxyribonucleotide analogs. Russ. J. Bioorg. Chem. 2019, 45, 709–718. [Google Scholar] [CrossRef]
- Skvortsova, Y.V.; Salina, E.G.; Burakova, E.A.; Bychenko, O.S.; Stetsenko, D.A.; Azhikina, T.L. A new antisense phosphoryl guanidine oligo-2′-O-methylribonucleotide penetrates into intracellular mycobacteria and suppresses target gene expression. Front. Pharmacol. 2019, 10, 1049. [Google Scholar] [CrossRef] [PubMed]
- Bondensgaard, K.; Petersen, M.; Singh, S.K.; Rajwanshi, V.K.; Kumar, R.; Wengel, J.; Jacobsen, J.P. Structural studies of LNA: RNA duplexes by NMR: Conformations and implications for RNase H activity. Chem.-Eur. J. 2000, 6, 2687–2695. [Google Scholar] [CrossRef]
- Stanton, R.; Sciabola, S.; Salatto, C.; Weng, Y.; Moshinsky, D.; Little, J.; Walters, E.; Kreeger, J.; DiMattia, D.; Chen, T. Chemical modification study of antisense gapmers. Nucleic Acid Ther. 2012, 22, 344–359. [Google Scholar] [CrossRef]
- Singh, S.K.; Koshkin, A.A.; Wengel, J.; Nielsen, P. LNA (locked nucleic acids): Synthesis and high-affinity nucleic acid recognition. Chem. Commun. 1998, 455–456. [Google Scholar] [CrossRef]
- Koshkin, A.A.; Singh, S.K.; Nielsen, P.; Rajwanshi, V.K.; Kumar, R.; Meldgaard, M.; Olsen, C.E.; Wengel, J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 1998, 54, 3607–3630. [Google Scholar] [CrossRef]
- Morita, K.; Hasegawa, C.; Kaneko, M.; Tsutsumi, S.; Sone, J.; Ishikawa, T.; Imanishi, T.; Koizumi, M. 2′-O, 4′-C-ethylene-bridged nucleic acids (ENA): Highly nuclease-resistant and thermodynamically stable oligonucleotides for antisense drug. Bioorg. Med. Chem. Lett. 2002, 12, 73–76. [Google Scholar] [CrossRef]
- Obika, S.; Nanbu, D.; Hari, Y.; Andoh, J.-i.; Morio, K.-i.; Doi, T.; Imanishi, T. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O, 4′-C-methyleneribonucleosides. Tetrahedron Lett. 1998, 39, 5401–5404. [Google Scholar] [CrossRef]
- Seth, P.P.; Vasquez, G.; Allerson, C.A.; Berdeja, A.; Gaus, H.; Kinberger, G.A.; Prakash, T.P.; Migawa, M.T.; Bhat, B.; Swayze, E.E. Synthesis and biophysical evaluation of 2′, 4′-constrained 2′ O-methoxyethyl and 2′, 4′-constrained 2′ O-ethyl nucleic acid analogues. J. Org. Chem. 2010, 75, 1569–1581. [Google Scholar] [CrossRef] [PubMed]
- Egli, M.; Minasov, G.; Tereshko, V.; Pallan, P.S.; Teplova, M.; Inamati, G.B.; Lesnik, E.A.; Owens, S.R.; Ross, B.S.; Prakash, T.P.; et al. Probing the influence of stereoelectronic effects on the biophysical properties of oligonucleotides: Comprehensive analysis of the RNA affinity, nuclease resistance, and crystal structure of ten 2’-O-ribonucleic acid modifications. Biochemistry 2005, 44, 9045–9057. [Google Scholar] [CrossRef] [PubMed]
- Teplova, M.; Minasov, G.; Tereshko, V.; Inamati, G.B.; Cook, P.D.; Manoharan, M.; Egli, M. Crystal structure and improved antisense properties of 2’-O-(2-methoxyethyl)-RNA. Nat. Struct. Biol. 1999, 6, 535–539. [Google Scholar] [CrossRef]
- Østergaard, M.E.; Southwell, A.L.; Kordasiewicz, H.; Watt, A.T.; Skotte, N.H.; Doty, C.N.; Vaid, K.; Villanueva, E.B.; Swayze, E.E.; Frank Bennett, C. Rational design of antisense oligonucleotides targeting single nucleotide polymorphisms for potent and allele selective suppression of mutant Huntingtin in the CNS. Nucleic Acids Res. 2013, 41, 9634–9650. [Google Scholar] [CrossRef] [PubMed]
- Seitz, O. Chemically Modified Antisense Oligonucleotides—Recent Improvements of RNA Binding and Ribonuclease H Recruitment. Angew. Chem. Int. Ed. 1999, 38, 3466–3469. [Google Scholar] [CrossRef]
- Ortega, J.-A.; Blas, J.R.; Orozco, M.; Grandas, A.; Pedroso, E.; Robles, J. Binding affinities of oligonucleotides and PNAs containing phenoxazine and G-clamp cytosine analogues are unusually sequence-dependent. Org. Lett. 2007, 9, 4503–4506. [Google Scholar] [CrossRef]
- Froehler, B.C.; Wadwani, S.; Terhorst, T.J.; Gerrard, S.R. Oligodeoxynucleotides containing C-5 propyne analogs of 2′-deoxyuridine and 2′-deoxycytidine. Tetrahedron Lett. 1992, 33, 5307–5310. [Google Scholar] [CrossRef]
