2D NMR Analysis as a Sensitive Tool for Evaluating the Higher-Order Structural Integrity of Monoclonal Antibody against COVID-19
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Nuclear Magnetic Resonance
2.3. Statistics on NMR Data
2.4. ELISA-Based Assay
3. Results
3.1. Comparison of Different mAb Batches and Stability over Time by NMR Technique
3.2. Stress Testing: Evaluation of HOS Perturbations after Incubation in H2O2 and at pH 10 by NMR and ELISA Methods
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andreano, E.; Nicastri, E.; Paciello, I.; Pileri, P.; Manganaro, N.; Piccini, G.; Manenti, A.; Pantano, E.; Kabanova, A.; Troisi, M.; et al. Extremely potent human monoclonal antibodies from COVID-19 convalescent patients. Cell 2021, 184, 1821–1835.e16. [Google Scholar] [CrossRef]
- Torres, J.L.; Ozorowski, G.; Andreano, E.; Liu, H.; Copps, J.; Piccini, G.; Donnici, L.; Conti, M.; Planchais, C.; Planas, D.; et al. Structural insights of a highly potent pan-neutralizing SARS-CoV-2 human monoclonal antibody. Proc. Natl. Acad. Sci. USA 2022, 119, e2120976119. [Google Scholar] [CrossRef]
