Overcoming Multidrug Resistance of Antibiotics via Nanodelivery Systems
Abstract
:1. Introduction
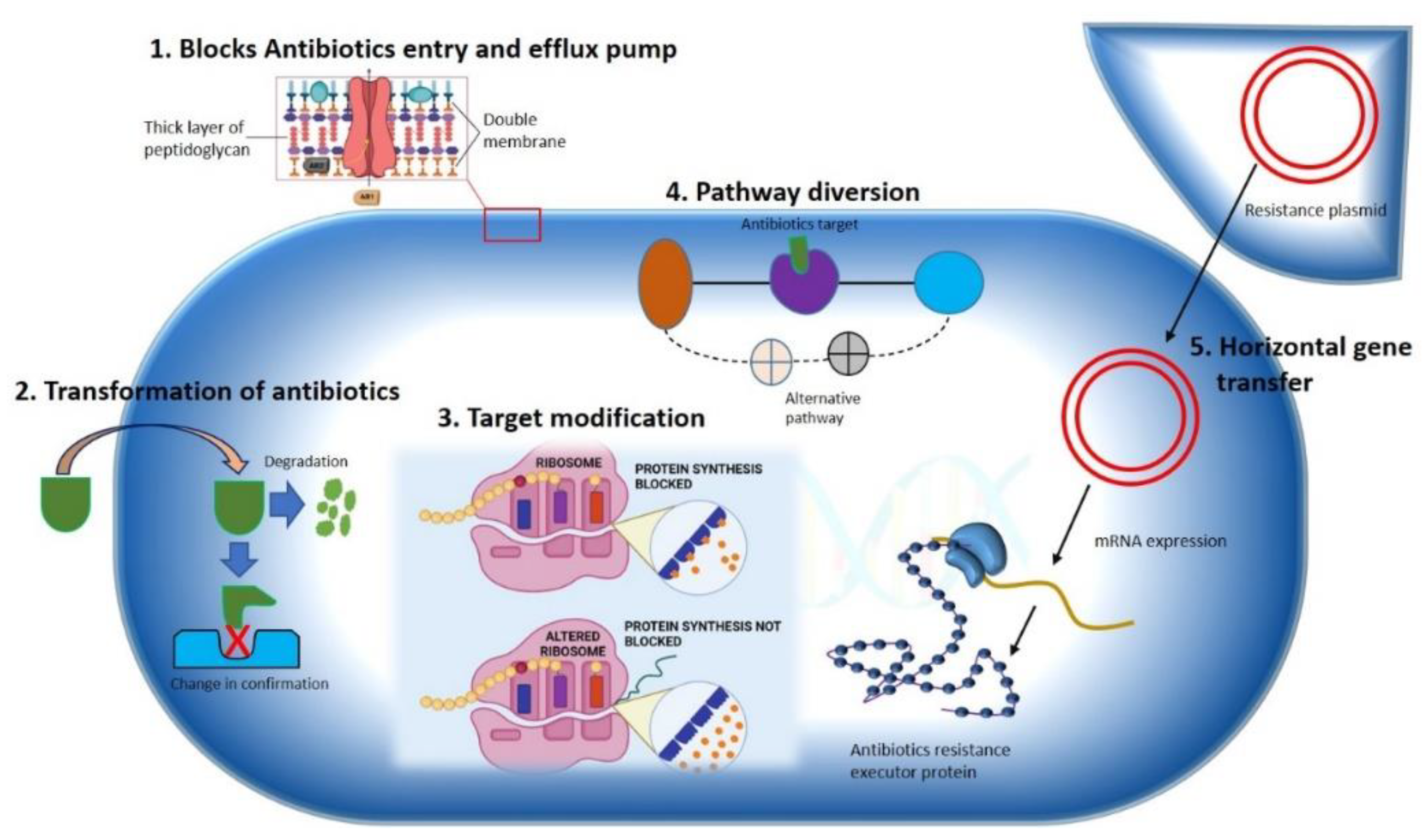
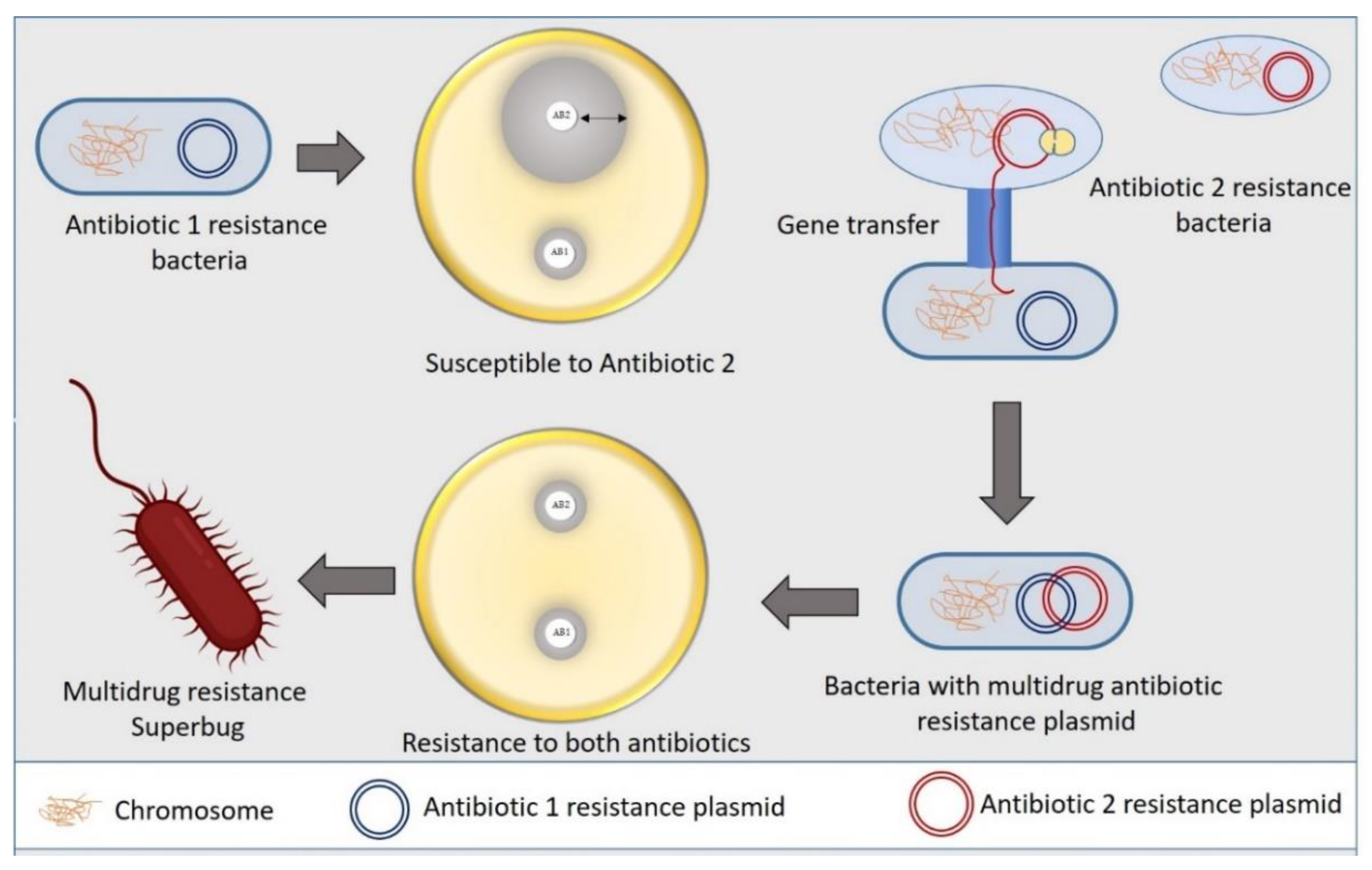
2. Antibiotic Resistance Mechanism in Bacteria
2.1. Constrained Antibiotic Entry and Efflux Pumps
2.2. Transformation or Termination of Antibiotics
2.3. Pathway Alteration to Avoid the Antibiotic Target Site
2.4. Antibiotics Target Modification
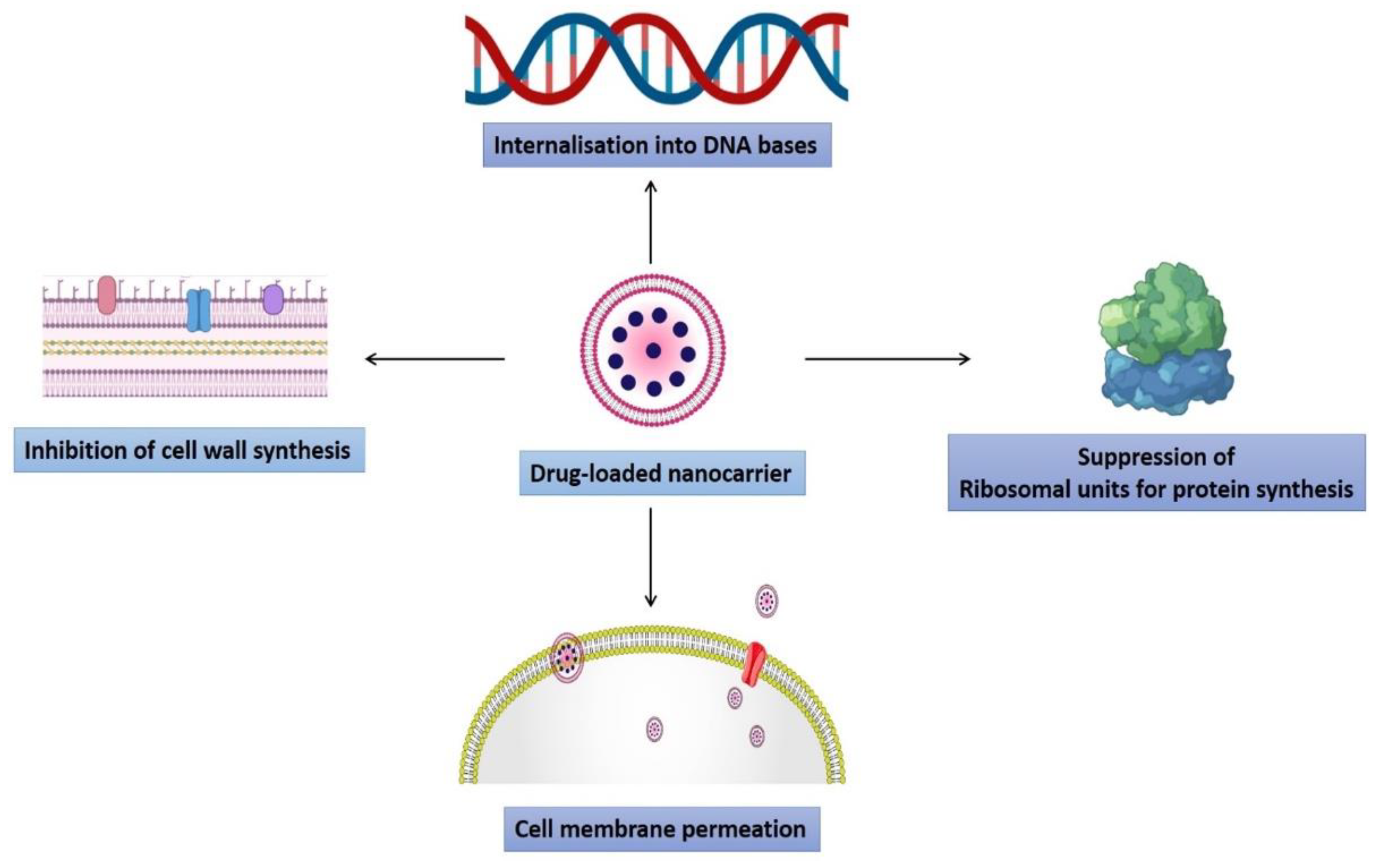
3. Mechanism of Advanced Nanotechnological Approaches against Antibiotic Resistance
3.1. Interaction with the Bacterial Cell Wall
3.2. Inhibition of Biofilm Formations
3.3. Activation of Innate as Well as Adaptive Host Immune Response
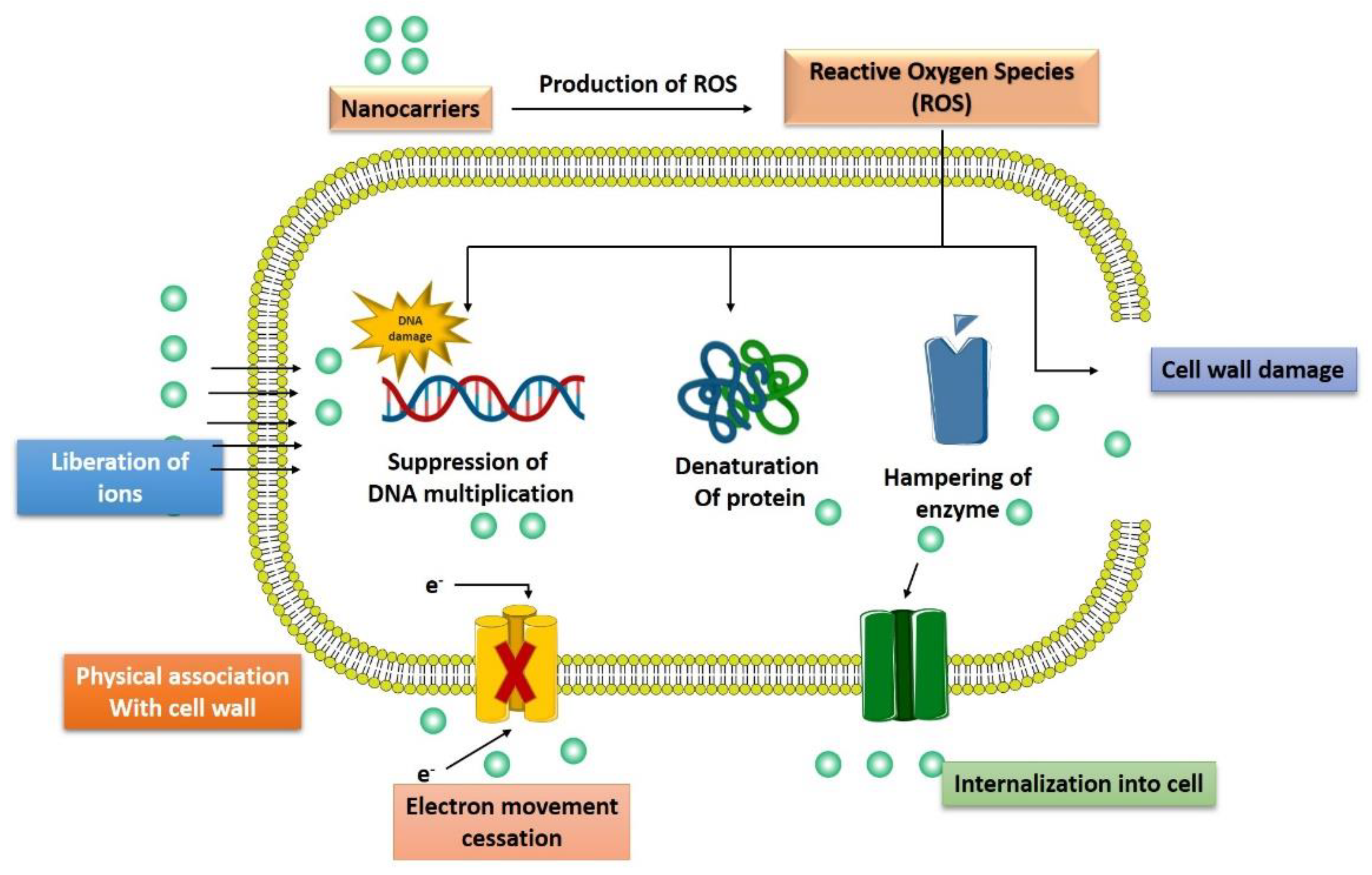
3.4. Generation of Reactive Oxygen Species
3.5. Induction of Intracellular Effects
4. Nanotechnology to Overcome Antimicrobial Resistance
4.1. Polymeric Nanoparticles
4.2. Liposomes
4.3. Solid Lipid Nanoparticles
4.4. Nanostructured Lipid Carriers
4.5. Dendrimers
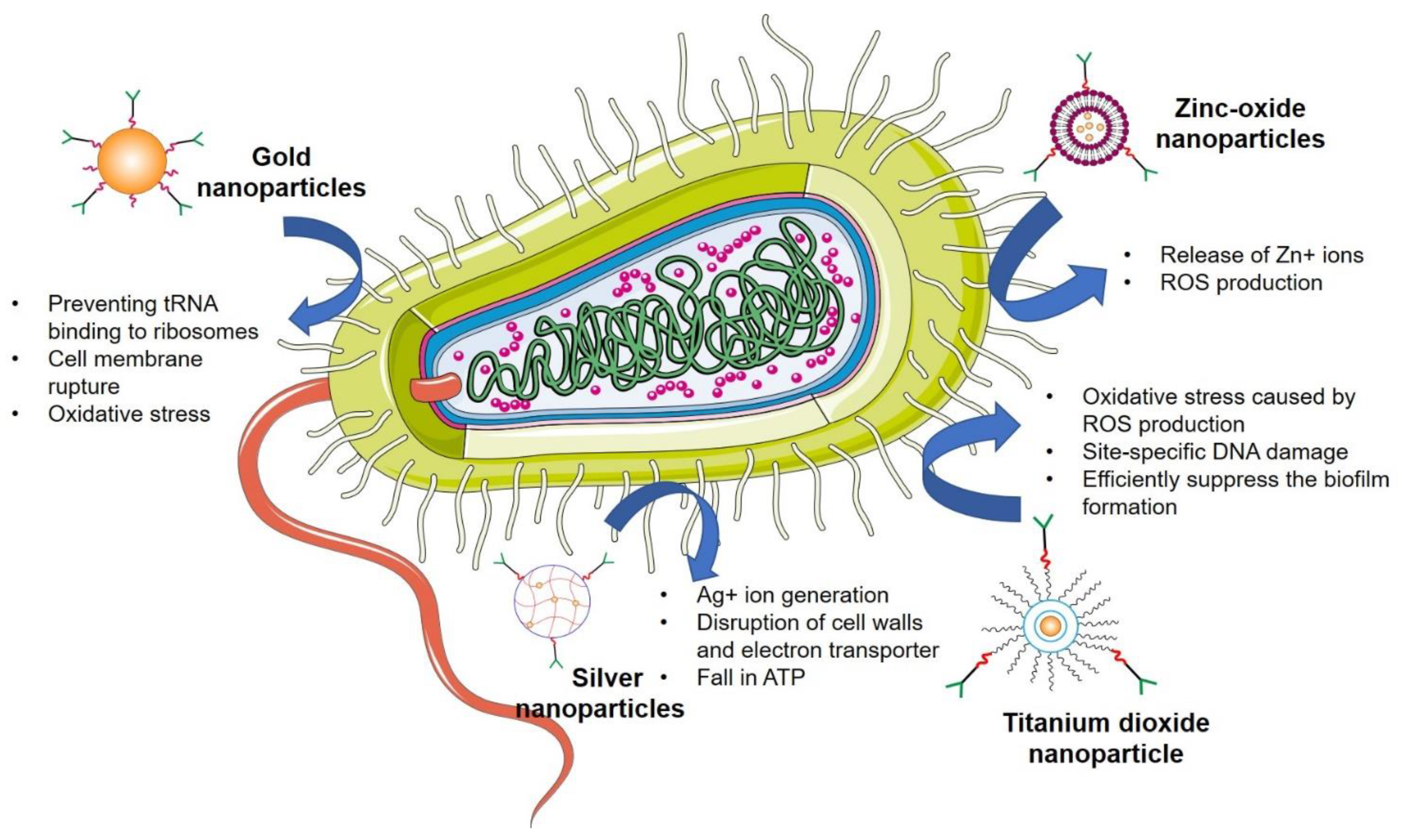
4.6. Inorganic Nanoparticles
4.6.1. Silver Nanoparticles
4.6.2. Gold Nanoparticles
4.6.3. Zinc Oxide Nanoparticles
4.6.4. Titanium Dioxide Nanoparticles
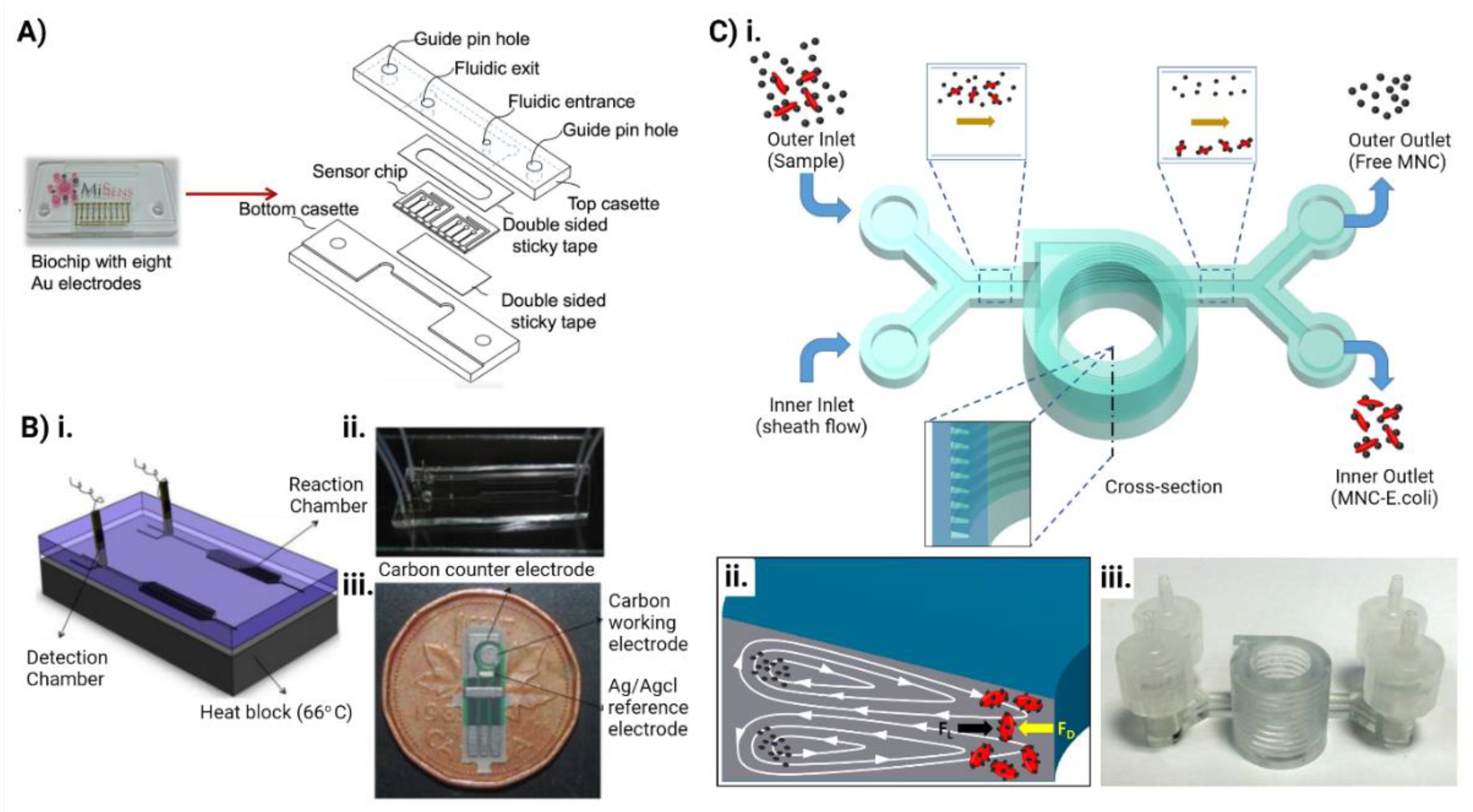
5. Microfluidics for Combating Antimicrobial Resistance