- Herdewijn, P. Heterocyclic modifications of oligonucleotides and antisense technology. Antisense Nucleic Acid Drug Dev. 2000, 10, 297–310. [Google Scholar] [CrossRef]
- Gutierrez, A.J.; Froehler, B.C. RNA duplex formation by oligodeoxynucleotides containing C-5 alkyne and C-5 thiazole substituted deoxyuridine analogs. Tetrahedron Lett. 1996, 37, 3959–3962. [Google Scholar] [CrossRef]
- Østergaard, M.E.; Kumar, P.; Nichols, J.; Watt, A.; Sharma, P.K.; Nielsen, P.; Seth, P.P. Allele-selective inhibition of mutant Huntingtin with 2-thio-and C5-triazolylphenyl-deoxythymidine-modified antisense oligonucleotides. Nucleic Acid Ther. 2015, 25, 266–274. [Google Scholar] [CrossRef]
- Dowdy, S.F. Overcoming cellular barriers for RNA therapeutics. Nat. Biotechnol. 2017, 35, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Vickers, T.A.; Liang, X.-h. Phosphorothioate modified oligonucleotide–protein interactions. Nucleic Acids Res. 2020, 48, 5235–5253. [Google Scholar] [CrossRef] [PubMed]
- Putney, S.D.; Benkovic, S.J.; Schimmel, P.R. A DNA fragment with an alpha-phosphorothioate nucleotide at one end is asymmetrically blocked from digestion by exonuclease III and can be replicated in vivo. Proc. Natl. Acad. Sci. USA 1981, 78, 7350–7354. [Google Scholar] [CrossRef] [PubMed]
- Laurent, Q.; Martinent, R.; Moreau, D.; Winssinger, N.; Sakai, N.; Matile, S. Oligonucleotide Phosphorothioates Enter Cells by Thiol-Mediated Uptake. Angew. Chem. 2021, 133, 19250–19254. [Google Scholar] [CrossRef]
- Eckstein, F. Phosphorothioate oligodeoxynucleotides: What is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev. 2000, 10, 117–121. [Google Scholar] [CrossRef]
- Iwamoto, N.; Butler, D.C.; Svrzikapa, N.; Mohapatra, S.; Zlatev, I.; Sah, D.W.; Standley, S.M.; Lu, G.; Apponi, L.H.; Frank-Kamenetsky, M. Control of phosphorothioate stereochemistry substantially increases the efficacy of antisense oligonucleotides. Nat. Biotechnol. 2017, 35, 845–851. [Google Scholar] [CrossRef]
- Chelobanov, B.P.; Burakova, E.A.; Prokhorova, D.V.; Fokina, A.A.; Stetsenko, D.A. New oligodeoxynucleotide derivatives containing N-(methanesulfonyl)-phosphoramidate (mesyl phosphoramidate) internucleotide group. Russ. J. Bioorganic Chem. 2017, 43, 664–668. [Google Scholar] [CrossRef]
- Prokhorova, D.; Chelobanov, B.; Burakova, E.; Fokina, A.; Stetsenko, D. New oligodeoxyribonucleotide derivatives bearing internucleotide N-tosyl phosphoramidate groups: Synthesis and complementary binding to DNA and RNA. Russ. J. Bioorganic Chem. 2017, 43, 38–42. [Google Scholar] [CrossRef]
- Zhang, L.; Liang, X.-h.; De Hoyos, C.L.; Migawa, M.; Nichols, J.G.; Freestone, G.; Tian, J.; Seth, P.P.; Crooke, S.T. The Combination of Mesyl-Phosphoramidate Inter-Nucleotide Linkages and 2′-O-Methyl in Selected Positions in the Antisense Oligonucleotide Enhances the Performance of RNaseH1 Active PS-ASOs. Nucleic Acid Ther. 2022. [Google Scholar] [CrossRef]
- Hagedorn, P.H.; Brown, J.M.; Easton, A.; Pierdomenico, M.; Jones, K.; Olson, R.E.; Mercer, S.E.; Li, D.; Loy, J.; Høg, A.M.; et al. Acute Neurotoxicity of Antisense Oligonucleotides After Intracerebroventricular Injection Into Mouse Brain Can Be Predicted from Sequence Features. Nucleic Acid Ther. 2022, 32, 151–162. [Google Scholar] [CrossRef]
- Patutina, O.A.; Gaponova, S.K.; Sen’kova, A.V.; Savin, I.A.; Gladkikh, D.V.; Burakova, E.A.; Fokina, A.A.; Maslov, M.A.; Shmendel’, E.V.; Wood, M.J. Mesyl phosphoramidate backbone modified antisense oligonucleotides targeting miR-21 with enhanced in vivo therapeutic potency. Proc. Natl. Acad. Sci. USA 2020, 117, 32370–32379. [Google Scholar] [CrossRef] [PubMed]
- Wave Life Science Provides Update on Phase 1b/2a PRECISION-HD Trials. Available online: https://ir.wavelifesciences.com/node/9126/pdf (accessed on 23 June 2022).
