Bioimaging Nucleic-Acid Aptamers with Different Specificities in Human Glioblastoma Tissues Highlights Tumoral Heterogeneity
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Western Blot
2.4. Flow Cytometry
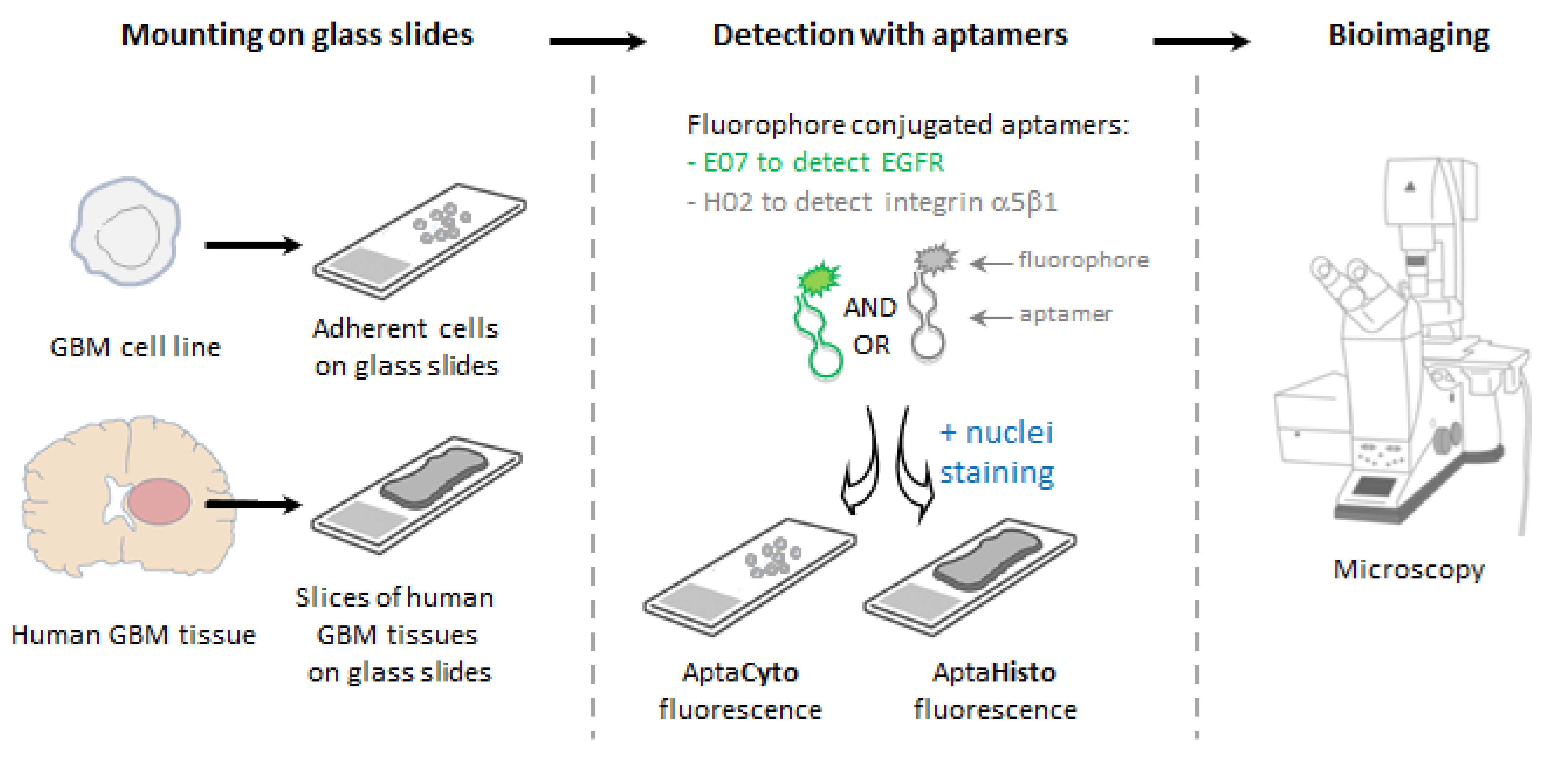
2.5. Fluorescence-Based Assays on Cell Lines
2.6. Human Tissue Samples
2.7. Fluorescence-Based Labeling Assays on Human Tissue Samples
2.8. EGFR Immunostaining of Human Tissue Samples
2.9. Imaging
3. Results
3.1. Validation of Target Expression and Aptamer Binding to Cell Lines
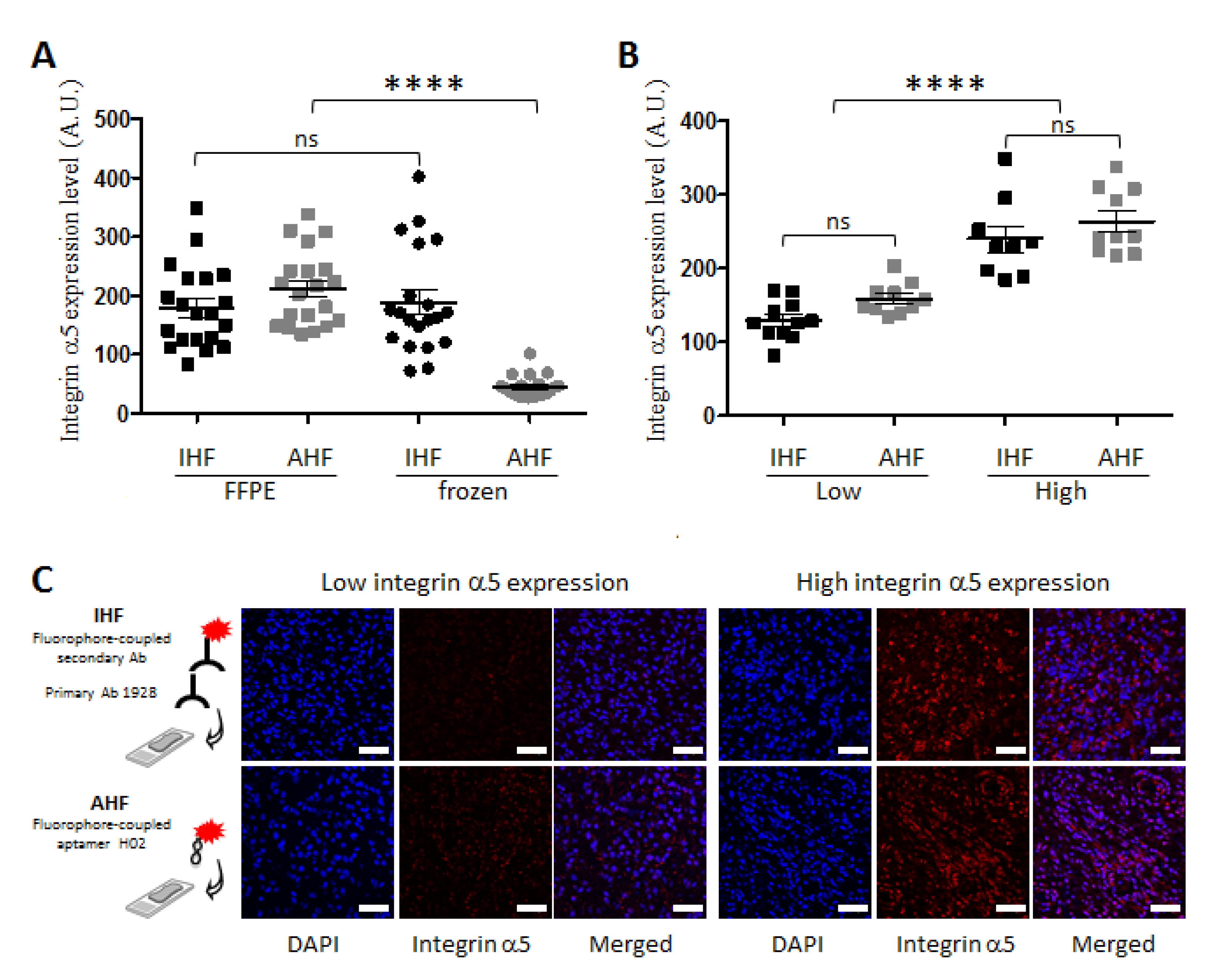
3.2. Apta- and Immunodetection of Integrin α5β1 in Paraffin-Embedded and Frozen Glioblastoma Sections
3.3. Detection of Integrin α5β1 Using Apta- and Immunohistofluorescence on FFPE GBM Sections Highlighted Inter-Tumoral Heterogeneity
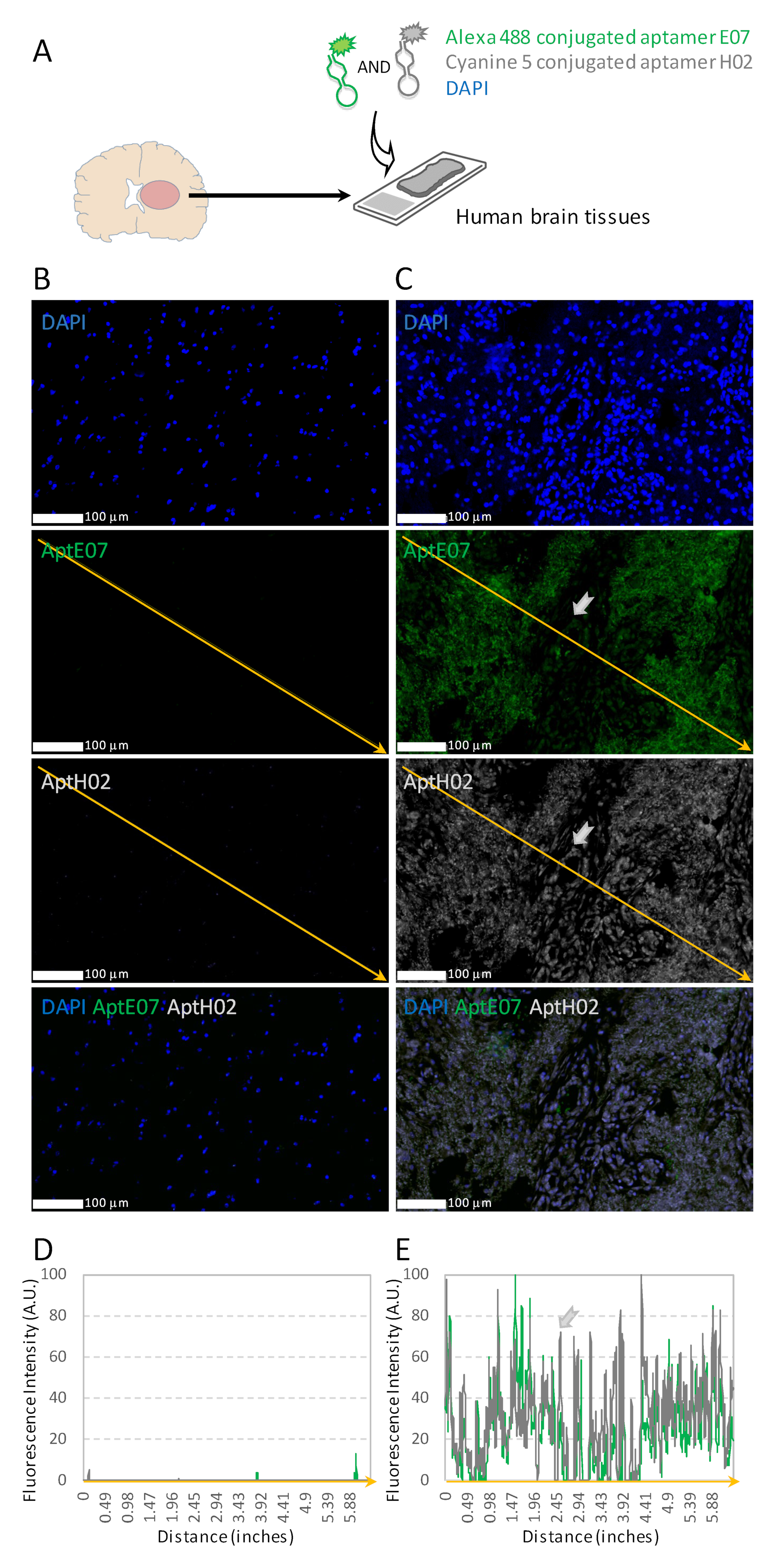
3.4. Aptahistofluorescence to Highlight Intra-Tumoral Heterogeneity
3.5. Multiplexing with Aptamers with Different Specificities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parra, E.R.; Francisco-Cruz, A.; Wistuba, I.I. State-of-the-Art of Profiling Immune Contexture in the Era of Multiplexed Staining and Digital Analysis to Study Paraffin Tumor Tissues. Cancers 2019, 11, 247. [Google Scholar] [CrossRef] [PubMed]
- Francisco-Cruz, A.; Parra, E.R.; Tetzlaff, M.T.; Wistuba, I.I. Multiplex Immunofluorescence Assays. In Biomarkers for Immunotherapy of Cancer. Methods in Molecular Biology; Humana: New York, NY, USA, 2020; Volume 2055, pp. 467–495. [Google Scholar] [CrossRef]
- Blom, S.; Paavolainen, L.; Bychkov, D.; Turkki, R.; Mäki-Teeri, P.; Hemmes, A.; Välimäki, K.; Lundin, J.; Kallioniemi, O.; Pellinen, T. Systems Pathology by Multiplexed Immunohistochemistry and Whole-Slide Digital Image Analysis. Sci. Rep. 2017, 7, 15580. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-W.; Ren, Y.J.; Marella, M.; Wang, M.; Hartke, J.; Couto, S.S. Multiplex Immunofluorescence Staining and Image Analysis Assay for Diffuse Large B Cell Lymphoma. J. Immunol. Methods 2020, 478, 112714. [Google Scholar] [CrossRef] [PubMed]