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gandra, S.; Tseng, K.K.; Arora, A.; Bhowmik, B.; Robinson, M.L.; Panigrahi, B.; Laxminarayan, R.; Klein, E.Y. The Mortality Burden of Multidrug-resistant Pathogens in India: A Retrospective, Observational Study. Clin. Infect. Dis. 2019, 69, 563–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.; Thamphiwatana, S.; Angsantikul, P.; Zhang, L. Nanoparticle approaches against bacterial infections. WIREs Nanomed. Nanobiotechnology 2014, 6, 532–547. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Lin, J.; Taggart, C.; Bengoechea, J.; Scott, C.J. Nanodelivery strategies for the treatment of multidrug-resistant bacterial infections. J. Interdiscip. Nanomed. 2018, 3, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Chia, P.Y.; Sengupta, S.; Kukreja, A.; Ponnampalavanar, S.S.; Ng, O.T.; Marimuthu, K. The role of hospital environment in transmissions of multidrug-resistant gram-negative organisms. Antimicrob. Resist. Infect. Control 2020, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Nnadozie, C.F.; Odume, O.N. Freshwater environments as reservoirs of antibiotic resistant bacteria and their role in the dissemination of antibiotic resistance genes. Environ. Pollut. 2019, 254, 113067. [Google Scholar] [CrossRef]
- Salyers, A.; Shoemaker, N.B. Reservoirs of Antibiotic Resistance Genes. Anim. Biotechnol. 2006, 17, 137–146. [Google Scholar] [CrossRef]
- Nikaido, H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [Green Version]
- Breijyeh, Z.; Jubeh, B.; Karaman, R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules 2020, 25, 1340. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.I. Antibiotic Resistance and Regulation of the Gram-Negative Bacterial Outer Membrane Barrier by Host Innate Immune Molecules. mBio 2016, 7, e01541–e01616. [Google Scholar] [CrossRef] [Green Version]