- Liu, Y.; Dodart, J.-C.; Tran, H.; Berkovitch, S.; Braun, M.; Byrne, M.; Durbin, A.F.; Hu, X.S.; Iwamoto, N.; Jang, H.G. Variant-selective stereopure oligonucleotides protect against pathologies associated with C9orf72-repeat expansion in preclinical models. Nat. Commun. 2021, 12, 847. [Google Scholar] [CrossRef] [PubMed]
- Kandasamy, P.; McClorey, G.; Shimizu, M.; Kothari, N.; Alam, R.; Iwamoto, N.; Kumarasamy, J.; Bommineni, G.R.; Bezigian, A.; Chivatakarn, O. Control of backbone chemistry and chirality boost oligonucleotide splice switching activity. Nucleic Acids Res. 2022, 50, 5443–5466. [Google Scholar] [CrossRef]
- Brunet de Courssou, J.-B.; Durr, A.; Adams, D.; Corvol, J.-C.; Mariani, L.-L. Antisense therapies in neurological diseases. Brain 2022, 145, 816–831. [Google Scholar] [CrossRef]
- Kupryushkin, M.; Pyshnyi, D.; Stetsenko, D. Phosphoryl guanidines: A new type of nucleic acid analogues. Acta Nat. 2014, 6, 116–118. [Google Scholar] [CrossRef]
- Toulme, J.; Di Primo, C.; Moreau, S. Modulation of RNA function by oligonucleotides recognizing RNA structure. Prog. Nucleic Acid Res. Mol. Biol. 2001, 69, 1–46. [Google Scholar]
- Lima, W.F.; Crooke, S.T. Binding affinity and specificity of Escherichia coli RNase H1: Impact on the kinetics of catalysis of antisense Oligonucleotide—RNA hybrids. Biochemistry 1997, 36, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Hayase, Y.; Iwai, S.; Ohtsuka, E. Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett. 1987, 215, 327–330. [Google Scholar] [CrossRef]
- Burmeister, P.E.; Lewis, S.D.; Silva, R.F.; Preiss, J.R.; Horwitz, L.R.; Pendergrast, P.S.; McCauley, T.G.; Kurz, J.C.; Epstein, D.M.; Wilson, C.; et al. Direct in vitro selection of a 2’-O-methyl aptamer to VEGF. Chem. Biol. 2005, 12, 25–33. [Google Scholar] [CrossRef] [PubMed]
- De Mesmaeker, A.; Haener, R.; Martin, P.; Moser, H.E. Antisense oligonucleotides. Acc. Chem. Res. 1995, 28, 366–374. [Google Scholar] [CrossRef]
- Hoy, S.M. Nusinersen: First Global Approval. Drugs 2017, 77, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Khorkova, O.; Hsiao, J.; Wahlestedt, C. Nucleic Acid–Based Therapeutics in Orphan Neurological Disorders: Recent Developments. Front. Mol. Biosci. 2021, 8, 643681. [Google Scholar] [CrossRef] [PubMed]
- Schobel, S.A. Preliminary results from GENERATION HD1, a phase III trial of tominersen in individuals with manifest HD. In Proceedings of the CHDI 16th Annual HD Therapeutics Conference, Online, 27–29 April 2021. [Google Scholar]
- Obika, S.; Nanbu, D.; Hari, Y.; Morio, K.-i.; In, Y.; Ishida, T.; Imanishi, T. Synthesis of 2′-O, 4′-C-methyleneuridine and-cytidine. Novel bicyclic nucleosides having a fixed C3,-endo sugar puckering. Tetrahedron Lett. 1997, 38, 8735–8738. [Google Scholar] [CrossRef]
- Nielsen, K.E.; Rasmussen, J.; Kumar, R.; Wengel, J.; Jacobsen, J.P.; Petersen, M. NMR studies of fully modified locked nucleic acid (LNA) hybrids: Solution structure of an LNA: RNA hybrid and characterization of an LNA: DNA hybrid. Bioconjug. Chem. 2004, 15, 449–457. [Google Scholar] [CrossRef]
- Swayze, E.E.; Siwkowski, A.M.; Wancewicz, E.V.; Migawa, M.T.; Wyrzykiewicz, T.K.; Hung, G.; Monia, B.P.; Bennett, C.F. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatotoxicity in animals. Nucleic Acids Res. 2007, 35, 687–700. [Google Scholar] [CrossRef]
- Seth, P.P.; Siwkowski, A.; Allerson, C.R.; Vasquez, G.; Lee, S.; Prakash, T.P.; Kinberger, G.; Migawa, M.T.; Gaus, H.; Bhat, B.; et al. Design, synthesis and evaluation of constrained methoxyethyl (cMOE) and constrained ethyl (cEt) nucleoside analogs. Nucleic Acids Symp. Ser. 2008, 553–554. [Google Scholar] [CrossRef]
- Krieg, A.M. Antiinfective applications of toll-like receptor 9 agonists. Proc. Am. Thorac. Soc. 2007, 4, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Varizhuk, A.M.; Zatsepin, T.S.; Golovin, A.V.; Belyaev, E.S.; Kostyukevich, Y.I.; Dedkov, V.G.; Shipulin, G.A.; Shpakovski, G.V.; Aralov, A.V. Synthesis of oligonucleotides containing novel G-clamp analogue with C8-tethered group in phenoxazine ring: Implication to qPCR detection of the low-copy Kemerovo virus dsRNA. Bioorg. Med. Chem. 2017, 25, 3597–3605. [Google Scholar] [CrossRef]
- Egli, M.; Manoharan, M. Re-engineering RNA molecules into therapeutic agents. Acc. Chem. Res. 2019, 52, 1036–1047. [Google Scholar] [CrossRef]
- Wilds, C.J.; Maier, M.A.; Tereshko, V.; Manoharan, M.; Egli, M. Direct observation of a cytosine analogue that forms five hydrogen bonds to guanosine: Guanidino G-clamp. Angew. Chem. Int. Ed. 2002, 41, 115–117. [Google Scholar] [CrossRef]
- Tabrizi, S.J.; Leavitt, B.R.; Landwehrmeyer, G.B.; Wild, E.J.; Saft, C.; Barker, R.A.; Blair, N.F.; Craufurd, D.; Priller, J.; Rickards, H. Targeting huntingtin expression in patients with Huntington’s disease. N. Engl. J. Med. 2019, 380, 2307–2316. [Google Scholar] [CrossRef] [PubMed]
- Stoker, T.B.; Andresen, K.E.; Barker, R.A. Hydrocephalus complicating intrathecal antisense oligonucleotide therapy for Huntington’s disease. Mov. Disord. 2021, 36, 263. [Google Scholar] [CrossRef] [PubMed]
- Kwon, D. Failure of genetic therapies for Huntington’s devastates community. Nature 2021, 593, 180. [Google Scholar] [CrossRef] [PubMed]
