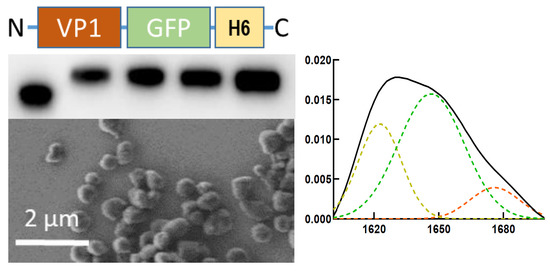
The Poly-Histidine Tag H6 Mediates Structural and Functional Properties of Disintegrating, Protein-Releasing Inclusion Bodies
Abstract
:1. Introduction
2. Materials and Methods
2.1. Proteins and IB Production
2.2. Field Emission Scanning Electron Microscopy (FESEM)
2.3. Dynamic Light Scattering
2.4. Fourier Transform Infrared Spectroscopy-Attenuated Total Reflectance (FT-IR ATR)
2.5. Protein Release from IBs
2.6. Statistical Analysis
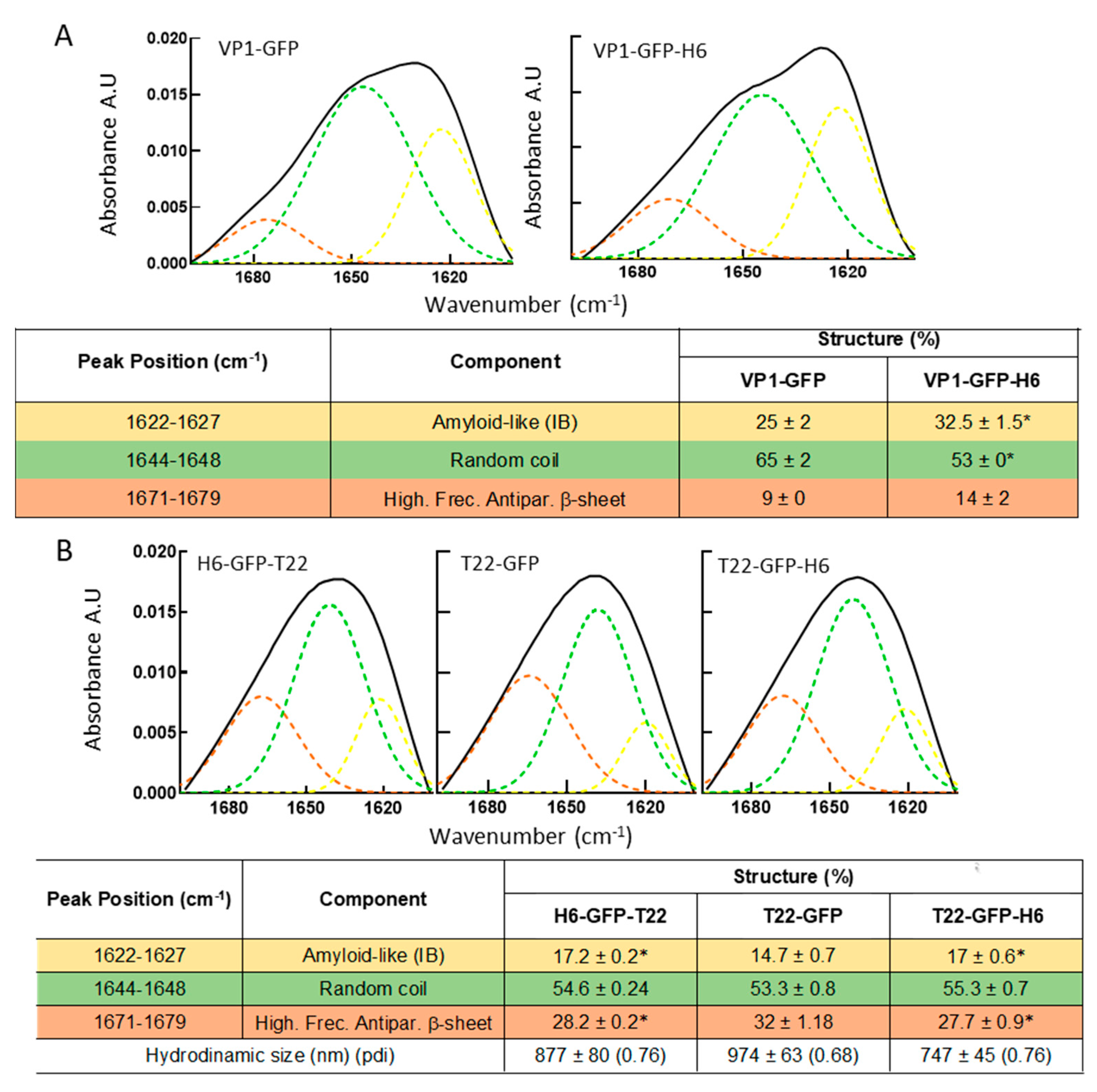
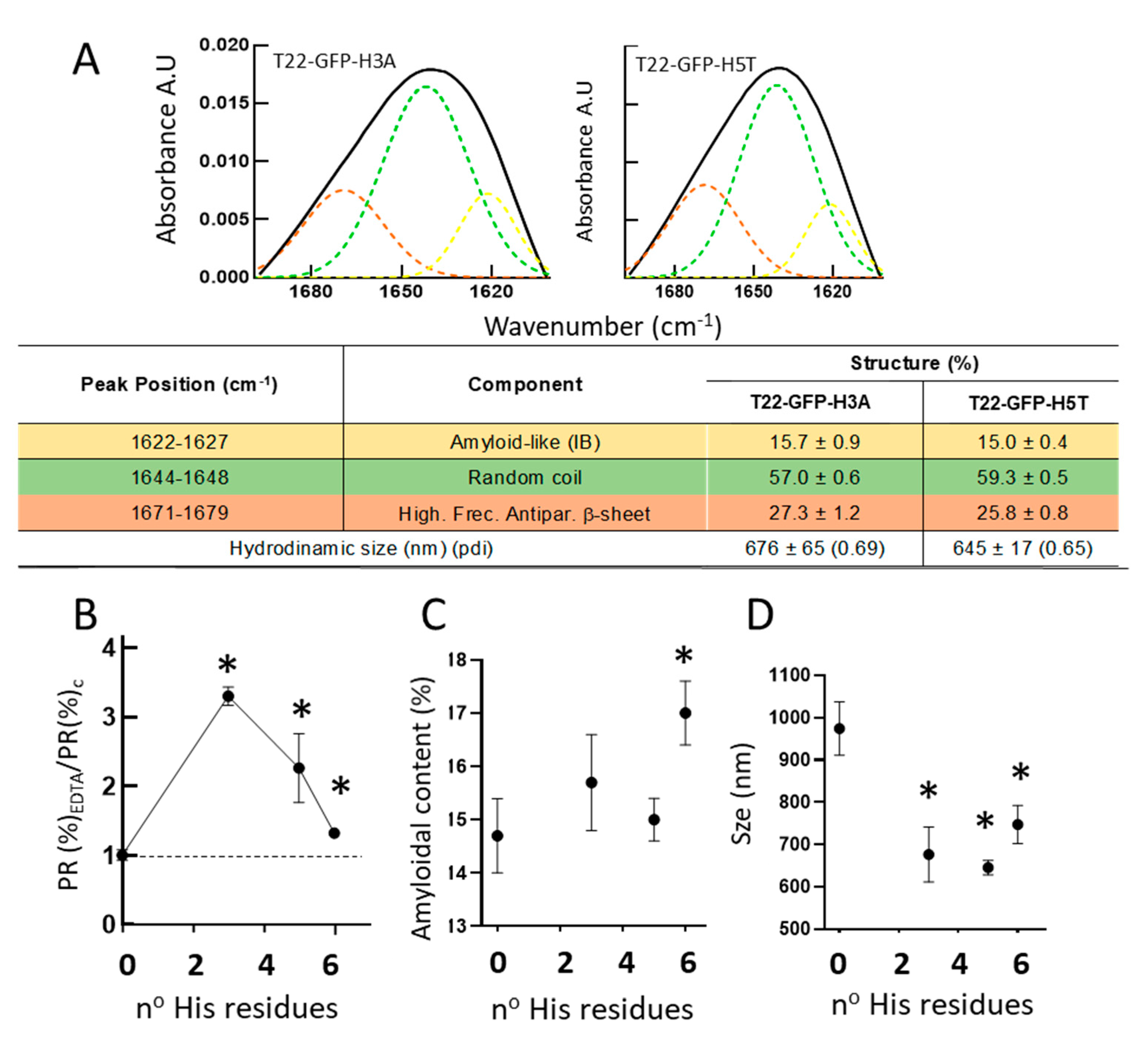
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ding, C.; Li, Z. A review of drug release mechanisms from nanocarrier systems. Mater. Sci. Eng. C 2017, 76, 1440–1453. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Qin, Y.; Lee, J.; Liao, H.; Wang, N.; Davis, T.P.; Qiao, R.; Ling, D. Stimuli-responsive nano-assemblies for remotely controlled drug delivery. J. Control. Release 2020, 322, 566–592. [Google Scholar] [CrossRef] [PubMed]
- Bayer, I.S. Hyaluronic Acid and Controlled Release: A Review. Molecules 2020, 25, 2649. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Alhareth, K.; Mignet, N. Advancement in nanogel formulations provides controlled drug release. Int. J. Pharm. 2020, 584, 119435. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Mooney, D.J. Designing hydrogels for controlled drug delivery. Nat. Reviews. Mater. 2016, 1, 16071. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Ching, Y.C.; Chuah, C.H. Preparation of aerogel beads and microspheres based on chitosan and cellulose for drug delivery: A review. Int. J. Biol. Macromol. 2021, 170, 751–767. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, N.; Pan, W.; Yu, Z.; Yang, L.; Tang, B. Hollow Mesoporous Silica Nanoparticles with Tunable Structures for Controlled Drug Delivery. ACS Appl. Mater. Interfaces 2017, 9, 2123–2129. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, T.; Peng, L.; Sun, Q.; Wei, Y.; Han, B. Advancements in Hydrogel-Based Drug Sustained Release Systems for Bone Tissue Engineering. Front. Pharmacol. 2020, 11, 622. [Google Scholar] [CrossRef]
- Zou, Q.; Li, J.; Niu, L.; Zuo, Y.; Li, J.; Li, Y. Modified n-HA/PA66 scaffolds with chitosan coating for bone tissue engineering: Cell stimulation and drug release. J. Biomater. Sci. Polym. Ed. 2017, 28, 1271–1285. [Google Scholar] [CrossRef]
- Wen, H.; Jung, H.; Li, X. Drug Delivery Approaches in Addressing Clinical Pharmacology-Related Issues: Opportunities and Challenges. AAPS J. 2015, 17, 1327–1340. [Google Scholar] [CrossRef]