- van Duijkeren, E.; Schink, A.-K.; Roberts, M.C.; Wang, Y.; Schwarz, S. Mechanisms of Bacterial Resistance to Antimicrobial Agents. Microbiol. Spectr. 2018, 6, 186–198. [Google Scholar] [CrossRef]
- Piddock, L.J.V. Clinically Relevant Chromosomally Encoded Multidrug Resistance Efflux Pumps in Bacteria. Clin. Microbiol. Rev. 2006, 19, 382–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, D.; Cook, D.N.; Alberti, M.; Pon, N.G.; Nikaido, H.; Hearst, J.E. Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 1993, 175, 6299–6313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Liu, D.; Wang, L.; Wang, Y.; Zang, Z.; Liu, Z.; Song, B.; Gu, L.; Fan, Z.; Yang, S.; et al. A Putative Efflux Transporter of the ABC Family, YbhFSR, in Escherichia Coli Functions in Tetracycline Efflux and Na+(Li+)/H+ Transport. Front. Microbiol. 2020, 11, 556. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, C.; Schwarz, S.; Liu, W.; Yang, Q.; Luan, T.; Wang, L.; Liu, S.; Zhang, W. Identification of a novel tetracycline resistance gene, tet(63), located on a multiresistance plasmid from Staphylococcus aureus. J. Antimicrob. Chemother. 2021, 76, 576–581. [Google Scholar] [CrossRef]
- Egorov, A.M.; Ulyashova, M.M.; Rubtsova, M.Y. Bacterial Enzymes and Antibiotic Resistance. Available online: https://pubmed.ncbi.nlm.nih.gov/30713760/ (accessed on 9 November 2021).