- Van Gijlswijk, R.P.M.; Zijlmans, H.J.M.A.A.; Wiegant, J.; Bobrow, M.N.; Erickson, T.J.; Adler, K.E.; Tanke, H.J.; Raap, A.K. Fluorochrome-Labeled Tyramides: Use in Immunocytochemistry and Fluorescence In Situ Hybridization. J. Histochem. Cytochem. 1997, 45, 375–382. [Google Scholar] [CrossRef]
- Bolognesi, M.M.; Manzoni, M.; Scalia, C.R.; Zannella, S.; Bosisio, F.M.; Faretta, M.; Cattoretti, G. Multiplex Staining by Sequential Immunostaining and Antibody Removal on Routine Tissue Sections. J. Histochem. Cytochem. 2017, 65, 431–444. [Google Scholar] [CrossRef]
- Zhou, G.; Wilson, G.; Hebbard, L.; Duan, W.; Liddle, C.; George, J.; Qiao, L. Aptamers: A Promising Chemical Antibody for Cancer Therapy. Oncotarget 2016, 7, 13446–13463. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as Targeted Therapeutics: Current Potential and Challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef]
- Bauer, M.; Strom, M.; Hammond, D.S.; Shigdar, S. Anything You Can Do, I Can Do Better: Can Aptamers Replace Antibodies in Clinical Diagnostic Applications? Molecules 2019, 24, 4377. [Google Scholar] [CrossRef]
- Roy, D.; Pascher, A.; Juratli, M.A.; Sporn, J.C. The Potential of Aptamer-Mediated Liquid Biopsy for Early Detection of Cancer. Int. J. Mol. Sci. 2021, 22, 5601. [Google Scholar] [CrossRef]
- Zamay, A.S.; Zamay, G.S.; Kolovskaya, O.S.; Zamay, T.N.; Berezovski, M.V. Aptamer-Based Methods for Detection of Circulating Tumor Cells and Their Potential for Personalized Diagnostics. Adv. Exp. Med. Biol. 2017, 994, 67–81. [Google Scholar] [CrossRef]
- Xiang, D.; Zheng, C.; Zhou, S.-F.; Qiao, S.; Tran, P.H.-L.; Pu, C.; Li, Y.; Kong, L.; Kouzani, A.Z.; Lin, J.; et al. Superior Performance of Aptamer in Tumor Penetration over Antibody: Implication of Aptamer-Based Theranostics in Solid Tumors. Theranostics 2015, 5, 1083–1097. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.; Macdonald, J.; Henri, J.; Duan, W.; Shigdar, S. The Application of Aptamers for Immunohistochemistry. Nucleic Acid Ther. 2016, 26, 120–126. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhang, P.; Zhao, N.; Sheehan, A.M.; Tung, C.-H.; Chang, C.-C.; Zu, Y. Using Oligonucleotide Aptamer Probes for Immunostaining of Formalin-Fixed and Paraffin-Embedded Tissues. Mod. Pathol. 2010, 23, 1553–1558. [Google Scholar] [CrossRef]
- Malicki, S.; Pucelik, B.; Żyła, E.; Benedyk-Machaczka, M.; Gałan, W.; Golda, A.; Sochaj-Gregorczyk, A.; Kamińska, M.; Encarnação, J.C.; Chruścicka, B.; et al. Imaging of Clear Cell Renal Carcinoma with Immune Checkpoint Targeting Aptamer-Based Probe. Pharm. Basel Switz. 2022, 15, 697. [Google Scholar] [CrossRef] [PubMed]
- Bukari, B.A.; Citartan, M.; Ch’ng, E.S.; Bilibana, M.P.; Rozhdestvensky, T.; Tang, T.-H. Aptahistochemistry in Diagnostic Pathology: Technical Scrutiny and Feasibility. Histochem. Cell Biol. 2017, 147, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Tan, W.; Zu, Y. Aptamers: Versatile Molecular Recognition Probes for Cancer Detection. Analyst 2016, 141, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Zamay, G.S.; Ivanchenko, T.I.; Zamay, T.N.; Grigorieva, V.L.; Glazyrin, Y.E.; Kolovskaya, O.S.; Garanzha, I.V.; Barinov, A.A.; Krat, A.V.; Mironov, G.G.; et al. DNA Aptamers for the Characterization of Histological Structure of Lung Adenocarcinoma. Mol. Ther. Nucleic Acids 2017, 6, 150–162. [Google Scholar] [CrossRef]
- Poon, M.T.C.; Sudlow, C.L.M.; Figueroa, J.D.; Brennan, P.M. Longer-Term (≥2 Years) Survival in Patients with Glioblastoma in Population-Based Studies Pre- and Post-2005: A Systematic Review and Meta-Analysis. Sci. Rep. 2020, 10, 11622. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Cruz Da Silva, E.; Mercier, M.-C.; Etienne-Selloum, N.; Dontenwill, M.; Choulier, L. A Systematic Review of Glioblastoma-Targeted Therapies in Phases II, III, IV Clinical Trials. Cancers 2021, 13, 1795. [Google Scholar] [CrossRef]