- Arnold, C. Record number of gene-therapy trials, despite setbacks. Nat. Med. 2021, 27, 1312–1315. [Google Scholar] [CrossRef]
- Bečanović, K.; Nørremølle, A.; Neal, S.J.; Kay, C.; Collins, J.A.; Arenillas, D.; Lilja, T.; Gaudenzi, G.; Manoharan, S.; Doty, C.N.; et al. A SNP in the HTT promoter alters NF-κB binding and is a bidirectional genetic modifier of Huntington disease. Nat. Neurosci. 2015, 18, 807–816. [Google Scholar] [CrossRef]
- Moore, L.R.; Keller, L.; Bushart, D.D.; Delatorre, R.G.; Li, D.; McLoughlin, H.S.; do Carmo Costa, M.; Shakkottai, V.G.; Smith, G.D.; Paulson, H.L. Antisense oligonucleotide therapy rescues aggresome formation in a novel spinocerebellar ataxia type 3 human embryonic stem cell line. Stem Cell Res. 2019, 39, 101504. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.R.; Rajpal, G.; Dillingham, I.T.; Qutob, M.; Blumenstein, K.G.; Gattis, D.; Hung, G.; Kordasiewicz, H.B.; Paulson, H.L.; McLoughlin, H.S. Evaluation of antisense oligonucleotides targeting ATXN3 in SCA3 mouse models. Mol. Ther.-Nucleic Acids 2017, 7, 200–210. [Google Scholar] [CrossRef]
- McLoughlin, H.S.; Moore, L.R.; Chopra, R.; Komlo, R.; McKenzie, M.; Blumenstein, K.G.; Zhao, H.; Kordasiewicz, H.B.; Shakkottai, V.G.; Paulson, H.L. Oligonucleotide therapy mitigates disease in spinocerebellar ataxia type 3 mice. Ann. Neurol. 2018, 84, 64–77. [Google Scholar] [CrossRef]
- Winklhofer, K.F.; Tatzelt, J.; Haass, C. The two faces of protein misfolding: Gain-and loss-of-function in neurodegenerative diseases. EMBO J. 2008, 27, 336–349. [Google Scholar] [CrossRef]
- Toonen, L.J.; Rigo, F.; van Attikum, H.; van Roon-Mom, W.M. Antisense oligonucleotide-mediated removal of the polyglutamine repeat in spinocerebellar ataxia type 3 mice. Mol. Ther.-Nucleic Acids 2017, 8, 232–242. [Google Scholar] [CrossRef]
- Evers, M.M.; Pepers, B.A.; van Deutekom, J.C.; Mulders, S.A.; den Dunnen, J.T.; Aartsma-Rus, A.; van Ommen, G.-J.B.; van Roon-Mom, W.M. Targeting several CAG expansion diseases by a single antisense oligonucleotide. PLoS ONE 2011, 6, e24308. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Matsui, M.; Gagnon, K.T.; Schwartz, J.C.; Gabillet, S.; Arar, K.; Wu, J.; Bezprozvanny, I.; Corey, D.R. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009, 27, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, K.T.; Pendergraff, H.M.; Deleavey, G.F.; Swayze, E.E.; Potier, P.; Randolph, J.; Roesch, E.B.; Chattopadhyaya, J.; Damha, M.J.; Bennett, C.F.; et al. Allele-Selective Inhibition of Mutant Huntingtin Expression with Antisense Oligonucleotides Targeting the Expanded CAG Repeat. Biochemistry 2010, 49, 10166–10178. [Google Scholar] [CrossRef]
- Kourkouta, E.; Weij, R.; González-Barriga, A.; Mulder, M.; Verheul, R.; Bosgra, S.; Groenendaal, B.; Puoliväli, J.; Toivanen, J.; van Deutekom, J.C. Suppression of mutant protein expression in SCA3 and SCA1 mice using a CAG repeat-targeting antisense oligonucleotide. Mol. Ther.-Nucleic Acids 2019, 17, 601–614. [Google Scholar] [CrossRef]
- Saudou, F.; Humbert, S. The biology of huntingtin. Neuron 2016, 89, 910–926. [Google Scholar] [CrossRef] [PubMed]
- Freier, S.M. Compositions and Their Uses Directed to Huntingtin. U.S. Patent 8952145B2, 10 February 2015. [Google Scholar]
- Brown, J.M.; Berkovitch, S.S.-M.; Iwamoto, N.; Vargeese, C.; Aklilu, K.M.; Frank-Kamenetsky, M.D.; Brown, D.P. Oligonucleotide Composistions and Methods Thereof. U.S. Patent 20220098585A1, 30 January 2020. [Google Scholar]
- Nolan, M.; Talbot, K.; Ansorge, O. Pathogenesis of FUS-associated ALS and FTD: Insights from rodent models. Acta Neuropathol. Commun. 2016, 4, 99. [Google Scholar] [CrossRef]
- Conte, A.; Lattante, S.; Zollino, M.; Marangi, G.; Luigetti, M.; Del Grande, A.; Servidei, S.; Trombetta, F.; Sabatelli, M. P525L FUS mutation is consistently associated with a severe form of juvenile amyotrophic lateral sclerosis. Neuromuscul. Disord. 2012, 22, 73–75. [Google Scholar] [CrossRef]
- Zhang, W.; Jiao, B.; Xiao, T.; Liu, X.; Liao, X.; Xiao, X.; Guo, L.; Yuan, Z.; Yan, X.; Tang, B.; et al. Association of rare variants in neurodegenerative genes with familial Alzheimer’s disease. Ann. Clin. Transl. Neurol. 2020, 7, 1985–1995. [Google Scholar] [CrossRef]
- Korobeynikov, V.A.; Lyashchenko, A.K.; Blanco-Redondo, B.; Jafar-Nejad, P.; Shneider, N.A. Antisense oligonucleotide silencing of FUS expression as a therapeutic approach in amyotrophic lateral sclerosis. Nat. Med. 2022, 28, 104–116. [Google Scholar] [CrossRef]
- McCord, J.M.; Fridovich, I. Superoxide dismutase: An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055. [Google Scholar] [CrossRef]
- Miller, T.; Cudkowicz, M.; Shaw, P.J.; Andersen, P.M.; Atassi, N.; Bucelli, R.C.; Genge, A.; Glass, J.; Ladha, S.; Ludolph, A.L.; et al. Phase 1–2 Trial of Antisense Oligonucleotide Tofersen for SOD1 ALS. N. Engl. J. Med. 2020, 383, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.F.; Kenneth, W.D. Antisense Modulation of Superoxide Dismutase 1, Soluble Expression. U.S. Patent 8993529B2, 21 June 2011. [Google Scholar]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.A.; Flynn, H.; Adamson, J. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Wave_Life_Sciences. Available online: https://ir.wavelifesciences.com/static-files/4d0a51d3-35bc-471e-b5d8-b395ac5c7702 (accessed on 24 May 2022).