- Collins, V.L.; Marchaim, D.; Pogue, J.M.; Moshos, J.; Bheemreddy, S.; Sunkara, B.; Shallal, A.; Chugh, N.; Eiseler, S.; Bhargava, P.; et al. Efficacy of Ertapenem for Treatment of Bloodstream Infections Caused by Extended-Spectrum-β-Lactamase-Producing Enterobacteriaceae. Antimicrob. Agents Chemother. 2012, 56, 2173–2177. [Google Scholar] [CrossRef] [Green Version]
- Zárate, S.G.; De La Cruz Claure, M.L.; Benito-Arenas, R.; Revuelta, J.; Santana, A.G.; Bastida, A. Overcoming Aminoglycoside Enzymatic Resistance: Design of Novel Antibiotics and Inhibitors. Molecules 2018, 23, 284. [Google Scholar] [CrossRef] [Green Version]
- Peterson, E.; Kaur, P. Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Front. Microbiol. 2018, 9, 2928. [Google Scholar] [CrossRef]
- Sköld, O. Sulfonamide resistance: Mechanisms and trends. Drug Resist. Updat. 2000, 3, 155–160. [Google Scholar] [CrossRef]
- Rådström, P.; Swedberg, G.; Sköld, O. Genetic analyses of sulfonamide resistance and its dissemination in gram-negative bacteria illustrate new aspects of R plasmid evolution. Antimicrob. Agents Chemother. 1991, 35, 1840–1848. [Google Scholar] [CrossRef] [Green Version]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 464–473. [Google Scholar] [CrossRef] [Green Version]
- Hiramatsu, K.; Ito, T.; Tsubakishita, S.; Sasaki, T.; Takeuchi, F.; Morimoto, Y.; Katayama, Y.; Matsuo, M.; Kuwahara-Arai, K.; Hishinuma, T.; et al. Genomic Basis for Methicillin Resistance inStaphylococcus aureus. Infect. Chemother. 2013, 45, 117–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaupp, R.; Lei, S.; Reed, J.M.; Peisker, H.; Boyle-Vavra, S.; Bayer, A.S.; Bischoff, M.; Herrmann, M.; Daum, R.S.; Powers, R.; et al. Staphylococcus aureus Metabolic Adaptations during the Transition from a Daptomycin Susceptibility Phenotype to a Daptomycin Nonsusceptibility Phenotype. Antimicrob. Agents Chemother. 2015, 59, 4226–4238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parrett, A.; Reed, J.M.; Gardner, S.G.; Mishra, N.N.; Bayer, A.S.; Powers, R.; Somerville, G.A. Metabolic changes associated with adaptive resistance to daptomycin in Streptococcus mitis-oralis. BMC Microbiol. 2020, 20, 162. [Google Scholar] [CrossRef] [PubMed]
- Floss, H.G.; Yu, T.-W. RifamycinMode of Action, Resistance, and Biosynthesis. Chem. Rev. 2005, 105, 621–632. [Google Scholar] [CrossRef]
- Campbell, E.; Korzheva, N.; Mustaev, A.; Murakami, K.; Nair, S.; Goldfarb, A.; Darst, S.A. Structural Mechanism for Rifampicin Inhibition of Bacterial RNA Polymerase. Cell 2001, 104, 901–912. [Google Scholar] [CrossRef]
- Weisblum, B. Erythromycin resistance by ribosome modification. Antimicrob. Agents Chemother. 1995, 39, 577–585. [Google Scholar] [CrossRef] [Green Version]
- Marshall, S.H.; Donskey, C.J.; Hutton-Thomas, R.; Salata, R.A.; Rice, L.B. Gene Dosage and Linezolid Resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 2002, 46, 3334–3336. [Google Scholar] [CrossRef] [Green Version]
- Tang, S.; Zheng, J. Antibacterial Activity of Silver Nanoparticles: Structural Effects. Adv. Healthc. Mater. 2018, 7, 1701503. [Google Scholar] [CrossRef]
- Wang, L.; Hu, C.; Shao, L. The antimicrobial activity of nanoparticles: Present situation and prospects for the future. Int. J. Nanomed. 2017, 12, 1227–1249. [Google Scholar] [CrossRef] [Green Version]
- Sarwar, A.; Katas, H.; Noradila Samsudin, S.; Mohamad Zin, N.; Malaysia, K.; Campus, K.L.; Abdul Aziz, J.R.M.; Lumpur, K.; Rittschof, D. Regioselective Sequential Modification of Chitosan via Azide-Alkyne Click Reaction: Synthesis, Characterization, and Antimicrobial Activity of Chitosan Derivatives and Nanoparticles. PLoS ONE 2015, 10, e0123084. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Zhang, W.; Li, Y.; Wang, G.; Yang, L.; Jin, J.; Chen, Q.; Huang, M. Synthesis, characterization, antimicrobial activity and mechanism of a novel hydroxyapatite whisker/nano zinc oxide biomaterial. Biomed. Mater. 2014, 10, 015001. [Google Scholar] [CrossRef] [PubMed]
- Hyldgaard, M.; Mygind, T.; Vad, B.S.; Stenvang, M.; Otzen, D.; Meyer, R.L. The Antimicrobial Mechanism of Action of Epsilon-Poly-L-Lysine. Appl. Environ. Microbiol. 2014, 80, 7758–7770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wehling, J.; Dringen, R.; Zare, R.N.; Maas, M.; Rezwan, K. Bactericidal Activity of Partially Oxidized Nanodiamonds. ACS Nano 2014, 8, 6475–6483. [Google Scholar] [CrossRef]
- Vestby, L.K.; Grønseth, T.; Simm, R.; Nesse, L.L. Bacterial Biofilm and its Role in the Pathogenesis of Disease. Antibiotics 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, T.; Putatunda, C.; Vyas, A.; Kumar, G. Zinc oxide nanoparticles inhibit bacterial biofilm formation via altering cell membrane permeability. Prep. Biochem. Biotechnol. 2020, 51, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Moser, D.; Han, F.; Leonhard, M.; Schneider-Stickler, B.; Tan, Y. Preparation and antibiofilm studies of curcumin loaded chitosan nanoparticles against polymicrobial biofilms of Candida albicans and Staphylococcus aureus. Carbohydr. Polym. 2020, 241, 116254. [Google Scholar] [CrossRef] [PubMed]
- Baelo, A.; Levato, R.; Julián, E.; Crespo, A.; Astola, J.; Gavaldà, J.; Engel, E.; Mateos-Timoneda, M.A.; Torrents, E. Disassembling bacterial extracellular matrix with DNase-coated nanoparticles to enhance antibiotic delivery in biofilm infections. J. Control. Release 2015, 209, 150–158. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Chai, M.; Li, X.; Deng, Y.; Jin, Q.; Ji, J. Size and Charge Adaptive Clustered Nanoparticles Targeting the Biofilm Microenvironment for Chronic Lung Infection Management. ACS Nano 2020, 14, 5686–5699. [Google Scholar] [CrossRef]
- Jamil, B.; Habib, H.; Abbasi, S.A.; Ihsan, A.; Nasir, H.; Imran, M. Development of Cefotaxime Impregnated Chitosan as Nano-antibiotics: De Novo Strategy to Combat Biofilm Forming Multi-drug Resistant Pathogens. Front. Microbiol. 2016, 7, 330. [Google Scholar] [CrossRef] [Green Version]
- Masood, F.; Yasin, T.; Bukhari, H.; Mujahid, M. Characterization and application of roxithromycin loaded cyclodextrin based nanoparticles for treatment of multidrug resistant bacteria. Mater. Sci. Eng. C 2016, 61, 1–7. [Google Scholar] [CrossRef]
- Pascoal, S.; Liu, X.; Ly, T.; Fang, Y.; Rockliffe, N.; Paterson, S.; Shirran, S.L.; Botting, C.H.; Bailey, N.W. Rapid evolution and gene expression: A rapidly evolving Mendelian trait that silences field crickets has widespread effects on mRNA and protein expression. J. Evol. Biol. 2016, 29, 1234–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, W.-L.; Lin, T.-T.; Sureshbabu, R.; Chia, W.-T.; Hsiao, H.-C.; Liu, H.-Y.; Yang, C.-M.; Sung, H.-W. A rapid drug release system with a NIR light-activated molecular switch for dual-modality photothermal/antibiotic treatments of subcutaneous abscesses. J. Control. Release 2015, 199, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Swartzwelter, B.; Fux, A.; Johnson, L.; Swart, E.; Hofer, S.; Hofstätter, N.; Geppert, M.; Italiani, P.; Boraschi, D.; Duschl, A.; et al. The Impact of Nanoparticles on Innate Immune Activation by Live Bacteria. Int. J. Mol. Sci. 2020, 21, 9695. [Google Scholar] [CrossRef] [PubMed]
