Drug Repurposing Using Modularity Clustering in Drug-Drug Similarity Networks Based on Drug–Gene Interactions
Abstract
1. Introduction
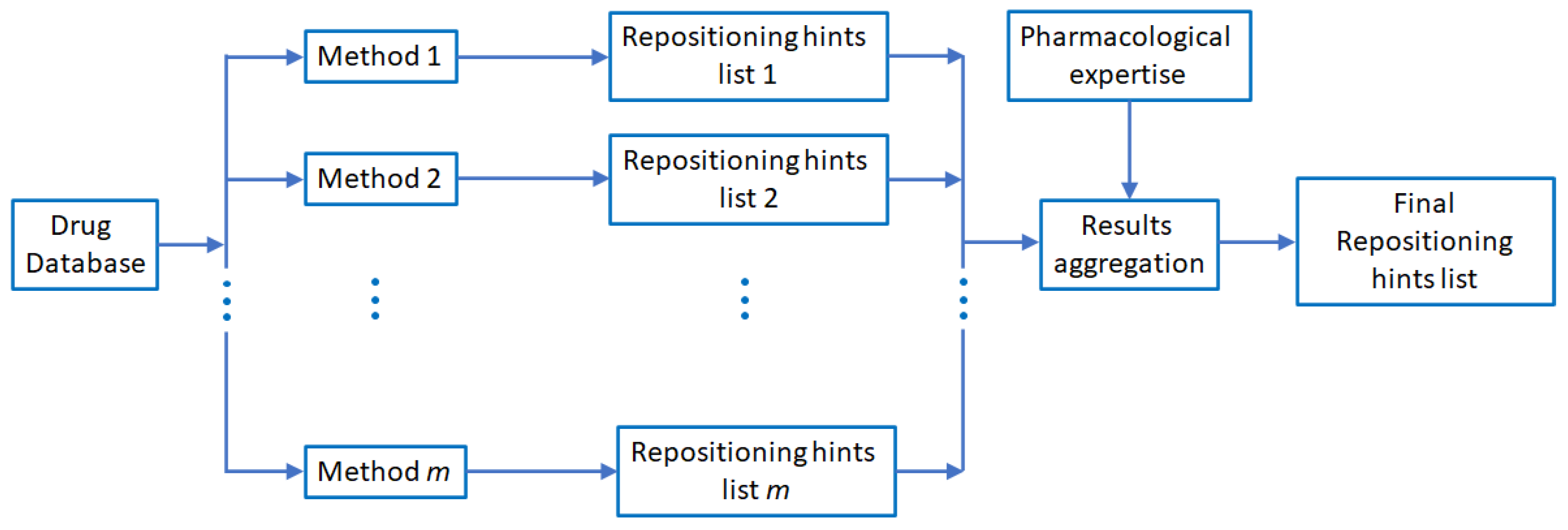
2. Materials and Methods
2.1. Databases
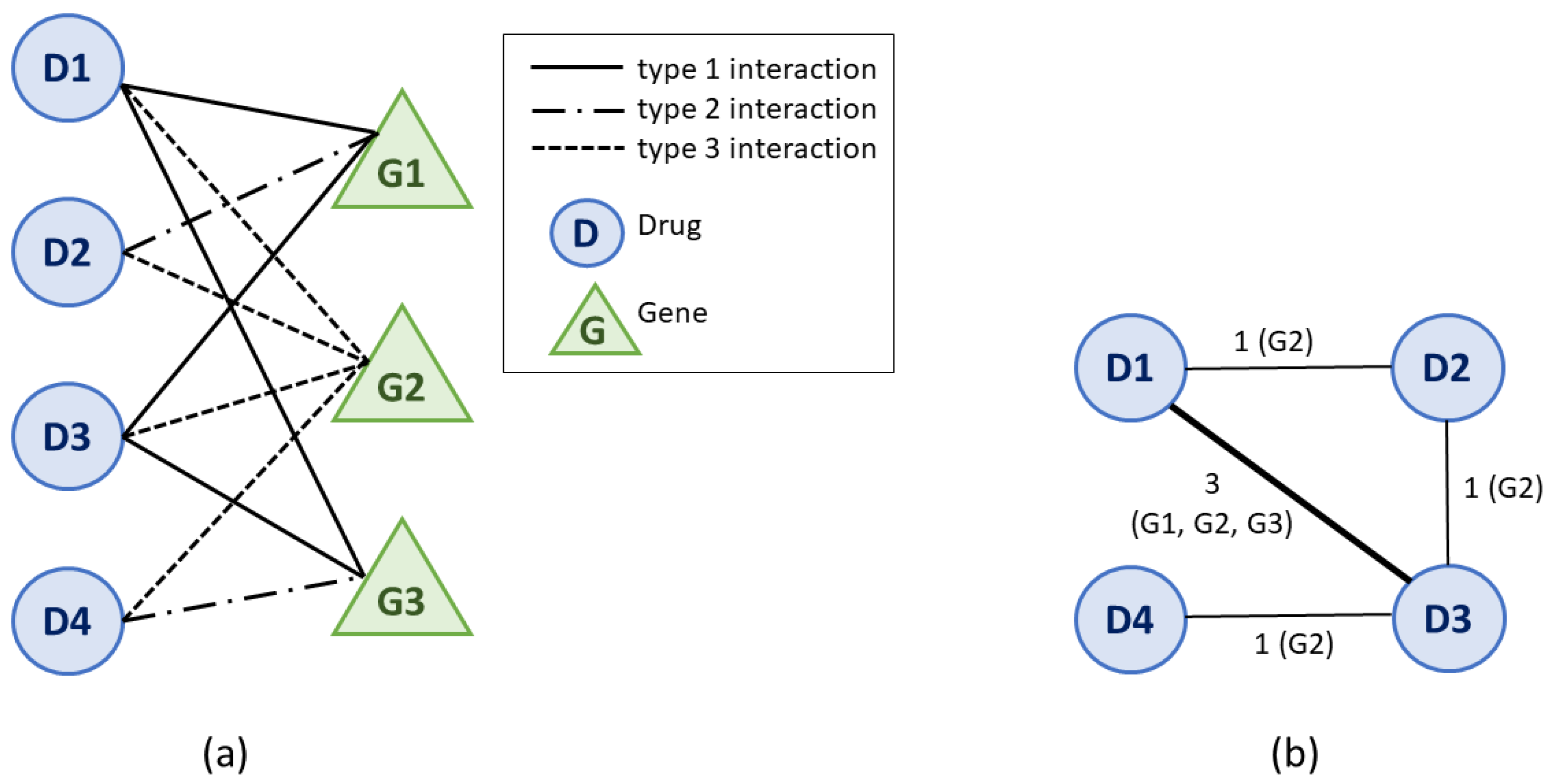
2.2. Building the Drug–Drug Similarity Network
2.3. Network Clustering Analysis
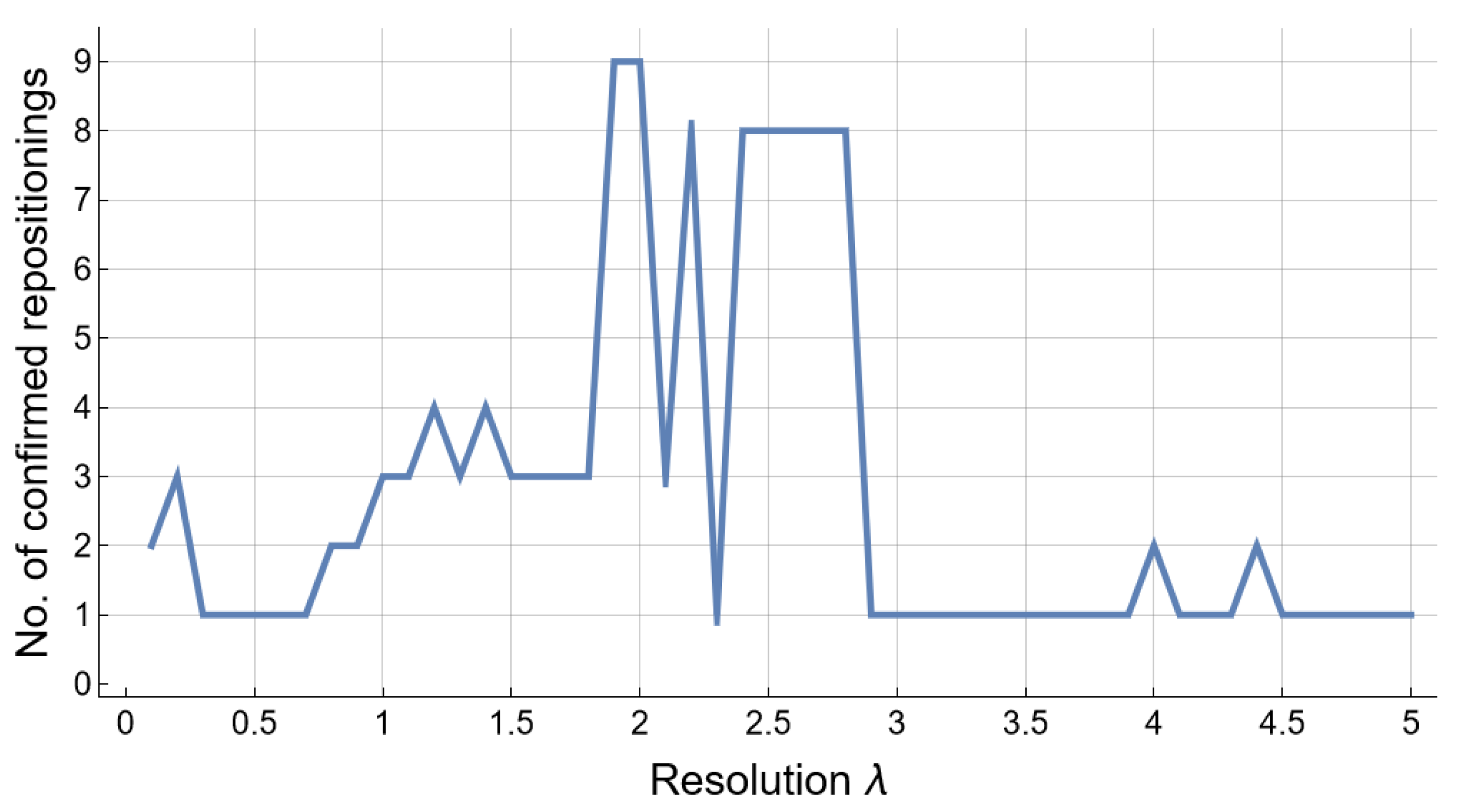
2.4. Tuning Resolution
| Algorithm 1 Find the parameter , such that the clustering of nodes/drugs in with modularity resolution (i.e., ) produces