- Lanini, S.; Milleri, S.; Andreano, E.; Nosari, S.; Paciello, I.; Piccini, G.; Gentili, A.; Phogat, A.; Hyseni, I.; Leonardi, M.; et al. Safety and serum distribution of anti-SARS-CoV-2 monoclonal antibody MAD0004J08 after intramuscular injection. Nat. Commun. 2022, 13, 2263. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, S.A.; Engen, J.R.; Mazzeo, J.R.; Jones, G.B. Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nat. Rev. Drug Discov. 2012, 11, 527–540. [Google Scholar] [CrossRef]
- Gabrielson, J.P.; Weiss, W.F. Technical decision-making with higher order structure data: Starting a new dialogue. J. Pharm. Sci. 2015, 104, 1240–1245. [Google Scholar] [CrossRef]
- Weiss, W.F.; Gabrielson, J.P.; Al-Azzam, W.; Chen, G.; Davis, D.L.; Das, T.K.; Hayes, D.B.; Houde, D.; Singh, S.K. Technical Decision Making with Higher Order Structure Data: Perspectives on Higher Order Structure Characterization from the Biopharmaceutical Industry. J. Pharm. Sci. 2016, 105, 3465–3470. [Google Scholar] [CrossRef]
- Verch, T.; Trausch, J.J.; Shank-Retzlaff, M. Principles of vaccine potency assays. Bioanalysis 2018, 10, 163–180. [Google Scholar] [CrossRef]