- Pareek, S.P.; Kumawat, S.; Sharma, V.; Sharma, D.; Rathore, D.S.; Agarwal, M. Review on sustained release technology. Int. J. Pharm. Biol. Sci. Arch. 2019, 7, 29–38. [Google Scholar]
- Li, C.; Wang, J.; Wang, Y.; Gao, H.; Wei, G.; Huang, Y.; Yu, H.; Gan, Y.; Wang, Y.; Mei, L.; et al. Recent progress in drug delivery. Acta Pharm. Sin. B 2019, 9, 1145–1162. [Google Scholar] [CrossRef] [PubMed]
- Rosen, H.; Abribat, T. The rise and rise of drug delivery. Nat. Rev. Drug Discov. 2005, 4, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Banerjee, P.; Leung, S.S.Y.; Yan, X. Application of Pharmacokinetic-Pharmacodynamic Modeling in Drug Delivery: Development and Challenges. Front. Pharmacol. 2020, 11, 997. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Wolfram, J.; Ferrari, M.; Shen, H. Taking the vehicle out of drug delivery. Mater. Today 2017, 20, 95–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacob, R.S.; Anoop, A.; Maji, S.K. Protein Nanofibrils as Storage Forms of Peptide Drugs and Hormones; Springer: Singapore, 2019. [Google Scholar] [CrossRef]
- Jacob, R.S.; Das, S.; Ghosh, S.; Anoop, A.; Jha, N.N.; Khan, T.; Singru, P.; Kumar, A.; Maji, S.K. Amyloid formation of growth hormone in presence of zinc: Relevance to its storage in secretory granules. Sci. Rep. 2016, 6, 23370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mankar, S.; Anoop, A.; Sen, S.; Maji, S.K. Nanomaterials: Amyloids reflect their brighter side. Nano Rev. 2011, 2, 6032. [Google Scholar] [CrossRef]
- Maji, S.K.; Perrin, M.H.; Sawaya, M.R.; Jessberger, S.; Vadodaria, K.; Rissman, R.A.; Singru, P.S.; Nilsson, K.P.R.; Simon, R.; Schubert, D.; et al. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 2009, 325, 328–332. [Google Scholar] [CrossRef] [Green Version]
- Rana, M.; Sharma, A.K. Cu and Zn interactions with Aβ peptides: Consequence of coordination on aggregation and formation of neurotoxic soluble Aβ oligomers. Metallomics 2019, 11, 64–84. [Google Scholar] [CrossRef]
- López-Laguna, H.; Sánchez, J.; Unzueta, U.; Mangues, R.; Vázquez, E.; Villaverde, A. Divalent Cations: A Molecular Glue for Protein Materials. Trends Biochem. Sci. 2020, 45, 992–1003. [Google Scholar] [CrossRef]
- Lautenschläger, J.; Stephens, A.D.; Fusco, G.; Ströhl, F.; Curry, N.; Zacharopoulou, M.; Michel, C.H.; Laine, R.; Nespovitaya, N.; Fantham, M.; et al. C-terminal calcium binding of α-synuclein modulates synaptic vesicle interaction. Nat. Commun. 2018, 9, 712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinas, U.; Garcia-Fruitos, E.; Corchero, J.L.; Vazquez, E.; Seras-Franzoso, J.; Villaverde, A. Bacterial Inclusion Bodies: Discovering Their Better Half. Trends Biochem. Sci. 2017, 42, 726–737. [Google Scholar] [CrossRef] [PubMed]