the biggest number of repositionings confirmed with the level 1 ATC codes in DrugBank 5.1.8. |
|
2.5. Generating New Repurposing Hints
| Algorithm 2 Generate the list of drug repurposing hints by clustering the DDSN with the tuned modularity resolution. |
|
3. Results
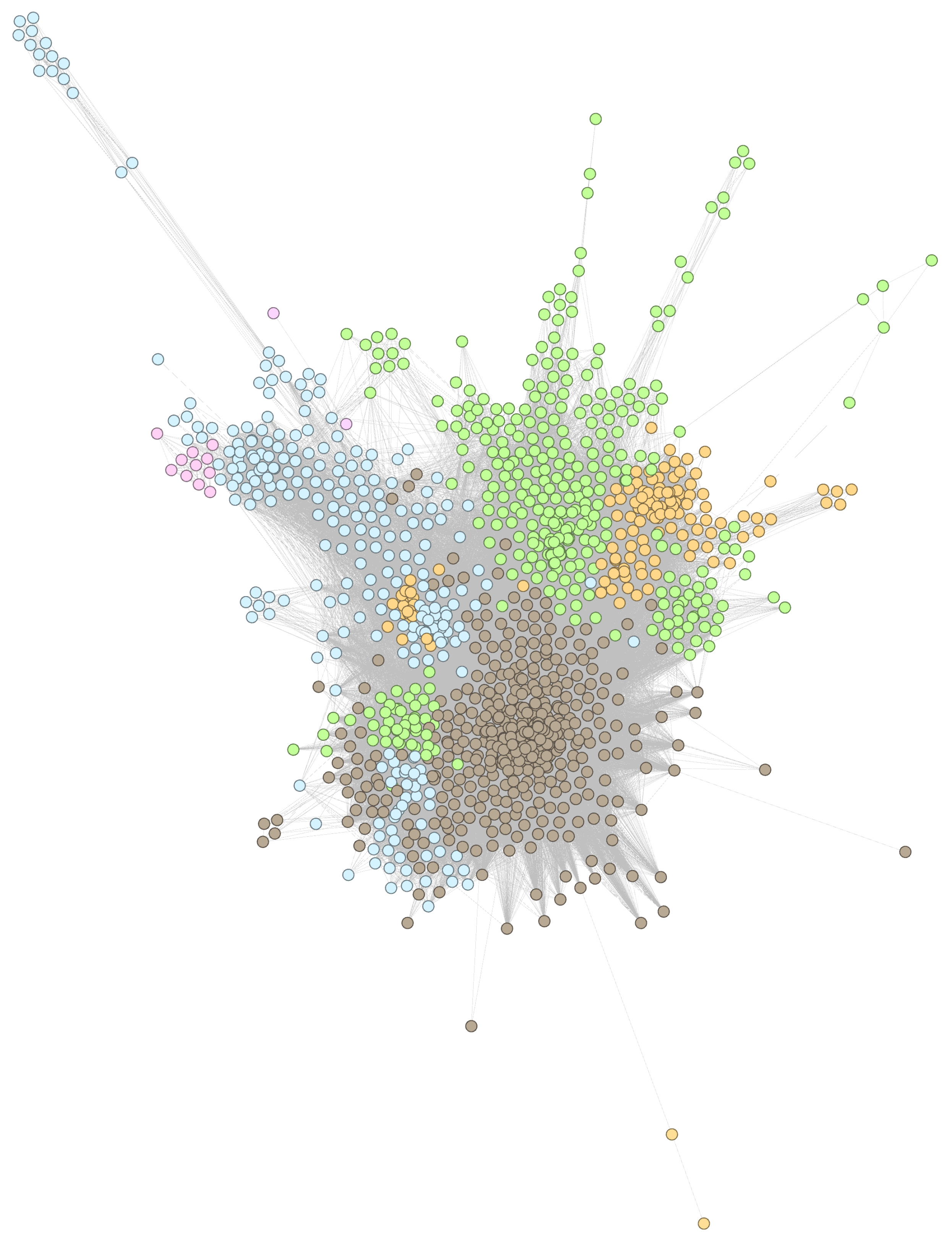
3.1. DDSN Using Drug–Gene Interactions from DrugBang 5.0.9
3.2. DDSN Using Drug–Gene Interactions from DrugBang 5.1.8
3.3. Repositioning Confirmations
3.3.1. Confirmed Drug Repositionings in DrugBank 5.0.9