- Lavagna, A.; Auger, J.-P.; Dumesnil, A.; Roy, D.; Girardin, S.E.; Gisch, N.; Segura, M.; Gottschalk, M. Interleukin-1 signaling induced by Streptococcus suis serotype 2 is strain-dependent and contributes to bacterial clearance and inflammation during systemic disease in a mouse model of infection. Veter Res. 2019, 50, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horn, C.M.; Kielian, T. Crosstalk Between Staphylococcus aureus and Innate Immunity: Focus on Immunometabolism. Front. Immunol. 2021, 11, 3767. [Google Scholar] [CrossRef] [PubMed]
- Dolgachev, V.A.; Yu, B.; Reinke, J.M.; Raghavendran, K.; Hemmila, M.R. Host susceptibility to gram-negative pneumonia after lung contusion. J. Trauma Inj. Infect. Crit. Care 2012, 72, 614–623. [Google Scholar] [CrossRef] [Green Version]
- Khan, F.; Manivasagan, P.; Lee, J.-W.; Pham, D.T.N.; Oh, J.; Kim, Y.-M. Fucoidan-Stabilized Gold Nanoparticle-Mediated Biofilm Inhibition, Attenuation of Virulence and Motility Properties in Pseudomonas aeruginosa PAO1. Mar. Drugs 2019, 17, 208. [Google Scholar] [CrossRef] [Green Version]
- Tyner, K.; Bancos, S.; Stevens, D. Effect of silica and gold nanoparticles on macrophage proliferation, activation markers, cytokine production, and phagocytosis in vitro. Int. J. Nanomed. 2014, 10, 183–206. [Google Scholar] [CrossRef] [Green Version]
- Kodali, V.; Littke, M.H.; Tilton, S.C.; Teeguarden, J.; Shi, L.; Frevert, C.W.; Wang, W.; Pounds, J.; Thrall, B.D. Dysregulation of Macrophage Activation Profiles by Engineered Nanoparticles. ACS Nano 2013, 7, 6997–7010. [Google Scholar] [CrossRef]
- Swartzwelter, B.J.; Barbero, F.; Verde, A.; Mangini, M.; Pirozzi, M.; De Luca, A.C.; Puntes, V.F.; Leite, L.C.C.; Italiani, P.; Boraschi, D. Gold Nanoparticles Modulate BCG-Induced Innate Immune Memory in Human Monocytes by Shifting the Memory Response towards Tolerance. Cells 2020, 9, 284. [Google Scholar] [CrossRef] [Green Version]
- Vimbela, G.V.; Ngo, S.M.; Fraze, C.; Yang, L.; Stout, D.A. Antibacterial properties and toxicity from metallic nanomaterials. Int. J. Nanomed. 2017, 12, 3941–3965. [Google Scholar] [CrossRef] [Green Version]
- Panth, N.; Paudel, K.R.; Parajuli, K. Reactive Oxygen Species: A Key Hallmark of Cardiovascular Disease. Adv. Med. 2016, 2016, 9152732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.-G.; Peng, Q.-L.; Gurunathan, S. Effects of Silver Nanoparticles on Multiple Drug-Resistant Strains of Staphylococcus aureus and Pseudomonas aeruginosa from Mastitis-Infected Goats: An Alternative Approach for Antimicrobial Therapy. Int. J. Mol. Sci. 2017, 18, 569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.; Cheng, S.; Singh, S. Oxidative stress-mediated genotoxic effect of zinc oxide nanoparticles on Deinococcus radiodurans. 3 Biotech 2020, 10, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díaz-García, D.; Ardiles, P.R.; Prashar, S.; Rodríguez-Diéguez, A.; Páez, P.L.; Gómez-Ruiz, S. Preparation and Study of the Antibacterial Applications and Oxidative Stress Induction of Copper Maleamate-Functionalized Mesoporous Silica Nanoparticles. Pharmaceutics 2019, 11, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wigginton, N.; de Titta, A.; Piccapietra, F.; Dobias, J.; Nesatyy, V.J.; Suter, M.J.-F.; Bernier-Latmani, R. Binding of Silver Nanoparticles to Bacterial Proteins Depends on Surface Modifications and Inhibits Enzymatic Activity. Environ. Sci. Technol. 2010, 44, 2163–2168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, W.K.; Koo, H.C.; Kim, K.W.; Shin, S.; Kim, S.H.; Park, Y.H. Antibacterial Activity and Mechanism of Action of the Silver Ion in Staphylococcus aureus and Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 2171–2178. [Google Scholar] [CrossRef] [Green Version]
- McQuillan, J.S.; Shaw, A. Differential gene regulation in the Ag nanoparticle and Ag+-induced silver stress response in Escherichia coli: A full transcriptomic profile. Nanotoxicology 2014, 8, 177–184. [Google Scholar] [CrossRef]
- Gou, N.; Onnis-Hayden, A.; Gu, A.Z. Mechanistic Toxicity Assessment of Nanomaterials by Whole-Cell-Array Stress Genes Expression Analysis. Environ. Sci. Technol. 2010, 44, 5964–5970. [Google Scholar] [CrossRef]
- Saleem, S.; Iqubal, M.K.; Garg, S.; Ali, J.; Baboota, S. Trends in nanotechnology-based delivery systems for dermal targeting of drugs: An enticing approach to offset psoriasis. Expert Opin. Drug Deliv. 2020, 17, 817–838. [Google Scholar] [CrossRef]
- Modi, D.; Nirmal, J.; Warsi, M.H.; Bhatia, M.; Hasan, N.; Kesharwani, P.; Jain, G.K. Formulation and development of tacrolimus-gellan gum nanoformulation for treatment of dry eye disease. Colloids Surf. B Biointerfaces 2021, 211, 112255. [Google Scholar] [CrossRef] [PubMed]
- Paudel, K.R.; Mehta, M.; Yin, G.H.S.; Yen, L.L.; Malyla, V.; Patel, V.K.; Panneerselvam, J.; Madheswaran, T.; MacLoughlin, R.; Jha, N.K.; et al. Berberine-loaded liquid crystalline nanoparticles inhibit non-small cell lung cancer proliferation and migration in vitro. Environ. Sci. Pollut. Res. 2022, 16, 1–18. [Google Scholar] [CrossRef]
- Roberts, M.S.; Cheruvu, H.S.; Mangion, S.E.; Alinaghi, A.; Benson, H.A.; Mohammed, Y.; Holmes, A.; van der Hoek, J.; Pastore, M.; Grice, J.E. Topical drug delivery: History, percutaneous absorption, and product development. Adv. Drug Deliv. Rev. 2021, 177, 113929. [Google Scholar] [CrossRef] [PubMed]
- Imran, M.; Saleem, S.; Chaudhuri, A.; Ali, J.; Baboota, S. Docetaxel: An update on its molecular mechanisms, therapeutic trajectory and nanotechnology in the treatment of breast, lung and prostate cancer. J. Drug Deliv. Sci. Technol. 2020, 60, 101959. [Google Scholar] [CrossRef]
- Paudel, K.R.; Mehta, M.; Shukla, S.D.; Panth, N.; Chellappan, D.K.; Dua, K.; Hansbro, P. Advancements in nanotherapeutics targeting senescence in chronic obstructive pulmonary disease. Nanomedicine 2022, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Devkota, H.P.; Paudel, K.R.; Jha, N.K.; Gupta, P.K.; Singh, S.K.; Chellappan, D.K.; Hansbro, P.M.; Dua, K. Applications of drug-delivery systems targeting inflammasomes in pulmonary diseases. Nanomedicine 2021, 16, 2407–2410. [Google Scholar] [CrossRef]
- Solanki, N.; Mehta, M.; Chellappan, D.K.; Gupta, G.; Hansbro, N.G.; Tambuwala, M.M.; Aljabali, A.A.; Paudel, K.R.; Liu, G.; Satija, S.; et al. Antiproliferative effects of boswellic acid-loaded chitosan nanoparticles on human lung cancer cell line A549. Futur. Med. Chem. 2020, 12, 2019–2034. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.; Misba, L.; Khan, A.U. Nano-therapeutics: A revolution in infection control in post antibiotic era. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 2281–2301. [Google Scholar] [CrossRef]
- Yeh, Y.-C.; Huang, T.-H.; Yang, S.-C.; Chen, C.-C.; Fang, J.-Y. Nano-Based Drug Delivery or Targeting to Eradicate Bacteria for Infection Mitigation: A Review of Recent Advances. Front. Chem. 2020, 8, 286. [Google Scholar] [CrossRef]
- Gupta, A.; Mumtaz, S.; Li, C.-H.; Hussain, I.; Rotello, V.M. Combatting antibiotic-resistant bacteria using nanomaterials. Chem. Soc. Rev. 2019, 48, 415–427. [Google Scholar] [CrossRef]