- Liu, Y.; Andreucci, A.; Iwamoto, N.; Yin, Y.; Yang, H.; Liu, F.; Bulychev, A.; Hu, X.S.; Lin, X.; Lamore, S. Preclinical evaluation of WVE-004, aninvestigational stereopure oligonucleotide forthe treatment of C9orf72-associated ALS or FTD. Mol. Ther.-Nucleic Acids 2022, 28, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, M.; Golatta, M.; Wüllner, U.; Lengauer, T. Structural and functional analysis of ataxin-2 and ataxin-3. Eur. J. Biochem. 2004, 271, 3155–3170. [Google Scholar] [CrossRef]
- IONIS. Available online: https://www.ionispharma.com/ionis-innovation/pipeline/ (accessed on 24 May 2022).
- Rademakers, R.; Cruts, M.; Van Broeckhoven, C. The role of tau (MAPT) in frontotemporal dementia and related tauopathies. Hum. Mutat. 2004, 24, 277–295. [Google Scholar] [CrossRef]
- Lane, R.M.; Kordasiewicz, H.B.; Smith, A.; Mignon, L.; Miller, T.; Narayanan, P.; Swayze, E.; Norris, D.; Fitzsimmons, B.; Bennett, C.F. Rationale for, and Development of, Ionis-MAPTRx, the First Tau-Lowering Antisense Oligonucleotide, in Patients with Mild AD. In Proceedings of the 142nd Annual Meeting of the American Neurological Association, San Diego, CA, USA, 15–17 October 2017; p. S151. [Google Scholar]
- Usmani, A.; Shavarebi, F.; Hiniker, A. The Cell Biology of LRRK2 in Parkinson’s Disease. Mol. Cell. Biol. 2021, 41, e00660-20. [Google Scholar] [CrossRef]
- Zhao, H.T.; John, N.; Delic, V.; Ikeda-Lee, K.; Kim, A.; Weihofen, A.; Swayze, E.E.; Kordasiewicz, H.B.; West, A.B.; Volpicelli-Daley, L.A. LRRK2 antisense oligonucleotides ameliorate α-synuclein inclusion formation in a Parkinson’s disease mouse model. Mol. Ther.-Nucleic Acids 2017, 8, 508–519. [Google Scholar] [CrossRef]
- Burré, J.; Sharma, M.; Tsetsenis, T.; Buchman, V.; Etherton, M.R.; Südhof, T.C. α-Synuclein Promotes SNARE-Complex Assembly in Vivo and in Vitro. Science 2010, 329, 1663–1667. [Google Scholar] [CrossRef]
- Alarcón-Arís, D.; Recasens, A.; Galofré, M.; Carballo-Carbajal, I.; Zacchi, N.; Ruiz-Bronchal, E.; Pavia-Collado, R.; Chica, R.; Ferrés-Coy, A.; Santos, M. Selective α-synuclein knockdown in monoamine neurons by intranasal oligonucleotide delivery: Potential therapy for Parkinson’s disease. Mol. Ther. 2018, 26, 550–567. [Google Scholar] [CrossRef]
- Hagemann, T.L. Alexander disease: Models, mechanisms, and medicine. Curr. Opin. Neurobiol. 2022, 72, 140–147. [Google Scholar] [CrossRef] [PubMed]
- Hagemann, T.L.; Powers, B.; Mazur, C.; Kim, A.; Wheeler, S.; Hung, G.; Swayze, E.; Messing, A. Antisense suppression of glial fibrillary acidic protein as a treatment for Alexander disease. Ann. Neurol. 2018, 83, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.A.; Miller, T.M.; Yamanaka, K.; Monia, B.P.; Condon, T.P.; Hung, G.; Lobsiger, C.S.; Ward, C.M.; McAlonis-Downes, M.; Wei, H.; et al. Antisense oligonucleotide therapy for neurodegenerative disease. J. Clin. Investig. 2006, 116, 2290–2296. [Google Scholar] [CrossRef]
- Donnelly, C.J.; Zhang, P.-W.; Pham, J.T.; Haeusler, A.R.; Mistry, N.A.; Vidensky, S.; Daley, E.L.; Poth, E.M.; Hoover, B.; Fines, D.M. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 2013, 80, 415–428. [Google Scholar] [CrossRef]
- Tran, H.; Moazami, M.P.; Yang, H.; McKenna-Yasek, D.; Douthwright, C.L.; Pinto, C.; Metterville, J.; Shin, M.; Sanil, N.; Dooley, C. Suppression of mutant C9orf72 expression by a potent mixed backbone antisense oligonucleotide. Nat. Med. 2022, 28, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Miller, T.M.; Pestronk, A.; David, W.; Rothstein, J.; Simpson, E.; Appel, S.H.; Andres, P.L.; Mahoney, K.; Allred, P.; Alexander, K. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: A phase 1, randomised, first-in-man study. Lancet Neurol. 2013, 12, 435–442. [Google Scholar] [CrossRef]
- DeVos, S.L.; Miller, R.L.; Schoch, K.M.; Holmes, B.B.; Kebodeaux, C.S.; Wegener, A.J.; Chen, G.; Shen, T.; Tran, H.; Nichols, B.; et al. Tau reduction prevents neuronal loss and reverses pathological tau deposition and seeding in mice with tauopathy. Sci. Transl. Med. 2017, 9, eaag0481. [Google Scholar] [CrossRef]