Modularity Cluster
Modularity Cluster
3.3.2. Drug Repositioning Hints in DrugBank 5.1.8
4. Discussion
4.1. Drug–Gene Interactions
4.2. Method Limitations
4.3. Labeling and Validation with ATC Codes
4.4. Method Application
5. Conclusions
- (i)
- A new method to build weighted drug–drug similarity networks based on drug–gene interactions;
- (ii)
- An automated procedure to optimize the modularity resolution such that network clustering maximizes the number of identified drug repurposings. A known/ confirmed drug repurposing is a drug with more level 1 ATC codes in the latest drug database, compared with the earlier database—used to generate the drug–drug similarity network;
- (iii)
- A new drug repurposing list was generated with our pipeline from the latest DrugBank 5.1.8 by analyzing the three most representative clusters.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ATC | Anatomical Therapeutic Chemical; |
| COPD | Chronic Obstructive Pulmonary Disease; |
| COX-2 | Cyclooxygenase-2; |
| DDSN | Drug–Drug Similarity Network; |
| NSCLC | Non-Small Cell Lung Cancer. |
Appendix A. Repositionings and Statistics for DrugBank 5.0.9 DDSN
Appendix A.1. DDSN Zoomed Details
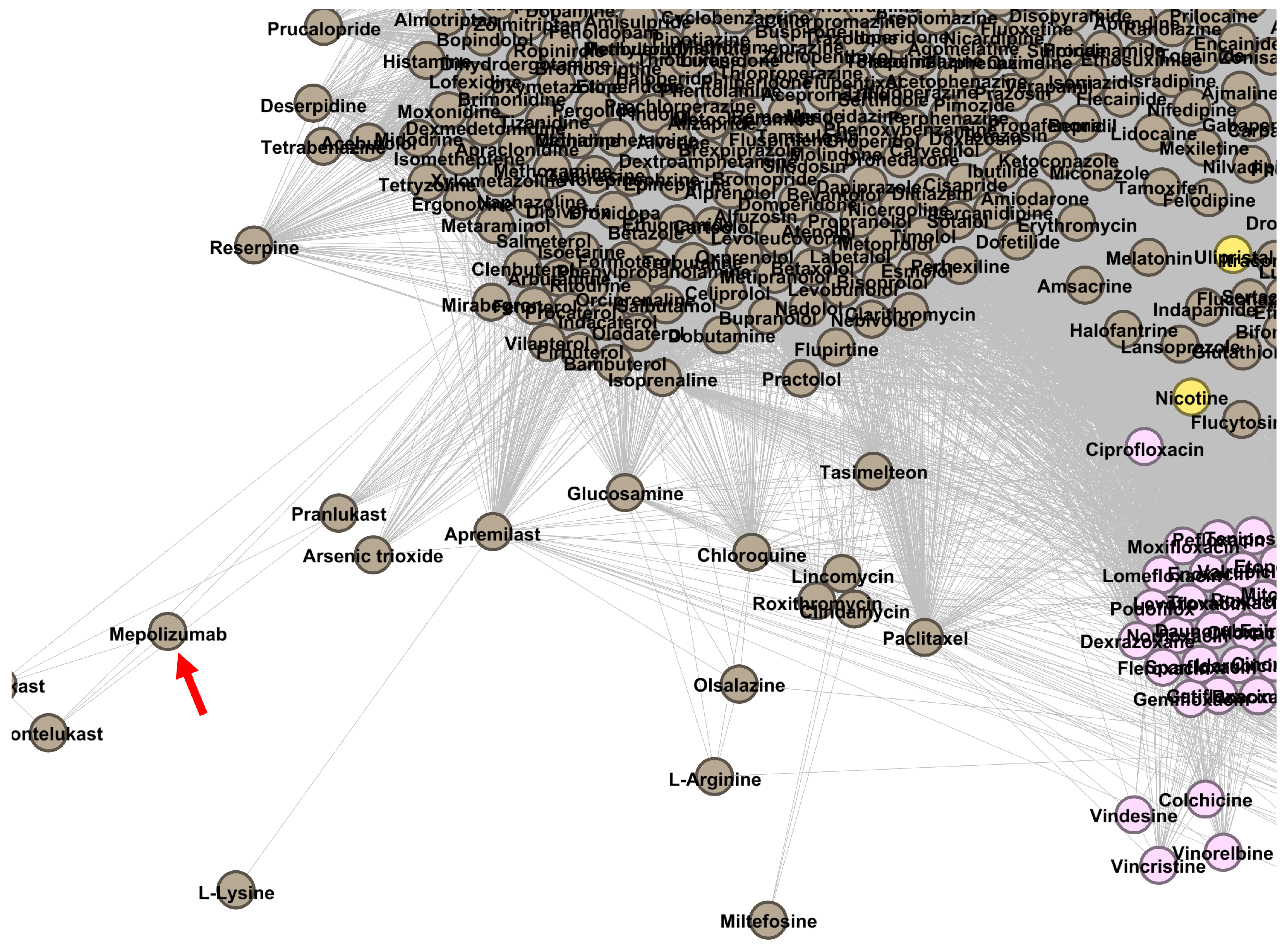


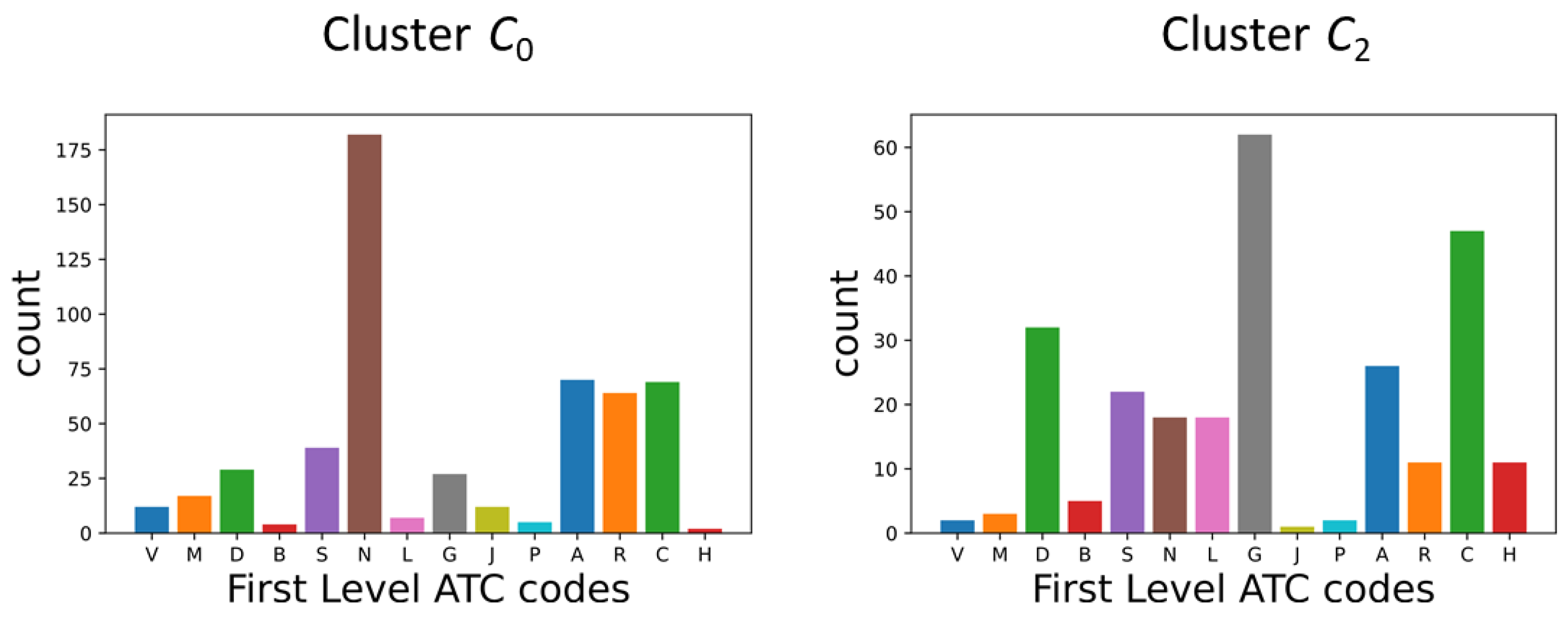
Appendix A.2. DDSN Cluster Histograms
Appendix B. Repositionings and Statistics for DrugBank 5.1.8 DDSN
Appendix B.1. DDSN Zoomed Details

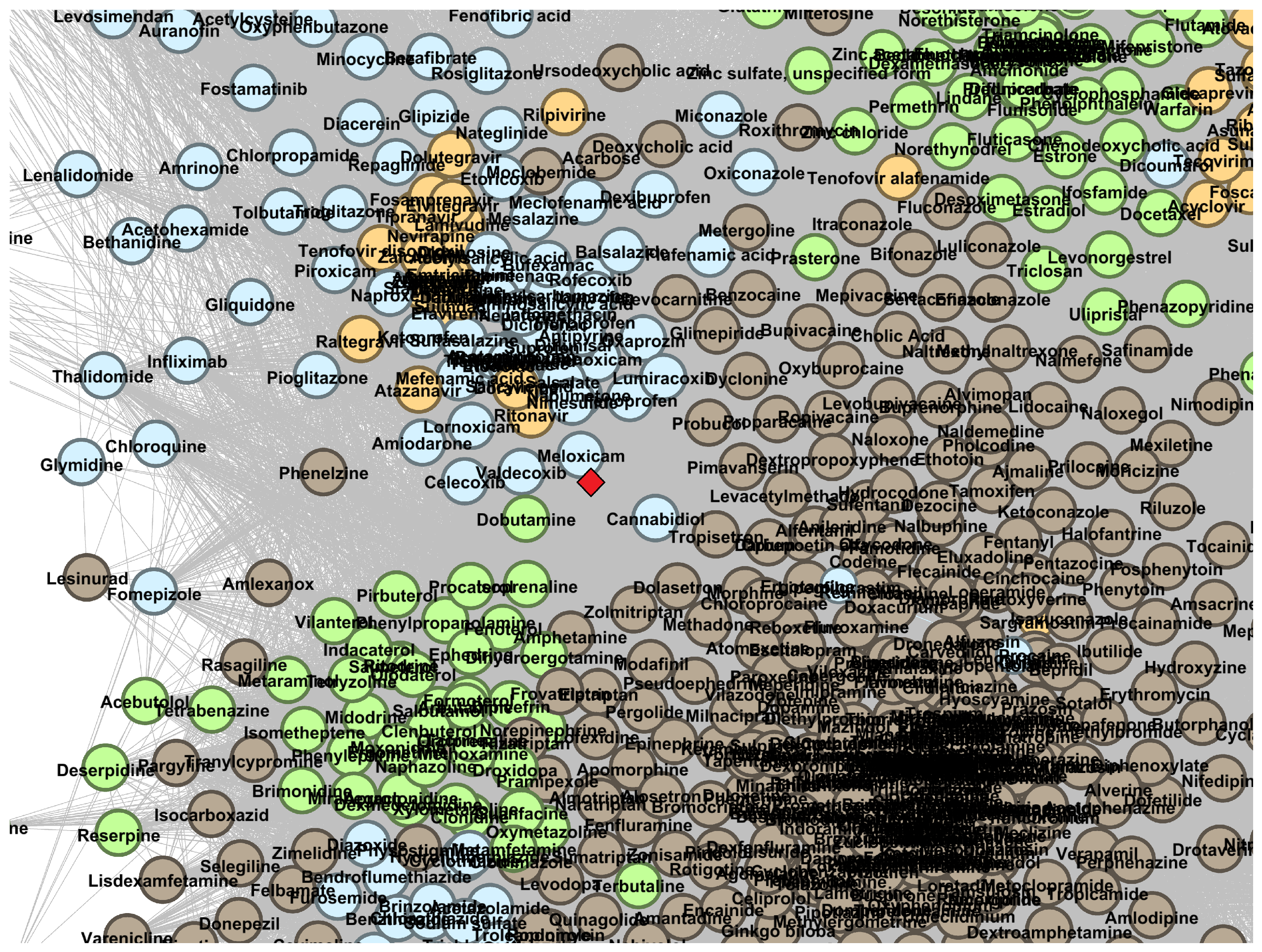


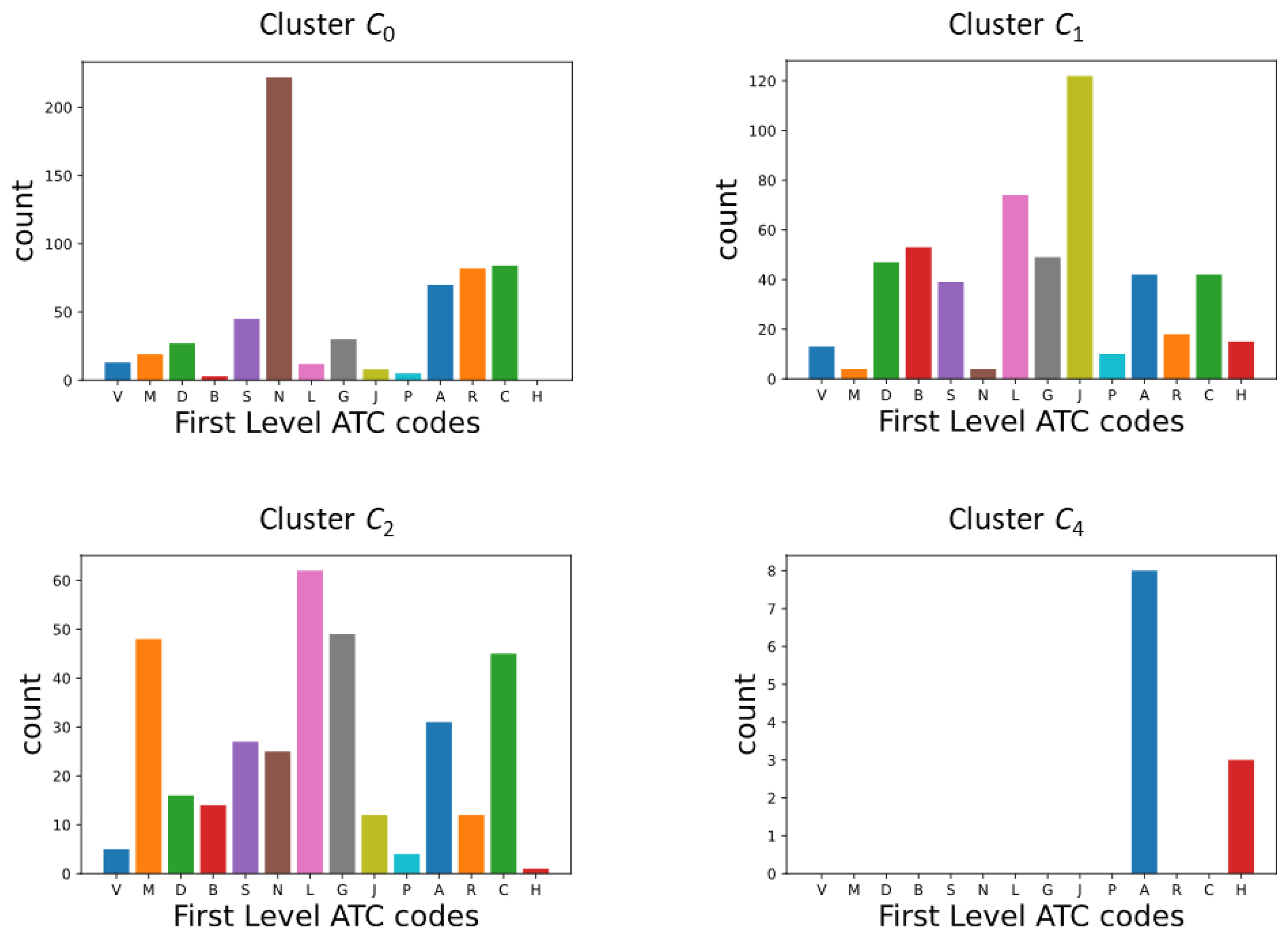
Appendix B.2. DDSN Cluster Histograms
References