- Lee, N.-Y.; Ko, W.-C.; Hsueh, P.-R. Nanoparticles in the Treatment of Infections Caused by Multidrug-Resistant Organisms. Front. Pharmacol. 2019, 10, 1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Arya, D.K.; Dua, M.; Chhatwal, G.S.; Johri, A.K. Nano-technology for targeted drug delivery to combat antibiotic resistance. Expert Opin. Drug Deliv. 2012, 9, 1325–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaikh, S.; Nazam, N.; Rizvi, S.M.D.; Ahmad, K.; Baig, M.H.; Lee, E.J.; Choi, I. Mechanistic Insights into the Antimicrobial Actions of Metallic Nanoparticles and Their Implications for Multidrug Resistance. Int. J. Mol. Sci. 2019, 20, 2468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, W.; Zhang, L. Nanomaterials arising amid antibiotic resistance. Nat. Rev. Genet. 2021, 19, 5–6. [Google Scholar] [CrossRef] [PubMed]
- Pelgrift, R.Y.; Friedman, A.J. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv. Drug Deliv. Rev. 2013, 65, 1803–1815. [Google Scholar] [CrossRef] [PubMed]
- Zielińska, A.; Carreiró, F.; Oliveira, A.; Neves, A.; Pires, B.; Venkatesh, D.; Durazzo, A.; Lucarini, M.; Eder, P.; Silva, A.; et al. Polymeric Nanoparticles: Production, Characterization, Toxicology and Ecotoxicology. Molecules 2020, 25, 3731. [Google Scholar] [CrossRef] [PubMed]
- Spirescu, V.; Chircov, C.; Grumezescu, A.; Andronescu, E. Polymeric Nanoparticles for Antimicrobial Therapies: An up-to-date Overview. Polymers 2021, 13, 724. [Google Scholar] [CrossRef]
- Blecher, K.; Nasir, A.; Friedman, A. The growing role of nanotechnology in combating infectious disease. Virulence 2011, 2, 395–401. [Google Scholar] [CrossRef] [Green Version]
- Caetano, G.F.; Frade, M.A.C.; Andrade, T.A.M.; Leite, M.N.; Bueno, C.Z.; Moraes, Â.M.; Ribeiro-Paes, J.T. Chitosan-alginate membranes accelerate wound healing. J. Biomed. Mater. Res. Part B Appl. Biomater. 2015, 103, 1013–1022. [Google Scholar] [CrossRef]
- Nehra, P.; Chauhan, R.; Garg, N.; Verma, K. Antibacterial and antifungal activity of chitosan coated iron oxide nanoparticles. Br. J. Biomed. Sci. 2017, 75, 13–18. [Google Scholar] [CrossRef]
- Piras, A.M.; Maisetta, G.; Sandreschi, S.; Gazzarri, M.; Bartoli, C.; Grassi, L.; Esin, S.; Chiellini, F.; Batoni, G. Chitosan nanoparticles loaded with the antimicrobial peptide temporin B exert a long-term antibacterial activity in vitro against clinical isolates of Staphylococcus epidermidis. Front. Microbiol. 2015, 6, 372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassetti, M.; Vena, A.; Russo, A.; Peghin, M. Inhaled Liposomal Antimicrobial Delivery in Lung Infections. Drugs 2020, 80, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.; Ogren, M.; Dias, J.N.R.; Silva, M.; Gil, S.; Tavares, L.; Aires-Da-silva, F.; Gaspar, M.M.; Aguiar, S.I. Liposomes as Antibiotic Delivery Systems: A Promising Nanotechnological Strategy against Antimicrobial Resistance. Molecules 2021, 26, 2047. [Google Scholar] [CrossRef]
- Hardwick, J.; Taylor, J.; Mehta, M.; Satija, S.; Paudel, K.R.; Hansbro, P.M.; Chellappan, D.K.; Bebawy, M.; Dua, K. Targeting Cancer using Curcumin Encapsulated Vesicular Drug Delivery Systems. Curr. Pharm. Des. 2021, 27, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Drulis-Kawa, Z.; Dorotkiewicz-Jach, A. Liposomes as delivery systems for antibiotics. Int. J. Pharm. 2010, 387, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Bakker-Woudenberg, I.A.; Ten Kate, M.T.; Guo, L.; Working, P.; Mouton, J.W. Improved Efficacy of Ciprofloxacin Administered in Polyethylene Glycol-Coated Liposomes for Treatment of Klebsiella pneumoniae Pneumonia in Rats. Antimicrob. Agents Chemother. 2001, 45, 1487–1492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Li, Z.; Wang, X.; Chen, Y.; Wu, F.; Men, K.; Xu, T.; Luo, Y.; Yang, L. Novel Antimicrobial Peptide-Modified Azithromycin-Loaded Liposomes against Methicillin-Resistant Staphylococcus Aureus. Int. J. Nanomed 2016, 11, 6781–6794. [Google Scholar] [CrossRef] [Green Version]
- Lakshminarayanan, R.; Ye, E.; Young, D.J.; Li, Z.; Loh, X.J. Recent Advances in the Development of Antimicrobial Nanoparticles for Combating Resistant Pathogens. Adv. Health Mater. 2018, 7, e1701400. [Google Scholar] [CrossRef] [Green Version]
- Imran, M.; Iqubal, M.K.; Imtiyaz, K.; Saleem, S.; Mittal, S.; Rizvi, M.M.A.; Ali, J.; Baboota, S. Topical nanostructured lipid carrier gel of quercetin and resveratrol: Formulation, optimization, in vitro and ex vivo study for the treatment of skin cancer. Int. J. Pharm. 2020, 587, 119705. [Google Scholar] [CrossRef]
- Hallan, S.; Kaur, P.; Kaur, V.; Mishra, N.; Vaidya, B. Lipid polymer hybrid as emerging tool in nanocarriers for oral drug delivery. Artif. Cells Nanomed. Biotechnol. 2016, 44, 334–349. [Google Scholar] [CrossRef]
- Briones, E.; Colino, C.I.; Lanao, J.M. Delivery systems to increase the selectivity of antibiotics in phagocytic cells. J. Control. Release 2008, 125, 210–227. [Google Scholar] [CrossRef] [PubMed]
- Khatak, S.; Mehta, M.; Awasthi, R.; Paudel, K.R.; Singh, S.K.; Gulati, M.; Hansbro, N.G.; Hansbro, P.M.; Dua, K.; Dureja, H. Solid lipid nanoparticles containing anti-tubercular drugs attenuate the Mycobacterium marinum infection. Tuberculosis 2020, 125, 102008. [Google Scholar] [CrossRef] [PubMed]
- Ghaderkhani, J.; Yousefimashouf, R.; Arabestani, M.; Roshanaei, G.; Asl, S.S.; Abbasalipourkabir, R. Improved antibacterial function of Rifampicin-loaded solid lipid nanoparticles on Brucella abortus. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1181–1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, B.; Vuddanda, P.R.; Vijayakumar, M.R.; Kumar, V.; Saxena, P.S.; Singh, S. Cefuroxime axetil loaded solid lipid nanoparticles for enhanced activity against S. aureus biofilm. Colloids Surf. B Biointerfaces 2014, 121, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Kalhapure, R.S.; Mocktar, C.; Sikwal, D.R.; Sonawane, S.J.; Kathiravan, M.K.; Skelton, A.; Govender, T. Ion pairing with linoleic acid simultaneously enhances encapsulation efficiency and antibacterial activity of vancomycin in solid lipid nanoparticles. Colloids Surf. B Biointerf. 2014, 117, 303–311. [Google Scholar] [CrossRef]
- Ghaffari, S.; Varshosaz, J.; Saadat, A.; Atyabi, F. Stability and antimicrobial effect of amikacin-loaded solid lipid nanoparticles. Int. J. Nanomed. 2011, 6, 35–43. [Google Scholar] [CrossRef] [Green Version]
- Mehta, M.; Malyla, V.; Paudel, K.R.; Chellappan, D.K.; Hansbro, P.M.; Oliver, B.G.; Dua, K. Berberine loaded liquid crystalline nanostructure inhibits cancer progression in adenocarcinomic human alveolar basal epithelial cells in vitro. J. Food Biochem. 2021, 45, e13954. [Google Scholar] [CrossRef]
- Hasan, N.; Imran, M.; Kesharwani, P.; Khanna, K.; Karwasra, R.; Sharma, N.; Rawat, S.; Sharma, D.; Ahmad, F.J.; Jain, G.K.; et al. Intranasal delivery of Naloxone-loaded solid lipid nanoparticles as a promising simple and non-invasive approach for the management of opioid overdose. Int. J. Pharm. 2021, 599, 120428. [Google Scholar] [CrossRef]
- Imran, M.; Iqubal, M.K.; Ahmad, S.; Ali, J.; Baboota, S. Stability-Indicating High-Performance Thin-Layer Chromatographic Method for the Simultaneous Determination of Quercetin and Resveratrol in the Lipid-Based Nanoformulation. JPC J. Planar Chromatogr. Mod. TLC 2019, 32, 393–400. [Google Scholar] [CrossRef]
- Cortesi, R.; Valacchi, G.; Muresan, X.M.; Drechsler, M.; Contado, C.; Esposito, E.; Grandini, A.; Guerrini, A.; Forlani, G.; Sacchetti, G. Nanostructured lipid carriers (NLC) for the delivery of natural molecules with antimicrobial activity: Production, characterisation and in vitro studies. J. Microencapsul. 2016, 34, 63–72. [Google Scholar] [CrossRef]