- Mignon, L.; Kordasiewicz, H.; Lane, R.; Smith, A.; Miller, T.; Narayanan, P.; Swayze, E.; Norris, D.; Fitzsimmons, B.; Bennett, F. Design of the first-in-human study of IONIS-MAPTRx, a tau-lowering antisense oligonucleotide, in patients with Alzheimer disease (S2. 006). Neurology 2018, 90, S2.006. [Google Scholar]
- Chang, J.L.; Hinrich, A.J.; Roman, B.; Norrbom, M.; Rigo, F.; Marr, R.A.; Norstrom, E.M.; Hastings, M.L. Targeting amyloid-β precursor protein, APP, splicing with antisense oligonucleotides reduces toxic amyloid-β production. Mol. Ther. 2018, 26, 1539–1551. [Google Scholar] [CrossRef]
- Hinrich, A.J.; Jodelka, F.M.; Chang, J.L.; Brutman, D.; Bruno, A.M.; Briggs, C.A.; James, B.D.; Stutzmann, G.E.; Bennett, D.A.; Miller, S.A.; et al. Therapeutic correction of ApoER2 splicing in Alzheimer’s disease mice using antisense oligonucleotides. EMBO Mol. Med. 2016, 8, 328–345. [Google Scholar] [CrossRef]
- Uehara, T.; Choong, C.-J.; Nakamori, M.; Hayakawa, H.; Nishiyama, K.; Kasahara, Y.; Baba, K.; Nagata, T.; Yokota, T.; Tsuda, H.; et al. Amido-bridged nucleic acid (AmNA)-modified antisense oligonucleotides targeting α-synuclein as a novel therapy for Parkinson’s disease. Sci. Rep. 2019, 9, 7567. [Google Scholar] [CrossRef] [PubMed]
- Cole, T.A.; Zhao, H.; Collier, T.J.; Sandoval, I.; Sortwell, C.E.; Steece-Collier, K.; Daley, B.F.; Booms, A.; Lipton, J.; Welch, M. α-Synuclein antisense oligonucleotides as a disease-modifying therapy for Parkinson’s disease. JCI Insight 2021, 6, e135633. [Google Scholar] [CrossRef] [PubMed]
- Carroll, J.B.; Warby, S.C.; Southwell, A.L.; Doty, C.N.; Greenlee, S.; Skotte, N.; Hung, G.; Bennett, C.F.; Freier, S.M.; Hayden, M.R. Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene/allele-specific silencing of mutant huntingtin. Mol. Ther. 2011, 19, 2178–2185. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.F.; Hayden, M.R.; Freier, S.M.; Greenlee, S.; Carroll, J.B.; Warby, S.C.; Swayze, E.E. Selective Reduction of Allelic Variants. U.S. Patent 20200377946, 14 January 2020. [Google Scholar]
- Southwell, A.L.; Kordasiewicz, H.B.; Langbehn, D.R.; Skotte, N.H.; Parsons, M.P.; Villanueva, E.B.; Caron, N.S.; Østergaard, M.E.; Anderson, L.M.; Xie, Y.; et al. Huntingtin suppression restores cognitive function in a mouse model of Huntington’s disease. Sci. Transl. Med. 2018, 10, eaar3959. [Google Scholar] [CrossRef] [PubMed]
- Southwell, A.L.; Skotte, N.H.; Kordasiewicz, H.B.; Østergaard, M.E.; Watt, A.T.; Carroll, J.B.; Doty, C.N.; Villanueva, E.B.; Petoukhov, E.; Vaid, K. In vivo evaluation of candidate allele-specific mutant huntingtin gene silencing antisense oligonucleotides. Mol. Ther. 2014, 22, 2093–2106. [Google Scholar] [CrossRef]
- Kay, C.; Collins, J.A.; Caron, N.S.; de Andrade Agostinho, L.; Findlay-Black, H.; Casal, L.; Sumathipala, D.; Dissanayake, V.H.; Cornejo-Olivas, M.; Baine, F. A comprehensive haplotype-targeting strategy for allele-specific HTT suppression in Huntington disease. Am. J. Hum. Genet. 2019, 105, 1112–1125. [Google Scholar] [CrossRef]
- Kay, C.; Collins, J.A.; Skotte, N.H.; Southwell, A.L.; Warby, S.C.; Caron, N.S.; Doty, C.N.; Nguyen, B.; Griguoli, A.; Ross, C.J. Huntingtin haplotypes provide prioritized target panels for allele-specific silencing in Huntington disease patients of European ancestry. Mol. Ther. 2015, 23, 1759–1771. [Google Scholar] [CrossRef]
- Shin, J.W.; Shin, A.; Park, S.S.; Lee, J.-M. Haplotype-specific insertion-deletion variations for allele-specific targeting in Huntington’s disease. Mol. Ther.-Methods Clin. Dev. 2022, 25, 84–95. [Google Scholar] [CrossRef]
- Gaspar, C.; Lopes-Cendes, I.; Hayes, S.; Goto, J.; Arvidsson, K.; Dias, A.; Silveira, I.; Maciel, P.; Coutinho, P.; Lima, M. Ancestral origins of the Machado-Joseph disease mutation: A worldwide haplotype study. Am. J. Hum. Genet. 2001, 68, 523–528. [Google Scholar] [CrossRef]
- Machiela, M.J.; Chanock, S.J. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef]
- Gruber, A.R.; Bernhart, S.H.; Lorenz, R. The ViennaRNA web services. In RNA Bioinformatics; Springer: Berlin/Heidelberg, Germany, 2015; pp. 307–326. [Google Scholar]