- Wen, J.; Batabyal, D.; Knutson, N.; Lord, H.; Wikström, M. A Comparison Between Emerging and Current Biophysical Methods for the Assessment of Higher-Order Structure of Biopharmaceuticals. J. Pharm. Sci. 2020, 109, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Hwang, T.L.; Batabyal, D.; Knutson, N.; Wikström, M. Use of the 2D (1)H-(13)C HSQC NMR Methyl Region to Evaluate the Higher Order Structural Integrity of Biopharmaceuticals. Molecules 2021, 26, 2714. [Google Scholar] [CrossRef]
- Brinson, R.G.; Marino, J.P.; Delaglio, F.; Arbogast, L.W.; Evans, R.M.; Kearsley, A.; Gingras, G.; Ghasriani, H.; Aubin, Y.; Pierens, G.K.; et al. Enabling adoption of 2D-NMR for the higher order structure assessment of monoclonal antibody therapeutics. MAbs 2019, 11, 94–105. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, D.; Cerofolini, L.; Giuntini, S.; Iozzino, L.; Pergola, C.; Sacco, F.; Palmese, A.; Ravera, E.; Luchinat, C.; Baroni, F.; et al. Epitope Mapping and Binding Assessment by Solid-State NMR Provide a Way for the Development of Biologics under the Quality by Design Paradigm. J. Am. Chem. Soc. 2022, 144, 10006–10016. [Google Scholar] [CrossRef]