- Vairo, C.; Basas, J.; Pastor, M.; Palau, M.; Gomis, X.; Almirante, B.; Gainza, E.; Hernandez, R.; Igartua, M.; Gavaldà, J.; et al. In vitro and in vivo antimicrobial activity of sodium colistimethate and amikacin-loaded nanostructured lipid carriers (NLC). Nanomed. Nanotechnol. Biol. Med. 2020, 29, 102259. [Google Scholar] [CrossRef] [PubMed]
- Svenson, S. Dendrimers as versatile platform in drug delivery applications. Eur. J. Pharm. Biopharm. 2009, 71, 445–462. [Google Scholar] [CrossRef]
- Chauhan, A.S. Dendrimers for Drug Delivery. Molecules 2018, 23, 938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huh, A.J.; Kwon, Y.J. “Nanoantibiotics”: A new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J. Control. Release 2011, 156, 128–145. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.S. Dendrimer nanotechnology for enhanced formulation and controlled delivery of resveratrol. Ann. N.Y. Acad. Sci. 2015, 1348, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Svenningsen, S.W.; Frederiksen, R.F.; Counil, C.; Ficker, M.; Leisner, J.J.; Christensen, J.B. Synthesis and Antimicrobial Properties of a Ciprofloxacin and PAMAM-dendrimer Conjugate. Molecules 2020, 25, 1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosnjakovic, A.; Mishra, M.K.; Ren, W.; Kurtoglu, Y.E.; Shi, T.; Fan, D.; Kannan, R.M. Poly(amidoamine) dendrimer-erythromycin conjugates for drug delivery to macrophages involved in periprosthetic inflammation. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 284–294. [Google Scholar] [CrossRef]
- Spirescu, V.; Chircov, C.; Grumezescu, A.; Vasile, B.; Andronescu, E. Inorganic Nanoparticles and Composite Films for Antimicrobial Therapies. Int. J. Mol. Sci. 2021, 22, 4595. [Google Scholar] [CrossRef]
- Kaur, R. Review Article on Inorganic Nanoparticles as Antibacterial Agents. Int. J. Res. Appl. Sci. Eng. Technol. 2020, 8, 944–948. [Google Scholar] [CrossRef]
- Miller, K.P.; Wang, L.; Benicewicz, B.C.; Decho, A.W. Inorganic nanoparticles engineered to attack bacteria. Chem. Soc. Rev. 2015, 44, 7787–7807. [Google Scholar] [CrossRef]
- Zhao, G.; Stevens, J.S.E. Multiple parameters for the comprehensive evaluation of the susceptibility of Escherichia coli to the silver ion. BioMetals 1998, 11, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Niyonshuti, I.I.; Krishnamurthi, V.R.; Okyere, D.; Song, L.; Benamara, M.; Tong, X.; Wang, Y.; Chen, J. Polydopamine Surface Coating Synergizes the Antimicrobial Activity of Silver Nanoparticles. ACS Appl. Mater. Interfaces 2020, 12, 40067–40077. [Google Scholar] [CrossRef] [PubMed]
- Hamed, S.; Emara, M.; Shawky, R.M.; El-Domany, R.A.; Youssef, T. Silver nanoparticles: Antimicrobial activity, cytotoxicity, and synergism with N-acetyl cysteine. J. Basic Microbiol. 2017, 57, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Dizaj, S.M.; Lotfipour, F.; Barzegar-Jalali, M.; Zarrintan, M.H.; Adibkia, K. Antimicrobial activity of the metals and metal oxide nanoparticles. Mater. Sci. Eng. C 2014, 44, 278–284. [Google Scholar] [CrossRef]
- Cui, Y.; Zhao, Y.; Tian, Y.; Zhang, W.; Lü, X.; Jiang, X. The molecular mechanism of action of bactericidal gold nanoparticles on Escherichia coli. Biomaterials 2012, 33, 2327–2333. [Google Scholar] [CrossRef]
- Tiwari, P.M.; Vig, K.; Dennis, V.A.; Singh, S.R. Functionalized Gold Nanoparticles and Their Biomedical Applications. Nanomaterials 2011, 1, 31–63. [Google Scholar] [CrossRef]
- Lima, E.; Guerra, R.; Lara, V.; Guzmán, A. Gold nanoparticles as efficient antimicrobial agents for Escherichia coli and Salmonella typhi. Chem. Central J. 2013, 7, 11. [Google Scholar] [CrossRef] [Green Version]
- Singh, S. Zinc oxide nanoparticles impacts: Cytotoxicity, genotoxicity, developmental toxicity, and neurotoxicity. Toxicol. Mech. Methods 2019, 29, 300–311. [Google Scholar] [CrossRef]
- Jain, D.; Shivani; Bhojiya, A.A.; Singh, H.; Daima, H.K.; Singh, M.; Mohanty, S.R.; Stephen, B.J.; Singh, A. Microbial Fabrication of Zinc Oxide Nanoparticles and Evaluation of Their Antimicrobial and Photocatalytic Properties. Front. Chem. 2020, 8, 778. [Google Scholar] [CrossRef]
- Azam, A.; Ahmed, A.S.; Oves, M.; Khan, M.S.; Habib, S.S.; Memic, A. Antimicrobial activity of metal oxide nanoparticles against Gram-positive and Gram-negative bacteria: A comparative study. Int. J. Nanomed. 2012, 7, 6003–6009. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; He, Y.; Irwin, P.L.; Jin, T.; Shi, X. Antibacterial Activity and Mechanism of Action of Zinc Oxide Nanoparticles against Campylobacter Jejuni. Appl. Environ. Microbiol. 2011, 77, 2325–2331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Magaye, R.; Castranova, V.; Zhao, J. Titanium dioxide nanoparticles: A review of current toxicological data. Part. Fibre Toxicol. 2013, 10, 15–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grande, F.; Tucci, P. Titanium Dioxide Nanoparticles: A Risk for Human Health? Mini-Rev. Med. Chem. 2016, 16, 762–769. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.S.; Parveen, A.; Koppalkar, A.R.; Prasad, M.V.N.A. Effect of Nano—Titanium Dioxide with Different Antibiotics against Methicillin-Resistant Staphylococcus Aureus. J. Biomater. Nanobiotechnol. 2010, 1, 37–41. [Google Scholar] [CrossRef] [Green Version]
- Haghighi, F.; Mohammadi, S.R.; Mohammadi, P.; Eskandari, M.; Hosseinkhani, S. The evaluation of Candida albicans biofilms formation on silicone catheter, PVC and glass coated with titanium dioxide nanoparticles by XTT method and ATPase assay. Bratisl. Lek. List. 2012, 113, 707–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boedicker, J.Q.; Li, L.; Kline, T.R.; Ismagilov, R.F. Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics. Lab Chip 2008, 8, 1265–1272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Bi, H.; Liu, B.; Qiao, L. Detection of Pathogenic Microorganisms by Microfluidics Based Analytical Methods. Anal. Chem. 2018, 90, 5512–5520. [Google Scholar] [CrossRef]
- Rahi, P.; Prakash, O.; Shouche, Y.S. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass-Spectrometry (MALDI-TOF MS) Based Microbial Identifications: Challenges and Scopes for Microbial Ecologists. Front. Microbiol. 2016, 7, 1359. [Google Scholar] [CrossRef] [Green Version]
- Hou, H.W.; Bhattacharyya, R.P.; Hung, D.T.; Han, J. Direct detection and drug-resistance profiling of bacteremias using inertial microfluidics. Lab Chip 2015, 15, 2297–2307. [Google Scholar] [CrossRef]
- Strommenger, B.; Kettlitz, C.; Werner, G.; Witte, W. Multiplex PCR Assay for Simultaneous Detection of Nine Clinically Relevant Antibiotic Resistance Genes in Staphylococcus aureus. J. Clin. Microbiol. 2003, 41, 4089–4094. [Google Scholar] [CrossRef] [Green Version]
- Sackmann, A.F.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yan, S.; Yuan, D.; Alici, G.; Nguyen, N.-T.; Warkiani, M.E.; Li, W. Fundamentals and applications of inertial microfluidics: A review. Lab Chip 2016, 16, 10–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mark, D.; Haeberle, S.; Roth, G.; Von Stetten, F.; Zengerle, R. Microfluidic Lab-on-a-Chip Platforms: Requirements, Characteristics and Applications. NATO Sci. Peace Secur. Ser. A Chem. Biol. 2010, 39, 305–376. [Google Scholar] [CrossRef]
- Nasseri, B.; Soleimani, N.; Rabiee, N.; Kalbasi, A.; Karimi, M.; Hamblin, M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018, 117, 112–128. [Google Scholar] [CrossRef]