- de Marco, A.; Ferrer-Miralles, N.; Garcia-Fruitos, E.; Mitraki, A.; Peternel, S.; Rinas, U.; Trujillo-Roldán, M.A.; Valdez-Cruz, N.A.; Vazquez, E.; Villaverde, A. Bacterial inclusion bodies are industrially exploitable amyloids. FEMS Microbiol. Rev. 2019, 43, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Mogk, A.; Bukau, B.; Kampinga, H.H. Cellular Handling of Protein Aggregates by Disaggregation Machines. Mol. Cell 2018, 69, 214–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mogk, A.; Kummer, E.; Bukau, B. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front. Mol. Biosci. 2015, 2, 22. [Google Scholar] [CrossRef] [Green Version]
- Mogk, A.; Haslberger, T.; Tessarz, P.; Bukau, B. Common and specific mechanisms of AAA+ proteins involved in protein quality control. Biochem. Soc. Trans. 2008, 36, 120–125. [Google Scholar] [CrossRef] [Green Version]
- Mogk, A.; Bukau, B. Molecular chaperones: Structure of a protein disaggregase. Curr. Biol. 2004, 14, R78–R80. [Google Scholar] [CrossRef]
- Weibezahn, J.; Bukau, B.; Mogk, A. Unscrambling an egg: Protein disaggregation by AAA+ proteins. Microb. Cell Factories 2004, 3, 1. [Google Scholar] [CrossRef] [Green Version]
- Stamm, A.; Strauß, S.; Vogt, P.; Scheper, T.; Pepelanova, I. Positive in vitro wound healing effects of functional inclusion bodies of a lipoxygenase from the Mexican axolotl. Microb. Cell Factories 2018, 17, 57. [Google Scholar] [CrossRef] [Green Version]
- Schetters, S.T.T.; Jong, W.S.P.; Kruijssen, L.J.W.; van den Berg van Saparoea, H.B.; Engels, S.; Unger, W.W.J.; Houben, D.; den Haan, J.M.M.; Luirink, J.; van Kooyk, Y. Bacterial inclusion bodies function as vehicles for dendritic cell-mediated T cell responses. Cell Mol. Immunol. 2020, 17, 415–417. [Google Scholar] [CrossRef] [Green Version]
- Wedrychowicz, H.; Kesik, M.; Kaliniak, M.; Kozak-Cieszczyk, M.; Jedlina-Panasiuk, L.; Jaros, S.; Plucienniczak, A. Vaccine potential of inclusion bodies containing cysteine proteinase of Fasciola hepatica in calves and lambs experimentally challenged with metacercariae of the fluke. Vet. Parasitol. 2007, 147, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Van Beek, L.F.; Langereis, J.D.; van den Berg van Saparoea, H.B.; Gillard, J.; Jong, W.S.P.; van Opzeeland, F.J.; Mesman, R.; van Niftrik, L.; Joosten, I.; Diavatopoulos, D.A.; et al. Intranasal vaccination with protein bodies elicit strong protection against Streptococcus pneumoniae colonization. Vaccine 2021, 39, 6920–6929. [Google Scholar] [CrossRef] [PubMed]