- Caillet-Saguy, C.; Piccioli, M.; Turano, P.; Izadi-Pruneyre, N.; Delepierre, M.; Bertini, I.; Lecroisey, A. Mapping the Interaction between the Hemophore HasA and its Outer Membrane Receptor HasR using CRINEPT-TROSY NMR Spectroscopy. J. Am. Chem. Soc. 2009, 131, 1736–1744. [Google Scholar] [CrossRef] [PubMed]
- Müller, L. Alternate HMQC experiments for recording HN and HC-correlation spectra in proteins at high throughput. J. Biomol. NMR 2008, 42, 129–137. [Google Scholar] [CrossRef]
- Koos, M.R.M.; Luy, B. Polarization recovery during ASAP and SOFAST/ALSOFAST-type experiments. J. Magn. Reson. 2019, 300, 61–75. [Google Scholar] [CrossRef]
- Keller, R. The Computer Aided Resonance Assignment Tutorial; CANTINA Verlag: Goldau, Switzerland, 2004. [Google Scholar]
- Brinson, R.G.; Arbogast, L.W.; Marino, J.P.; Delaglio, F. Best Practices in Utilization of 2D-NMR Spectral Data as the Input for Chemometric Analysis in Biopharmaceutical Applications. J. Chem. Inf. Model. 2020, 60, 2339–2355. [Google Scholar] [CrossRef]