- Wu, Z.; Willing, B.; Bjerketorp, J.; Jansson, J.K.; Hjort, K. Soft inertial microfluidics for high throughput separation of bacteria from human blood cells. Lab Chip 2009, 9, 1193–1199. [Google Scholar] [CrossRef] [Green Version]
- Hou, H.W.; Gan, H.Y.; Bhagat, A.A.S.; Li, L.D.; Lim, C.T.; Han, J. A microfluidics approach towards high-throughput pathogen removal from blood using margination. Biomicrofluidics 2012, 6, 24115. [Google Scholar] [CrossRef] [Green Version]
- Boehm, D.A.; Gottlieb, P.A.; Hua, S.Z. On-chip microfluidic biosensor for bacterial detection and identification. Sens. Actuators B Chem. 2007, 126, 508–514. [Google Scholar] [CrossRef]
- Altintas, Z.; Akgun, M.; Kokturk, G.; Uludag, Y. A fully automated microfluidic-based electrochemical sensor for real-time bacteria detection. Biosens. Bioelectron. 2018, 100, 541–548. [Google Scholar] [CrossRef]
- Yung, C.W.; Fiering, J.; Mueller, A.J.; Ingber, D.E. Micromagnetic–microfluidic blood cleansing device. Lab Chip 2009, 9, 1171–1177. [Google Scholar] [CrossRef]
- Kang, J.H.; Super, M.; Yung, C.W.; Cooper, R.M.; Domansky, K.; Graveline, A.; Mammoto, T.; Hicks-Berthet, J.; Tobin, H.; Cartwright, M.J.; et al. An extracorporeal blood-cleansing device for sepsis therapy. Nat. Med. 2014, 20, 1211–1216. [Google Scholar] [CrossRef]
- Breadmore, M.C.; Wolfe, K.A.; Arcibal, I.G.; Leung, W.K.; Dickson, D.; Giordano, B.C.; Power, M.E.; Ferrance, J.P.; Feldman, S.H.; Norris, P.M.; et al. Microchip-Based Purification of DNA from Biological Samples. Anal. Chem. 2003, 75, 1880–1886. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Fauvart, M.; Wiederkehr, R.S.; Jones, B.; Cools, P.; Goos, P.; Vaneechoutte, M.; Stakenborg, T. Ultra-fast, sensitive and quantitative on-chip detection of group B streptococci in clinical samples. Talanta 2019, 192, 220–225. [Google Scholar] [CrossRef] [PubMed]
- Oblath, E.A.; Henley, W.H.; Alarie, J.P.; Ramsey, J.M. A microfluidic chip integrating DNA extraction and real-time PCR for the detection of bacteria in saliva. Lab Chip 2013, 13, 1325–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azizi, M.; Zaferani, M.; Cheong, S.H.; Abbaspourrad, A. Pathogenic Bacteria Detection Using RNA-Based Loop-Mediated Isothermal-Amplification-Assisted Nucleic Acid Amplification via Droplet Microfluidics. ACS Sens. 2019, 4, 841–848. [Google Scholar] [CrossRef]
- Shang, Y.; Sun, J.; Ye, Y.; Zhang, J.; Zhang, Y.; Sun, X. Loop-mediated isothermal amplification-based microfluidic chip for pathogen detection. Crit. Rev. Food Sci. Nutr. 2020, 60, 201–224. [Google Scholar] [CrossRef]
- Jin, Z.; Ding, G.; Yang, G.; Li, G.; Zhang, W.; Yang, L.; Li, W. Rapid detection of antibiotic resistance genes in lactic acid bacteria using PMMA-based microreactor arrays. Appl. Microbiol. Biotechnol. 2020, 104, 6375–6383. [Google Scholar] [CrossRef]
- Guo, P.-L.; Tang, M.; Hong, S.-L.; Yu, X.; Pang, D.-W.; Zhang, Z.-L. Combination of dynamic magnetophoretic separation and stationary magnetic trap for highly sensitive and selective detection of Salmonella typhimurium in complex matrix. Biosens. Bioelectron. 2015, 74, 628–636. [Google Scholar] [CrossRef]
- Feng, X.; Liu, B.-F.; Li, J.; Liu, X. Advances in coupling microfluidic chips to mass spectrometry. Mass Spectrom. Rev. 2014, 34, 535–557. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Low, L.A.; Mummery, C.; Berridge, B.R.; Austin, C.P.; Tagle, D.A. Organs-on-chips: Into the next decade. Nat. Rev. Drug Discov. 2021, 20, 345–361. [Google Scholar] [CrossRef]
- Shrestha, J.; Ryan, S.T.; Mills, O.; Zhand, S.; Bazaz, S.R.; Hansbro, P.M.; Ghadiri, M.; Warkiani, M.E. A 3D-printed microfluidic platform for simulating the effects of CPAP on the nasal epithelium. Biofabrication 2021, 13, 035028. [Google Scholar] [CrossRef] [PubMed]
- Bein, A.; Shin, W.; Jalili-Firoozinezhad, S.; Park, M.H.; Sontheimer-Phelps, A.; Tovaglieri, A.; Chalkiadaki, A.; Kim, H.J.; Ingber, D.E. Microfluidic Organ-on-a-Chip Models of Human Intestine. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 659–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Tian, T. Recent advances in an organ-on-a-chip: Biomarker analysis and applications. Anal. Methods 2018, 10, 3122–3130. [Google Scholar] [CrossRef]
- Shrestha, J.; Bazaz, S.R.; Es, H.A.; Azari, D.Y.; Thierry, B.; Warkiani, M.E.; Ghadiri, M. Lung-on-a-chip: The future of respiratory disease models and pharmacological studies. Crit. Rev. Biotechnol. 2020, 40, 213–230. [Google Scholar] [CrossRef]
- Dai, J.; Hamon, M.; Jambovane, S. Microfluidics for Antibiotic Susceptibility and Toxicity Testing. Bioengineering 2016, 3, 25. [Google Scholar] [CrossRef] [Green Version]
- Safavieh, M.; Ahmed, M.U.; Tolba, M.; Zourob, M. Microfluidic electrochemical assay for rapid detection and quantification of Escherichia coli. Biosens. Bioelectron. 2012, 31, 523–528. [Google Scholar] [CrossRef]
- Lee, W.; Kwon, D.; Choi, W.; Jung, G.Y.; Au, A.K.; Folch, A.; Jeon, S. 3D-Printed Microfluidic Device for the Detection of Pathogenic Bacteria Using Size-based Separation in Helical Channel with Trapezoid Cross-Section. Sci. Rep. 2015, 5, 7717. [Google Scholar] [CrossRef]
| SI. No | Nanoparticles | Microorganism Targeted | Inference |
|---|---|---|---|
| 1 | Solid lipid nanoparticles | B. abortus; S. aureus |
|
| 2 | Liposomes | Klebsiella pneumoniae; Pseudomonas aeruginosa pneumonia; Methicillin-resistant Staphylococcus aureus |
|
| 3 | Polymeric nanoparticles | Candida albicans; Aspergillus niger; Bacillus subtilis Fusarium solani; Staphylococcus epidermidis |
|
| 4 | Dendrimers | Gram-positive; Gram-negative |
|
| 5 | Inorganic nanoparticles i.Ag-NPs ii.Au-NPs iii.ZnO NPs iv.TiO2 NPs | E. coli Staphylococcus aureus, Pseudomonas aeruginosa, Escherichia coli, Aspergillus fumigates, and Aspergillus niger Gram positive; Gram-negative bacteria Candida albicans Staphylococcus aureus. Candida albicans |
|
| 6 | NO-NPs | Pseudomonas aeruginosa; Burkholderia cepacian; Staphylococcus aureus; Methicillin-resistant S. aureus |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imran, M.; Jha, S.K.; Hasan, N.; Insaf, A.; Shrestha, J.; Shrestha, J.; Devkota, H.P.; Khan, S.; Panth, N.; Warkiani, M.E.; et al. Overcoming Multidrug Resistance of Antibiotics via Nanodelivery Systems. Pharmaceutics 2022, 14, 586. https://doi.org/10.3390/pharmaceutics14030586
Imran M, Jha SK, Hasan N, Insaf A, Shrestha J, Shrestha J, Devkota HP, Khan S, Panth N, Warkiani ME, et al. Overcoming Multidrug Resistance of Antibiotics via Nanodelivery Systems. Pharmaceutics. 2022; 14(3):586. https://doi.org/10.3390/pharmaceutics14030586
Chicago/Turabian StyleImran, Mohammad, Saurav Kumar Jha, Nazeer Hasan, Areeba Insaf, Jitendra Shrestha, Jesus Shrestha, Hari Prasad Devkota, Salman Khan, Nisha Panth, Majid Ebrahimi Warkiani, and et al. 2022. "Overcoming Multidrug Resistance of Antibiotics via Nanodelivery Systems" Pharmaceutics 14, no. 3: 586. https://doi.org/10.3390/pharmaceutics14030586
APA StyleImran, M., Jha, S. K., Hasan, N., Insaf, A., Shrestha, J., Shrestha, J., Devkota, H. P., Khan, S., Panth, N., Warkiani, M. E., Dua, K., Hansbro, P. M., Paudel, K. R., & Mohammed, Y. (2022). Overcoming Multidrug Resistance of Antibiotics via Nanodelivery Systems. Pharmaceutics, 14(3), 586. https://doi.org/10.3390/pharmaceutics14030586