- Pesarrodona, M.; Jauset, T.; Díaz-Riascos, Z.V.; Sánchez-Chardi, A.; Beaulieu, M.-E.; Seras-Franzoso, J.; Sánchez-García, L.; Baltà-Foix, R.; Mancilla, S.; Fernández, Y.; et al. Targeting Antitumoral Proteins to Breast Cancer by Local Administration of Functional Inclusion Bodies. Adv. Sci. 2019, 6, 1900849. [Google Scholar] [CrossRef]
- Céspedes, M.V.; Cano-Garrido, O.; Álamo, P.; Sala, R.; Gallardo, A.; Serna, N.; Falgàs, A.; Voltà-Durán, E.; Casanova, I.; Sánchez-Chardi, A.; et al. Engineering Secretory Amyloids for Remote and Highly Selective Destruction of Metastatic Foci. Adv. Mater. 2020, 32, 1907348. [Google Scholar] [CrossRef] [PubMed]
- Gifre-Renom, L.; Carratalá, J.V.; Parés, S.; Sánchez-García, L.; Ferrer-Miralles, N.; Villaverde, A.; Bach, A.; Garcia-Fruitós, E.; Arís, A. Potential of MMP-9 based nanoparticles at optimizing the cow dry period: Pulling apart the effects of MMP-9 and nanoparticles. Sci. Rep. 2020, 10, 11299. [Google Scholar] [CrossRef] [PubMed]
- Carratalá, J.V.; Cano-Garrido, O.; Sánchez, J.; Membrado, C.; Pérez, E.; Conchillo-Solé, O.; Daura, X.; Sánchez-Chardi, A.; Villaverde, A.; Arís, A.; et al. Aggregation-prone peptides modulate activity of bovine interferon gamma released from naturally occurring protein nanoparticles. New Biotechnol. 2020, 57, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Thwaite, R.; Ji, J.; Torrealba, D.; Coll, J.; Sabés, M.; Villaverde, A.; Roher, N. Protein Nanoparticles Made of Recombinant Viral Antigens: A Promising Biomaterial for Oral Delivery of Fish Prophylactics. Front. Immunol. 2018, 9, 1652. [Google Scholar] [CrossRef]
- Torrealba, D.; Seras-Franzoso, J.; Mamat, U.; Wilke, K.; Villaverde, A.; Roher, N.; Garcia-Fruitós, E. Complex Particulate Biomaterials as Immunostimulant-Delivery Platforms. PLoS ONE 2016, 11, e0164073. [Google Scholar] [CrossRef] [Green Version]
- Torrealba, D.; Parra, D.; Seras-Franzoso, J.; Vallejos-Vidal, E.; Yero, D.; Gibert, I.; Villaverde, A.; Garcia-Fruitós, E.; Roher, N. Nanostructured recombinant cytokines: A highly stable alternative to short-lived prophylactics. Biomaterials 2016, 107, 102–114. [Google Scholar] [CrossRef] [Green Version]
- Roca-Pinilla, R.; Fortuna, S.; Natalello, A.; Sánchez-Chardi, A.; Ami, D.; Arís, A.; Garcia-Fruitós, E. Exploring the use of leucine zippers for the generation of a new class of inclusion bodies for pharma and biotechnological applications. Microb. Cell Factories 2020, 19, 175. [Google Scholar] [CrossRef]