- Yandrofski, K.; Mouchahoir, T.; De Leoz, M.L.; Duewer, D.; Hudgens, J.W.; Anderson, K.W.; Arbogast, L.; Delaglio, F.; Brinson, R.G.; Marino, J.P.; et al. Interlaboratory Studies Using the NISTmAb to Advance Biopharmaceutical Structural Analytics. Front. Mol. Biosci. 2022, 9, 876780. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
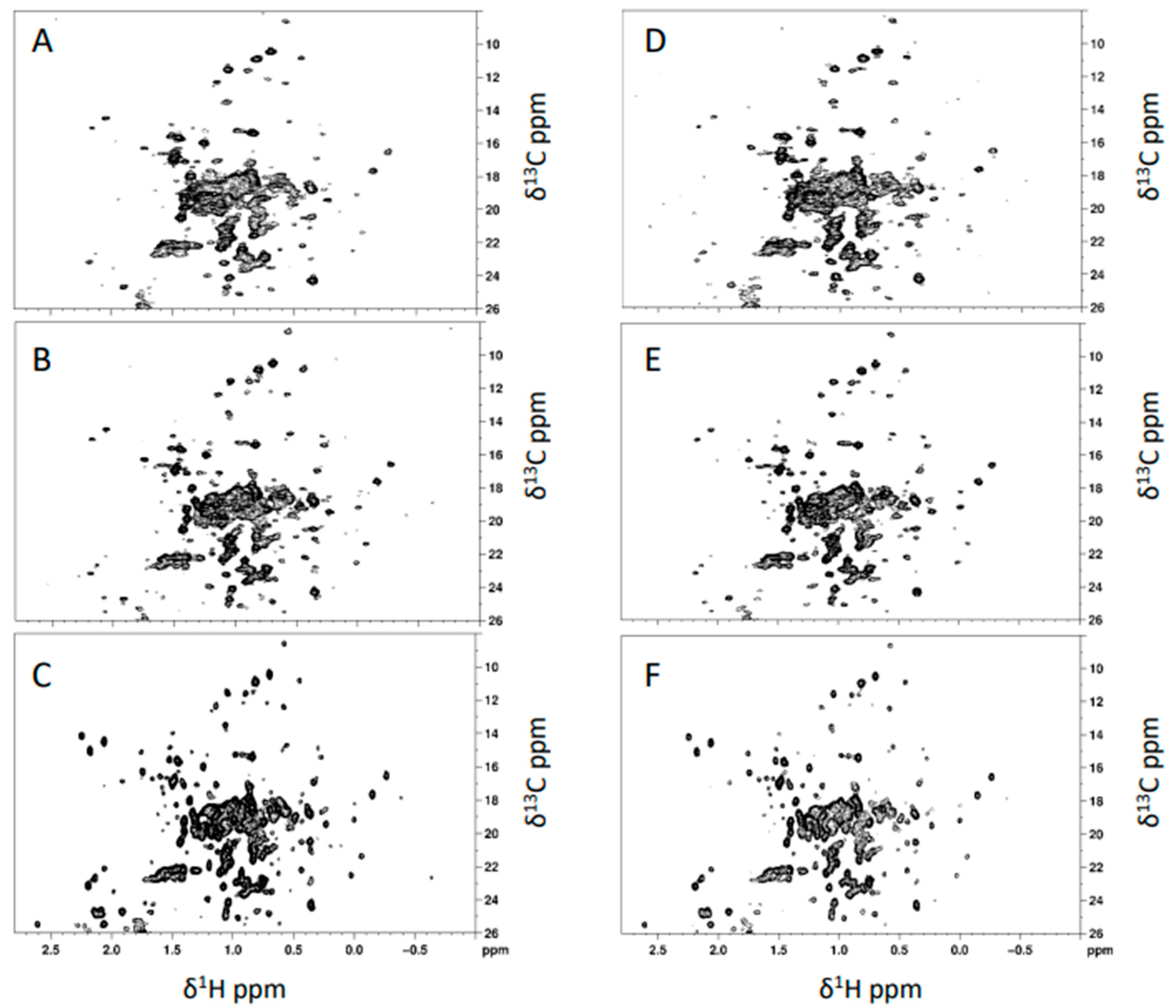

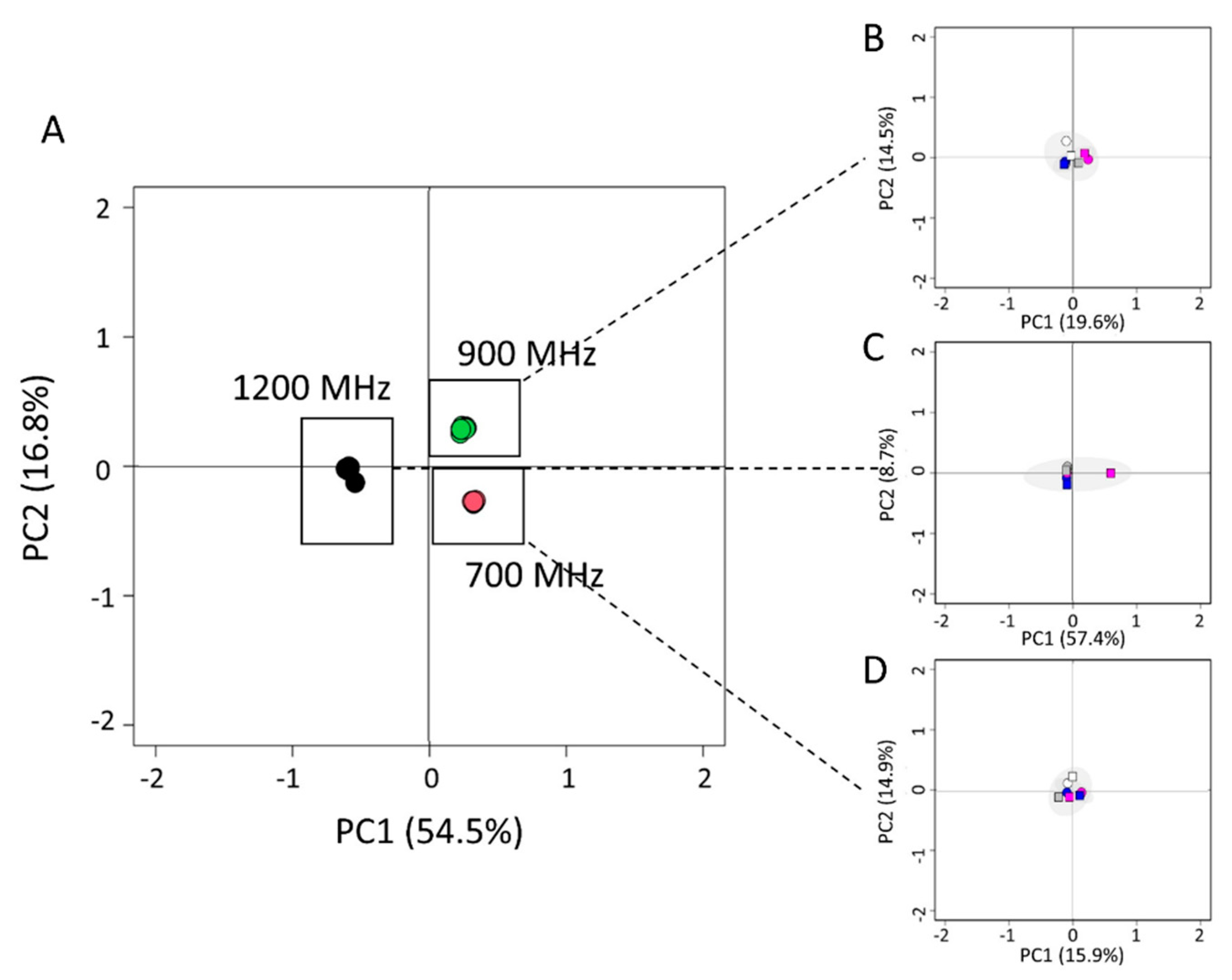

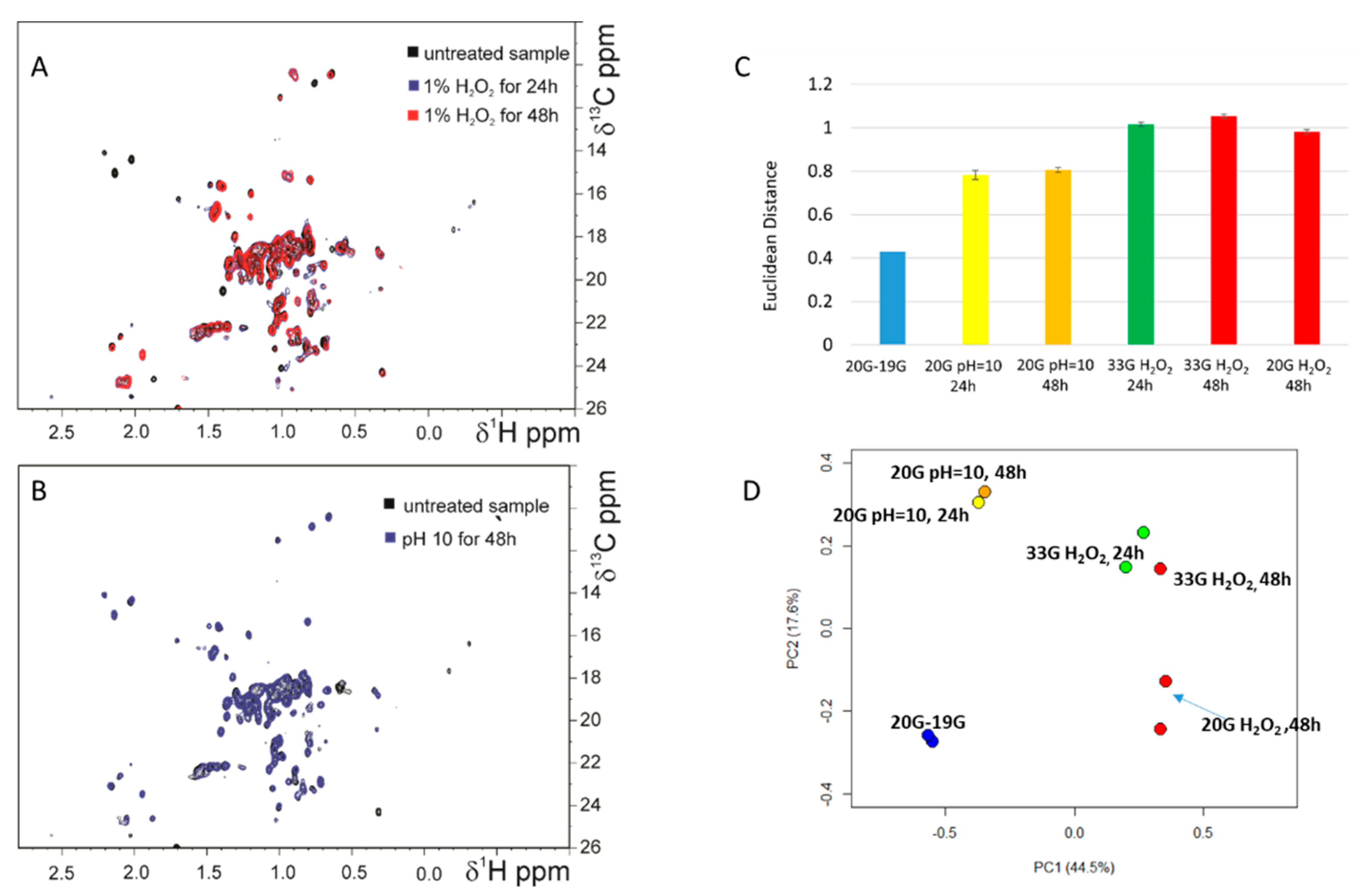

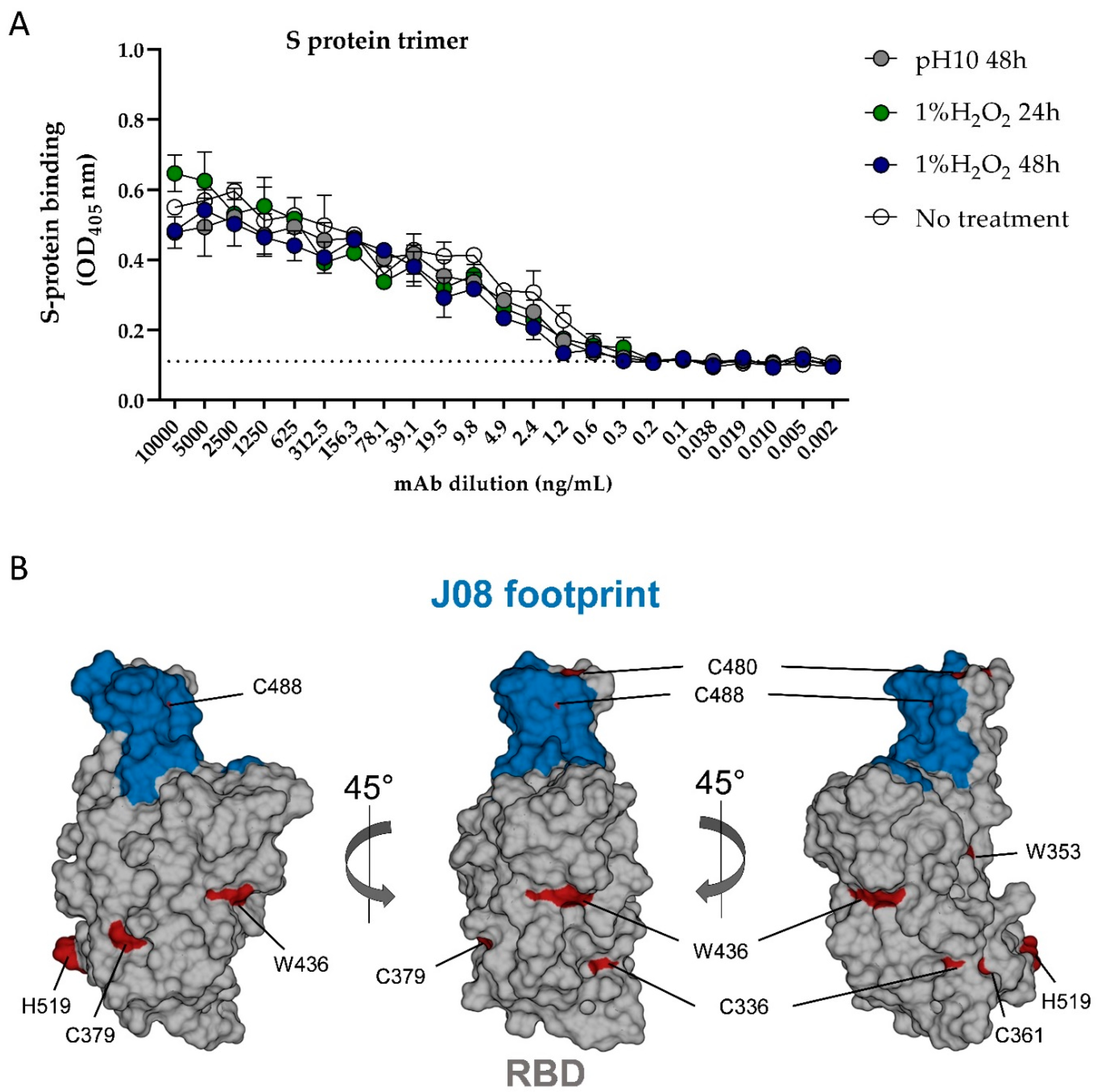
| Average CCSD (ppb) | mAb 19G T0 | mAb 19G T7 | mAb 19G T30 | mAb 19G T60 |
| mAb 19G T0 | 0.00 | 2.36 | 3.91 | 4.96 |
| mAb 19G T7 | 2.36 | 0.00 | 3.18 | 5.60 |
| mAb 19G T30 | 3.91 | 3.18 | 0.00 | 6.52 |
| mAb 19G T60 | 4.96 | 5.60 | 6.52 | 0.00 |
| Average CCSD (ppb) | mAb 20G T0 | mAb 20G T7 | mAb 20G T30 | mAb 20G T60 |
| mAb 20G T0 | 0.00 | 2.38 | 2.08 | 1.91 |
| mAb 20G T7 | 2.38 | 0.00 | 2.16 | 2.61 |
| mAb 20G T30 | 2.08 | 2.16 | 0.00 | 1.71 |
| mAb 20G T60 | 1.91 | 2.61 | 1.71 | 0.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cantini, F.; Andreano, E.; Paciello, I.; Ghini, V.; Berti, F.; Rappuoli, R.; Banci, L. 2D NMR Analysis as a Sensitive