- Kros, J.M. Grading of Gliomas: The Road from Eminence to Evidence. J. Neuropathol. Exp. Neurol. 2011, 70, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Cloughesy, T.F.; Cavenee, W.K.; Mischel, P.S. Glioblastoma: From Molecular Pathology to Targeted Treatment. Annu. Rev. Pathol. 2014, 9, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Jacobi, N.; Seeboeck, R.; Hofmann, E.; Eger, A. ErbB Family Signalling: A Paradigm for Oncogene Addiction and Personalized Oncology. Cancers 2017, 9, 33. [Google Scholar] [CrossRef]
- Olayioye, M.A.; Neve, R.M.; Lane, H.A.; Hynes, N.E. The ErbB Signaling Network: Receptor Heterodimerization in Development and Cancer. EMBO J. 2000, 19, 3159–3167. [Google Scholar] [CrossRef]
- Frederick, L.; Wang, X.-Y.; Eley, G.; James, C.D. Diversity and Frequency of Epidermal Growth Factor Receptor Mutations in Human Glioblastomas. Cancer Res. 2000, 60, 1383–1387. [Google Scholar]
- Eskilsson, E.; Røsland, G.V.; Solecki, G.; Wang, Q.; Harter, P.N.; Graziani, G.; Verhaak, R.G.W.; Winkler, F.; Bjerkvig, R.; Miletic, H. EGFR Heterogeneity and Implications for Therapeutic Intervention in Glioblastoma. Neuro-Oncology 2018, 20, 743–752. [Google Scholar] [CrossRef]
- Lowell, C.A.; Mayadas, T.N. Overview-Studying Integrins in Vivo. In Integrin and Cell Adhesion Molecules. Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2011; Volume 757, pp. 369–397. [Google Scholar] [CrossRef]
- Blandin, A.-F.; Renner, G.; Lehmann, M.; Lelong-Rebel, I.; Martin, S.; Dontenwill, M. Β1 Integrins as Therapeutic Targets to Disrupt Hallmarks of Cancer. Front. Pharmacol. 2015, 6, 279. [Google Scholar] [CrossRef]
- Cruz da Silva, E.; Dontenwill, M.; Choulier, L.; Lehmann, M. Role of Integrins in Resistance to Therapies Targeting Growth Factor Receptors in Cancer. Cancers 2019, 11, 692. [Google Scholar] [CrossRef]
- Hamidi, H.; Ivaska, J. Every Step of the Way: Integrins in Cancer Progression and Metastasis. Nat. Rev. Cancer 2018, 18, 533–548. [Google Scholar] [CrossRef] [PubMed]
- Desgrosellier, J.S.; Cheresh, D.A. Integrins in Cancer: Biological Implications and Therapeutic Opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Harburger, D.S.; Calderwood, D.A. Integrin Signalling at a Glance. J. Cell Sci. 2009, 122, 159–163. [Google Scholar] [CrossRef]