- Matveeva, O.; Tsodikov, A.; Giddings, M.; Freier, S.; Wyatt, J.; Spiridonov, A.a.; Shabalina, S.; Gesteland, R.; Atkins, J. Identification of sequence motifs in oligonucleotides whose presence is correlated with antisense activity. Nucleic Acids Res. 2000, 28, 2862–2865. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wick, L.M.; Rouillard, J.M.; Whittam, T.S.; Gulari, E.; Tiedje, J.M.; Hashsham, S.A. On-chip non-equilibrium dissociation curves and dissociation rate constants as methods to assess specificity of oligonucleotide probes. Nucleic Acids Res. 2006, 34, e26. [Google Scholar] [CrossRef] [PubMed]
- Naiser, T.; Ehler, O.; Kayser, J.; Mai, T.; Michel, W.; Ott, A. Impact of point-mutations on the hybridization affinity of surface-bound DNA/DNA and RNA/DNA oligonucleotide-duplexes: Comparison of single base mismatches and base bulges. BMC Biotechnol. 2008, 8, 48. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, N.; Abdollahii, S.; Kouhbanani, M.A.J.; Hassanzadeh, A. Induced pluripotent stem cells (iPSCs) as game-changing tools in the treatment of neurodegenerative disease: Mirage or reality? J. Cell. Physiol. 2020, 235, 9166–9184. [Google Scholar] [CrossRef]
- Pasteuning-Vuhman, S.; de Jongh, R.; Timmers, A.; Pasterkamp, R.J. Towards advanced iPSC-based drug development for neurodegenerative disease. Trends Mol. Med. 2021, 27, 263–279. [Google Scholar] [CrossRef]
- Gonçalves, N.; Simões, A.T.; Cunha, R.A.; de Almeida, L.P. Caffeine and adenosine A2A receptor inactivation decrease striatal neuropathology in a lentiviral-based model of Machado–Joseph disease. Ann. Neurol. 2013, 73, 655–666. [Google Scholar] [CrossRef]
- Nobre, R.J.; Lobo, D.D.; Henriques, C.; Duarte, S.P.; Lopes, S.M.; Silva, A.C.; Lopes, M.M.; Mariet, F.; Schwarz, L.K.; Baatje, M. miRNA-Mediated Knockdown of ATXN3 Alleviates Molecular Disease Hallmarks in a Mouse Model for Spinocerebellar Ataxia Type 3. Nucleic Acid Ther. 2022, 32, 194–205. [Google Scholar] [CrossRef]
- Grenier, K.; Kao, J.; Diamandis, P. Three-dimensional modeling of human neurodegeneration: Brain organoids coming of age. Mol. Psychiatry 2020, 25, 254–274. [Google Scholar] [CrossRef]
- Slanzi, A.; Iannoto, G.; Rossi, B.; Zenaro, E.; Constantin, G. In vitro models of neurodegenerative diseases. Front. Cell Dev. Biol. 2020, 8, 328. [Google Scholar] [CrossRef]
- Venkataraman, L.; Fair, S.R.; McElroy, C.A.; Hester, M.E.; Fu, H. Modeling neurodegenerative diseases with cerebral organoids and other three-dimensional culture systems: Focus on Alzheimer’s disease. Stem Cell Rev. Rep. 2022, 18, 696–717. [Google Scholar] [CrossRef]
- Hu, J.; Liu, J.; Narayanannair, K.J.; Lackey, J.G.; Kuchimanchi, S.; Rajeev, K.G.; Manoharan, M.; Swayze, E.E.; Lima, W.F.; Prakash, T.P. Allele-selective inhibition of mutant atrophin-1 expression by duplex and single-stranded RNAs. Biochemistry 2014, 53, 4510–4518. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Gagnon, K.T.; Liu, J.; Watts, J.K.; Syeda-Nawaz, J.; Bennett, C.F.; Swayze, E.E.; Randolph, J.; Chattopadhyaya, J.; Corey, D.R. Allele-selective inhibition of ataxin-3 (ATX3) expression by antisense oligomers and duplex RNAs. Biol. Chem. 2011, 392, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Gonsior, K.; Kaucher, G.A.; Pelz, P.; Schumann, D.; Gansel, M.; Kuhs, S.; Klockgether, T.; Forlani, S.; Durr, A.; Hauser, S. PolyQ-expanded ataxin-3 protein levels in peripheral blood mononuclear cells correlate with clinical parameters in SCA3: A pilot study. J. Neurol. 2021, 268, 1304–1315. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Pu, F.; Huang, Z.; Ren, J.; Qu, X. A Quadruplex-Based, Label-Free, and Real-Time Fluorescence Assay for RNase H Activity and Inhibition. Chem.-Eur. J. 2010, 16, 2605–2610. [Google Scholar] [CrossRef]
- Lee, C.Y.; Jang, H.; Park, K.S.; Park, H.G. A label-free and enzyme-free signal amplification strategy for a sensitive RNase H activity assay. Nanoscale 2017, 9, 16149–16153. [Google Scholar] [CrossRef]
- Kono, N.; Arakawa, K. Nanopore sequencing: Review of potential applications in functional genomics. Dev. Growth Differ. 2019, 61, 316–326. [Google Scholar] [CrossRef]