- Sánchez, J.M.; López-Laguna, H.; Álamo, P.; Serna, N.; Sánchez-Chardi, A.; Nolan, V.; Cano-Garrido, O.; Casanova, I.; Unzueta, U.; Vazquez, E.; et al. Artificial inclusion bodies for clinical development. Adv. Sci. 2020, 7, 1902420. [Google Scholar] [CrossRef]
- López-Laguna, H.; Voltà-Durán, E.; Parladé, E.; Villaverde, A.; Vázquez, E.; Unzueta, U. Insights on the emerging biotechnology of histidine-rich peptides. Biotechnol. Adv. 2021, 54, 107817. [Google Scholar] [CrossRef] [PubMed]
- Tamamura, H.; Arakaki, R.; Funakoshi, H.; Imai, M.; Otaka, A.; Ibuka, T.; Nakashima, H.; Murakami, T.; Waki, M.; Matsumoto, A.; et al. Effective lowly cytotoxic analogs of an HIV-cell fusion inhibitor, T22 ([Tyr5,12, Lys7]-polyphemusin II). Bioorganic Med. Chem. 1998, 6, 231–238. [Google Scholar] [CrossRef]
- Tamamura, H.; Imai, M.; Ishihara, T.; Masuda, M.; Funakoshi, H.; Oyake, H.; Murakami, T.; Arakaki, R.; Nakashima, H.; Otaka, A.; et al. Pharmacophore identification of a chemokine receptor (CXCR4) antagonist, T22 ([Tyr5,12, Lys7]-polyphemusin II), which specifically blocks T cell-line-tropic HIV-1 infection. Bioorganic Med. Chem. 1998, 6, 1033–1041. [Google Scholar] [CrossRef]
- Rueda, F.; Céspedes, M.V.; Conchillo-Solé, O.; Sánchez-Chardi, A.; Seras-Franzoso, J.; Cubarsi, R.; Gallardo, A.; Pesarrodona, M.; Ferrer-Miralles, N.; Daura, X.; et al. Bottom-Up Instructive Quality Control in the Biofabrication of Smart Protein Materials. Adv. Mater. 2015, 27, 7816–7822. [Google Scholar] [CrossRef] [Green Version]
- García-Fruitós, E.; González-Montalbán, N.; Morell, M.; Vera, A.; Ferraz, R.M.; Arís, A.; Ventura, S.; Villaverde, A. Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins. Microb. Cell Factories 2005, 4, 27. [Google Scholar] [CrossRef] [Green Version]
- Carrió, M.; González-Montalbán, N.; Vera, A.; Villaverde, A.; Ventura, S. Amyloid-like properties of bacterial inclusion bodies. J. Mol. Biol. 2005, 347, 1025–1037. [Google Scholar] [CrossRef]
- López-Laguna, H.; Unzueta, U.; Conchillo-Solé, O.; Sánchez-Chardi, A.; Pesarrodona, M.; Cano-Garrido, O.; Voltà, E.; Sánchez-García, L.; Serna, N.; Saccardo, P.; et al. Assembly of histidine-rich protein materials controlled through divalent cations. Acta Biomater. 2019, 83, 257–264. [Google Scholar] [CrossRef]