- Paolillo, M.; Serra, M.; Schinelli, S. Integrins in Glioblastoma: Still an Attractive Target? Pharmacol. Res. 2016, 113, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Gingras, M.C.; Roussel, E.; Bruner, J.M.; Branch, C.D.; Moser, R.P. Comparison of Cell Adhesion Molecule Expression between Glioblastoma Multiforme and Autologous Normal Brain Tissue. J. Neuroimmunol. 1995, 57, 143–153. [Google Scholar] [CrossRef]
- Janouskova, H.; Maglott, A.; Leger, D.Y.; Bossert, C.; Noulet, F.; Guerin, E.; Guenot, D.; Pinel, S.; Chastagner, P.; Plenat, F.; et al. Integrin A5β1 Plays a Critical Role in Resistance to Temozolomide by Interfering with the P53 Pathway in High-Grade Glioma. Cancer Res. 2012, 72, 3463–3470. [Google Scholar] [CrossRef] [PubMed]
- Freije, W.A.; Castro-Vargas, F.E.; Fang, Z.; Horvath, S.; Cloughesy, T.; Liau, L.M.; Mischel, P.S.; Nelson, S.F. Gene Expression Profiling of Gliomas Strongly Predicts Survival. Cancer Res. 2004, 64, 6503–6510. [Google Scholar] [CrossRef]
- Phillips, H.S.; Kharbanda, S.; Chen, R.; Forrest, W.F.; Soriano, R.H.; Wu, T.D.; Misra, A.; Nigro, J.M.; Colman, H.; Soroceanu, L.; et al. Molecular Subclasses of High-Grade Glioma Predict Prognosis, Delineate a Pattern of Disease Progression, and Resemble Stages in Neurogenesis. Cancer Cell 2006, 9, 157–173. [Google Scholar] [CrossRef]
- Etienne-Selloum, N.; Prades, J.; Bello-Roufai, D.; Boone, M.; Sevestre, H.; Trudel, S.; Caillet, P.; Coutte, A.; Desenclos, C.; Constans, J.-M.; et al. Expression Analysis of A5 Integrin Subunit Reveals Its Upregulation as a Negative Prognostic Biomarker for Glioblastoma. Pharmaceuticals 2021, 14, 882. [Google Scholar] [CrossRef]
- Wen, P.Y.; Weller, M.; Lee, E.Q.; Alexander, B.M.; Barnholtz-Sloan, J.S.; Barthel, F.P.; Batchelor, T.T.; Bindra, R.S.; Chang, S.M.; Chiocca, E.A.; et al. Glioblastoma in Adults: A Society for Neuro-Oncology (SNO) and European Society of Neuro-Oncology (EANO) Consensus Review on Current Management and Future Directions. Neuro-Oncology 2020, 22, 1073–1113. [Google Scholar] [CrossRef]