Tool for Evaluating the Higher-Order Structural Integrity of Monoclonal Antibody against COVID-19. Pharmaceutics 2022, 14, 1981. https://doi.org/10.3390/pharmaceutics14101981
Cantini F, Andreano E, Paciello I, Ghini V, Berti F, Rappuoli R, Banci L. 2D NMR Analysis as a Sensitive Tool for Evaluating the Higher-Order Structural Integrity of Monoclonal Antibody against COVID-19. Pharmaceutics. 2022; 14(10):1981. https://doi.org/10.3390/pharmaceutics14101981
Chicago/Turabian StyleCantini, Francesca, Emanuele Andreano, Ida Paciello, Veronica Ghini, Francesco Berti, Rino Rappuoli, and Lucia Banci. 2022. "2D NMR Analysis as a Sensitive Tool for Evaluating the Higher-Order Structural Integrity of Monoclonal Antibody against COVID-19" Pharmaceutics 14, no. 10: 1981. https://doi.org/10.3390/pharmaceutics14101981
APA StyleCantini, F., Andreano, E., Paciello, I., Ghini, V., Berti, F., Rappuoli, R., & Banci, L. (2022). 2D NMR Analysis as a Sensitive Tool for Evaluating the Higher-Order Structural Integrity of Monoclonal Antibody against COVID-19. Pharmaceutics, 14(10), 1981. https://doi.org/10.3390/pharmaceutics14101981