- Voltà-Durán, E.; Cano-Garrido, O.; Serna, N.; López-Laguna, H.; Sánchez-García, L.; Pesarrodona, M.; Sánchez-Chardi, A.; Mangues, R.; Villaverde, A.; Vázquez, E.; et al. Controlling self-assembling and tumor cell-targeting of protein-only nanoparticles through modular protein engineering. Sci. China Mater. 2019, 63, 147–156. [Google Scholar] [CrossRef] [Green Version]
- López-Laguna, H.; Sala, R.; Sánchez, J.M.; Álamo, P.; Unzueta, U.; Sánchez-Chardi, A.; Serna, N.; Sánchez-García, L.; Voltà-Durán, E.; Mangues, R.; et al. Nanostructure Empowers Active Tumor Targeting in Ligand-Based Molecular Delivery. Part. Part. Charact. Syst. 2019, 36, 1900304. [Google Scholar] [CrossRef]
- Unzueta, U.; Cespedes, M.V.; Sala, R.; Alamo, P.; Sánchez-Chardi, A.; Pesarrodona, M.; Sánchez-García, L.; Cano-Garrido, O.; Villaverde, A.; Vázquez, E.; et al. Release of targeted protein nanoparticles from functional bacterial amyloids: A death star-like approach. J. Control. Release 2018, 279, 29–39. [Google Scholar] [CrossRef]
- Nolan, V.; Sánchez, J.M.; Perillo, M.A. PEG-induced molecular crowding leads to a relaxed conformation, higher thermal stability and lower catalytic efficiency of Escherichia coli β-galactosidase. Colloids Surfaces. B Biointerfaces 2015, 136, 1202–1206. [Google Scholar] [CrossRef]
- Gatti-Lafranconi, P.; Natalello, A.; Ami, D.; Doglia, S.M.; Lotti, M. Concepts and tools to exploit the potential of bacterial inclusion bodies in protein science and biotechnology. FEBS J. 2011, 278, 2408–2418. [Google Scholar] [CrossRef] [Green Version]
- Doglia, S.M.; Ami, D.; Natalello, A.; Gatti-Lafranconi, P.; Lotti, M. Fourier transform infrared spectroscopy analysis of the conformational quality of recombinant proteins within inclusion bodies. Biotechnol. J. 2008, 3, 193–201. [Google Scholar] [CrossRef]
- Ami, D.; Natalello, A.; Gatti-Lafranconi, P.; Lotti, M.; Doglia, S.M. Kinetics of inclusion body formation studied in intact cells by FT-IR spectroscopy. FEBS Lett. 2005, 579, 3433–3436. [Google Scholar] [CrossRef] [Green Version]
- Unzueta, U.; Seras-Franzoso, J.; Céspedes, M.V.; Saccardo, P.; Cortés, F.; Rueda, F.; Garcia-Fruitos, E.; Ferrer-Miralles, N.; Mangues, R.; Vazquez, E.; et al. Engineering tumor cell targeting in nanoscale amyloidal materials. Nanotechnology 2017, 28, 015102. [Google Scholar] [CrossRef]
- Rueda, F.; Gasser, B.; Sánchez-Chardi, A.; Roldán, M.; Villegas, S.; Puxbaum, V.; Ferrer-Miralles, N.; Unzueta, U.; Vázquez, E.; Garcia-Fruitós, E.; et al. Functional inclusion bodies produced in the yeast Pichia pastoris. Microb. Cell Factories 2016, 15, 166. [Google Scholar] [CrossRef] [Green Version]