- Bonavia, R.; Inda, M.M.; Vandenberg, S.; Cheng, S.-Y.; Nagane, M.; Hadwiger, P.; Tan, P.; Sah, D.W.Y.; Cavenee, W.K.; Furnari, F.B. EGFRvIII Promotes Glioma Angiogenesis and Growth through the NF-ΚB, Interleukin-8 Pathway. Oncogene 2012, 31, 4054–4066. [Google Scholar] [CrossRef] [PubMed]
- Fechter, P.; Cruz Da Silva, E.; Mercier, M.-C.; Noulet, F.; Etienne-Seloum, N.; Guenot, D.; Lehmann, M.; Vauchelles, R.; Martin, S.; Lelong-Rebel, I.; et al. RNA Aptamers Targeting Integrin A5β1 as Probes for Cyto- and Histofluorescence in Glioblastoma. Mol. Ther. Nucleic Acids 2019, 17, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Nguyen, H.H.; Byrom, M.; Ellington, A.D. Inhibition of Cell Proliferation by an Anti-EGFR Aptamer. PLoS ONE 2011, 6, e20299. [Google Scholar] [CrossRef] [PubMed]
- Schaffner, F.; Ray, A.M.; Dontenwill, M. Integrin A5β1, the Fibronectin Receptor, as a Pertinent Therapeutic Target in Solid Tumors. Cancers 2013, 5, 27–47. [Google Scholar] [CrossRef]
- Torp, S.H.; Bringedal, K.; Dalen, A. Immunohistochemical Detection of Epidermal Growth Factor Receptor in Human High-Grade Astrocytomas--a Comparison between Frozen- and Paraffin Sections. J. Exp. Clin. Cancer Res. 2005, 24, 89–92. [Google Scholar]
- Kratschmer, C.; Levy, M. Targeted Delivery of Auristatin-Modified Toxins to Pancreatic Cancer Using Aptamers. Mol. Ther. - Nucleic Acids 2018, 10, 227–236. [Google Scholar] [CrossRef]
- Becker, A.P.; Sells, B.E.; Haque, S.J.; Chakravarti, A. Tumor Heterogeneity in Glioblastomas: From Light Microscopy to Molecular Pathology. Cancers 2021, 13, 761. [Google Scholar] [CrossRef]
- Tan, W.C.C.; Nerurkar, S.N.; Cai, H.Y.; Ng, H.H.M.; Wu, D.; Wee, Y.T.F.; Lim, J.C.T.; Yeong, J.; Lim, T.K.H. Overview of Multiplex Immunohistochemistry/Immunofluorescence Techniques in the Era of Cancer Immunotherapy. Cancer Commun. Lond. Engl. 2020, 40, 135–153. [Google Scholar] [CrossRef]