- Echigoya, Y.; Lim, K.R.Q.; Melo, D.; Bao, B.; Trieu, N.; Mizobe, Y.; Maruyama, R.; Mamchaoui, K.; Tanihata, J.; Aoki, Y. Exons 45–55 skipping using mutation-tailored cocktails of antisense morpholinos in the DMD gene. Mol. Ther. 2019, 27, 2005–2017. [Google Scholar] [CrossRef]

| Modification | Tm per Nucleotide | References | Specific | General |
|---|---|---|---|---|
| Backbone | ||||
| Phosphorothioate (PS) | 0.45 to 1 °C | [27,34,35,36,37,38,39] |
|
|
| Mesylphosphoramidate (MsPA) | 1.3 to +1.1 °C (with respect to PS) | [39,40] |
| |
| Phosphoryl guanidines (PG) | 1.2 to 0 °C | [41,42,43,44] |
| |
| Sugar | ||||
| Locked nucleic acids (LNA) | +1.5 to +9.1 °C | [27,45,46,47,48,49,50,51] |
|
|
| 2’O-methyl (2’-OMe) | 0 to +1.3 °C | [27,34,38,46] | ||
| 2’-O-methoxyethyl (2’-MOE) | +0.9 to +1.9 °C | [34,46,51,52,53] |
| |
| 2’4’-constrained 2’-O-ethyl (2’-cEt) | +4.7 to +6.1 °C | [51,54] |
| |
| 2’-O,4’-C-ethylene-bridged nucleic acid (ENA) | +5.2 °C | [49] |
| |
| Base | ||||
| G-clamp | +4 to +18 °C | [46,55,56] |
|
|
| C5-propyne C | +1.5 to 1.6 °C | [55,57,58,59] |
| |
| C5-propyne T | +0.9 to +2.6 °C | [34,54,55,57,58,59] |
| |
| 2-thio-thymidine | +0.3 to +1.8 °C | [54,60] |
| |
| 5’-thiazole analogues | +1.7 to 2.2 °C | [58,59] |
| |
| 5-Methyl cytosine | 0 to +1.1 °C | [25,34] | ||
| Disease 1 Target | Cellular Function | ASO | Phase | ASO Type/Modifications | Ref./Clinical Trial |
|---|---|---|---|---|---|
| HD–HTT | Brain development, involved in vesicle trafficking and recycling, cell division, ciliogenesis, autophagy, development [107] | Tominersen, IONIS-HTTRx | Phase III halted (03/21) | Non-allele-specific, PS 2′-MOE | [2,84,108] NCT03842969 |
| HD–HTT | WVE-003 (WVE-120101 & 120102: suspended) | Phase I/II | Allele-specific, PS stereopure | [2,72,109] NCT05032196 | |
| ALS/FTD–FUS | DNA/RNA metabolism [110] | Jacifusen/ION36 | Phase III | Mutation-specific (p.P525L), PS 2′-MOE | [111,112,113] NCT04768972 |
| ALS–SOD1 | Antioxidant [114] | Tofersen/IONIS-SOD1Rx (BIIB067) | Phase III | Non-allele-specific, PS 2′-MOE | [115,116] NCT02623699 NCT03070119 |
| ALS/FTD–C9ORF72 | Repeat in noncoding region [117] | IONIS-C9Rx (BIIB078) | Phase I discontinued (03/22) | Non-allele specific, PS 2′-MOE | NCT03626012 NCT04288856 |
| ALS/FTD–C9ORF72 | WVE004 | Phase I/II | Allele-specific (Targeting V1 and V3 transcript), PS PG stereopure | [118,119] NCT04931862 | |
| ALS/SCA2–ATXN2 | RNA metabolism [120] | ION541 (BIIB105) | Phase I/II | PS 2′-MOE | [83,121] NCT04494256 |
| AD/FTD–MAPT (TAU) | Stabilizing & promotion of microtubule assembly [122] | IONIS-MAPTRx (BIIB080) | Phase II | PS 2′-MOE | [123] |
| SCA3–ATXN3 | Deubiquitinase [120] | ION260 (BIIB132) | Phase I | Non-allele-specific, PS 2′-MOE | [100,121] NCT05160558 |
| PD–LRRK2 | Kinase involved in lysosomal processes, autophagy, mitophagy, vesicle trafficking [124] | ION859 (BIIB094) | Phase I/II | PS 2′-MOE | [125] NCT03976349 |
| PD–SNCA | Presynaptic protein, involved in SNARE complex assembly [126] | ION464 (BIIB101) | Phase II | PS 2′-MOE | [127] NCT04165486 |
| Alexander disease–GFAP | Intermediate filament [128] | Zilganersen, ION373 | Phase II | Non-allele-specific, PS 2′-MOE | [129] NCT04849741 CAS2305355-56-8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Helm, J.; Schöls, L.; Hauser, S. Towards Personalized Allele-Specific Antisense Oligonucleotide Therapies for Toxic Gain-of-Function Neurodegenerative Diseases. Pharmaceutics 2022, 14, 1708. https://doi.org/10.3390/pharmaceutics14081708
Helm J, Schöls L, Hauser S. Towards Personalized Allele-Specific Antisense Oligonucleotide Therapies for Toxic Gain-of-Function Neurodegenerative Diseases. Pharmaceutics. 2022; 14(8):1708. https://doi.org/10.3390/pharmaceutics14081708
Chicago/Turabian StyleHelm, Jacob, Ludger Schöls, and Stefan Hauser. 2022. "Towards Personalized Allele-Specific Antisense Oligonucleotide Therapies for Toxic Gain-of-Function Neurodegenerative Diseases" Pharmaceutics 14, no. 8: 1708. https://doi.org/10.3390/pharmaceutics14081708
APA StyleHelm, J., Schöls, L., & Hauser, S. (2022). Towards Personalized Allele-Specific Antisense Oligonucleotide Therapies for Toxic Gain-of-Function Neurodegenerative Diseases. Pharmaceutics, 14(8), 1708. https://doi.org/10.3390/pharmaceutics14081708