- Faller, P.; Hureau, C.; Berthoumieu, O. Role of metal ions in the self-assembly of the Alzheimer’s amyloid-β peptide. Inorg. Chem. 2013, 52, 12193–12206. [Google Scholar] [CrossRef]
- Dannies, P.S. Prolactin and growth hormone aggregates in secretory granules: The need to understand the structure of the aggregate. Endocr. Rev. 2012, 33, 254–270. [Google Scholar] [CrossRef] [Green Version]
- Józsa, É.; Ősz, K.; Kállay, C.; de Bona, P.; Damante, C.A.; Pappalardo, G.; Rizzarelli, E.; Sóvágó, I. Nickel(II) and mixed metal complexes of amyloid-β N-terminus. Dalton Trans. 2010, 39, 7046–7053. [Google Scholar] [CrossRef]
- Speed, M.A.; Wang, D.I.C.; King, J. Specific aggregation of partially folded polypeptide chains: The molecular basis of inclusion body composition. Nat. Biotechnol. 1996, 14, 1283–1287. [Google Scholar] [CrossRef]
- Morell, M.; Bravo, R.; Espargaró, A.; Sisquella, X.; Aviles, F.X.; Fernàndez-Busquets, X.; Ventura, S. Inclusion bodies: Specificity in their aggregation process and amyloid-like structure. BBA Mol. Cell Res. 2008, 1783, 1815–1825. [Google Scholar] [CrossRef] [Green Version]
- Diaz-Espinoza, R.; Nova, E.; Monasterio, O. Overcoming electrostatic repulsions during amyloid assembly: Effect of pH and interaction with divalent metals using model peptides. Arch. Biochem. Biophys. 2017, 621, 46–53. [Google Scholar] [CrossRef]
- Hane, F.; Leonenko, Z. Effect of metals on kinetic pathways of amyloid-β aggregation. Biomolecules 2014, 4, 101–116. [Google Scholar] [CrossRef] [Green Version]
- Grenács, Á.; Sóvágó, I. Copper(II), nickel(II) and zinc(II) complexes of the N-terminal nonapeptide fragment of amyloid-β and its derivatives. J. Inorg. Biochem. 2014, 139, 49–56. [Google Scholar] [CrossRef]
- Calabrese, M.F.; Miranker, A.D. Metal binding sheds light on mechanisms of amyloid assembly. Prion 2009, 3, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Zechel, S.; Hager, M.D.; Priemel, T.; Harrington, M.J. Healing through Histidine: Bioinspired Pathways to Self-Healing Polymers via Imidazole–Metal Coordination. Biomimetics 2019, 4, 20. [Google Scholar] [CrossRef] [Green Version]
- Jehle, F.; Fratzl, P.; Harrington, M.J. Metal-Tunable Self-Assembly of Hierarchical Structure in Mussel-Inspired Peptide Films. ACS Nano 2018, 12, 2160–2168. [Google Scholar] [CrossRef]
- Munch, H.K.; Nygaard, J.; Christensen, N.J.; Engelbrekt, C.; Østergaard, M.; Porsgaard, T.; Hoeg-Jensen, T.; Zhang, J.; Arleth, L.; Thulstrup, P.W.; et al. Construction of Insulin 18-mer Nanoassemblies Driven by Coordination to Iron(II) and Zinc(II) Ions at Distinct Sites. Angew. Chem. Int. Ed. 2016, 55, 2378–2381. [Google Scholar] [CrossRef] [Green Version]