- Bradbury, A.; Plückthun, A. Reproducibility: Standardize Antibodies Used in Research. Nature 2015, 518, 27–29. [Google Scholar] [CrossRef]
- Bradbury, A.R.M.; Plückthun, A. Getting to Reproducible Antibodies: The Rationale for Sequenced Recombinant Characterized Reagents. Protein Eng. Des. Sel. PEDS 2015, 28, 303–305. [Google Scholar] [CrossRef]
- Kelly, L.; Maier, K.E.; Yan, A.; Levy, M. A Comparative Analysis of Cell Surface Targeting Aptamers. Nat. Commun. 2021, 12, 6275. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Mercier, M.-C.; Dontenwill, M.; Choulier, L. Selection of Nucleic Acid Aptamers Targeting Tumor Cell-Surface Protein Biomarkers. Cancers 2017, 9, 69. [Google Scholar] [CrossRef]
- Li, S.; Xu, H.; Ding, H.; Huang, Y.; Cao, X.; Yang, G.; Li, J.; Xie, Z.; Meng, Y.; Li, X.; et al. Identification of an Aptamer Targeting HnRNP A1 by Tissue Slide-Based SELEX. J. Pathol. 2009, 218, 327–336. [Google Scholar] [CrossRef]
- Gomes de Castro, M.A.; Höbartner, C.; Opazo, F. Aptamers Provide Superior Stainings of Cellular Receptors Studied under Super-Resolution Microscopy. PLoS ONE 2017, 12, e0173050. [Google Scholar] [CrossRef]
- Whittington, N.C.; Wray, S. Suppression of Red Blood Cell Autofluorescence for Immunocytochemistry on Fixed Embryonic Mouse Tissue. Curr. Protoc. Neurosci. 2017, 81, 2–28. [Google Scholar] [CrossRef]
- Zhang, P.; Zhao, N.; Zeng, Z.; Chang, C.-C.; Zu, Y. Combination of an Aptamer Probe to CD4 and Antibodies for Multicolored Cell Phenotyping. Am. J. Clin. Pathol. 2010, 134, 586–593. [Google Scholar] [CrossRef]
- Zamay, G.S.; Kolovskaya, O.S.; Zamay, T.N.; Glazyrin, Y.E.; Krat, A.V.; Zubkova, O.; Spivak, E.; Wehbe, M.; Gargaun, A.; Muharemagic, D.; et al. Aptamers Selected to Postoperative Lung Adenocarcinoma Detect Circulating Tumor Cells in Human Blood. Mol. Ther. 2015, 23, 1486–1496. [Google Scholar] [CrossRef]