- López-Laguna, H.; Sánchez, J.M.; Carratalá, J.V.; Rojas-Peña, M.; Sánchez-García, L.; Parladé, E.; Sánchez-Chardi, A.; Voltà-Durán, E.; Serna, N.; Cano-Garrido, O.; et al. Biofabrication of functional protein nanoparticles through simple His-tag engineering. ACS Sustain. Chem. Eng. 2021, 9, 12341–12354. [Google Scholar] [CrossRef]
- López-Laguna, H.; Sánchez-García, L.; Serna, N.; Voltà-Durán, E.; Sánchez, J.M.; Sánchez-Chardi, A.; Unzueta, U.; Łoś, M.; Villaverde, A.; Vázquez, E. Engineering protein nanoparticles out from components of the human microbiome. Small 2020, 16, 2001885. [Google Scholar] [CrossRef] [PubMed]
- Villaverde, A.; García-Fruitós, E.; Rinas, U.; Seras-Franzoso, J.; Kosoy, A.; Corchero, J.L.; Vazquez, E. Packaging protein drugs as bacterial inclusion bodies for therapeutic applications. Microb. Cell Factories 2012, 11, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez, J.M.; Carratalá, J.V.; Serna, N.; Unzueta, U.; Nolan, V.; Sánchez-Chardi, A.; Voltà-Durán, E.; López-Laguna, H.; Ferrer-Miralles, N.; Villaverde, A.; et al. The Poly-Histidine Tag H6 Mediates Structural and Functional Properties of Disintegrating, Protein-Releasing Inclusion Bodies. Pharmaceutics 2022, 14, 602. https://doi.org/10.3390/pharmaceutics14030602
Sánchez JM, Carratalá JV, Serna N, Unzueta U, Nolan V, Sánchez-Chardi A, Voltà-Durán E, López-Laguna H, Ferrer-Miralles N, Villaverde A, et al. The Poly-Histidine Tag H6 Mediates Structural and Functional Properties of Disintegrating, Protein-Releasing Inclusion Bodies. Pharmaceutics. 2022; 14(3):602. https://doi.org/10.3390/pharmaceutics14030602
Chicago/Turabian StyleSánchez, Julieta María, José Vicente Carratalá, Naroa Serna, Ugutz Unzueta, Verónica Nolan, Alejandro Sánchez-Chardi, Eric Voltà-Durán, Hèctor López-Laguna, Neus Ferrer-Miralles, Antonio Villaverde, and et al. 2022. "The Poly-Histidine Tag H6 Mediates Structural and Functional Properties of Disintegrating, Protein-Releasing Inclusion Bodies" Pharmaceutics 14, no. 3: 602. https://doi.org/10.3390/pharmaceutics14030602
APA StyleSánchez, J. M., Carratalá, J. V., Serna, N., Unzueta, U., Nolan, V., Sánchez-Chardi, A., Voltà-Durán, E., López-Laguna, H., Ferrer-Miralles, N., Villaverde, A., & Vazquez, E. (2022). The Poly-Histidine Tag H6 Mediates Structural and Functional Properties of Disintegrating, Protein-Releasing Inclusion Bodies. Pharmaceutics, 14(3), 602. https://doi.org/10.3390/pharmaceutics14030602