- Blandin, A.-F.; Cruz Da Silva, E.; Mercier, M.-C.; Glushonkov, O.; Didier, P.; Dedieu, S.; Schneider, C.; Devy, J.; Etienne-Selloum, N.; Dontenwill, M.; et al. Gefitinib Induces EGFR and A5β1 Integrin Co-Endocytosis in Glioblastoma Cells. Cell. Mol. Life Sci. 2021, 78, 2949–2962. [Google Scholar] [CrossRef]


| Aptamer | Target | Glioblastoma Cell Lines | KD | Reference |
|---|---|---|---|---|
| H02 | Integrin α5β1 | U87MG α5+ (expressing α5 integrin) | 277.8 ± 51.8 nM | [44] |
| E07 | EGFR | U87MG EGFR WT (expressing EGFR) | 208.7 ± 45.6 nM | Current study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz Da Silva, E.; Foppolo, S.; Lhermitte, B.; Ingremeau, M.; Justiniano, H.; Klein, L.; Chenard, M.-P.; Vauchelles, R.; Abdallah, B.; Lehmann, M.; et al. Bioimaging Nucleic-Acid Aptamers with Different Specificities in Human Glioblastoma Tissues Highlights Tumoral Heterogeneity. Pharmaceutics 2022, 14, 1980. https://doi.org/10.3390/pharmaceutics14101980
Cruz Da Silva E, Foppolo S, Lhermitte B, Ingremeau M, Justiniano H, Klein L, Chenard M-P, Vauchelles R, Abdallah B, Lehmann M, et al. Bioimaging Nucleic-Acid Aptamers with Different Specificities in Human Glioblastoma Tissues Highlights Tumoral Heterogeneity. Pharmaceutics. 2022; 14(10):1980. https://doi.org/10.3390/pharmaceutics14101980
Chicago/Turabian StyleCruz Da Silva, Elisabete, Sophie Foppolo, Benoît Lhermitte, Marina Ingremeau, Hélène Justiniano, Lorraine Klein, Marie-Pierre Chenard, Romain Vauchelles, Basma Abdallah, Maxime Lehmann, and et al. 2022. "Bioimaging Nucleic-Acid Aptamers with Different Specificities in Human Glioblastoma Tissues Highlights Tumoral Heterogeneity" Pharmaceutics 14, no. 10: 1980. https://doi.org/10.3390/pharmaceutics14101980
APA StyleCruz Da Silva, E., Foppolo, S., Lhermitte, B., Ingremeau, M., Justiniano, H., Klein, L., Chenard, M.-P., Vauchelles, R., Abdallah, B., Lehmann, M., Etienne-Selloum, N., Dontenwill, M., & Choulier, L. (2022). Bioimaging Nucleic-Acid Aptamers with Different Specificities in Human Glioblastoma Tissues Highlights Tumoral Heterogeneity. Pharmaceutics, 14(10), 1980. https://doi.org/10.3390/pharmaceutics14101980