- Munos, B. Lessons from 60 years of pharmaceutical innovation. Nat. Rev. Drug Discov. 2009, 8, 959–968. [Google Scholar] [CrossRef] [PubMed]
- Dickson, M.; Gagnon, J.P. The cost of new drug discovery and development. Discov. Med. 2009, 4, 172–179. [Google Scholar]
- Chen, X.Q.; Antman, M.D.; Gesenberg, C.; Gudmundsson, O.S. Discovery pharmaceutics—Challenges and opportunities. AAPS J. 2006, 8, E402–E408. [Google Scholar] [CrossRef]
- Pammolli, F.; Magazzini, L.; Riccaboni, M. The productivity crisis in pharmaceutical R&D. Nat. Rev. Drug Discov. 2011, 10, 428–438. [Google Scholar] [PubMed]
- Lombardino, J.G.; Lowe, J.A. The role of the medicinal chemist in drug discovery—Then and now. Nat. Rev. Drug Discov. 2004, 3, 853–862. [Google Scholar] [CrossRef]
- Gysi, D.M.; Do Valle, Í.; Zitnik, M.; Ameli, A.; Gan, X.; Varol, O.; Ghiassian, S.D.; Patten, J.; Davey, R.A.; Loscalzo, J.; et al. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc. Natl. Acad. Sci. USA 2021, 118, e2025581118. [Google Scholar] [CrossRef] [PubMed]
- Meganck, R.M.; Baric, R.S. Developing therapeutic approaches for twenty-first-century emerging infectious viral diseases. Nat. Med. 2021, 27, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Zhang, S. Polypharmacology: Drug discovery for the future. Expert Rev. Clin. Pharmacol. 2013, 6, 41–47. [Google Scholar] [CrossRef]
- Pinzi, L.; Tinivella, A.; Caporuscio, F.; Rastelli, G. Drug repurposing and polypharmacology to fight SARS-CoV-2 through inhibition of the main protease. Front. Pharmacol. 2021, 12, 84. [Google Scholar] [CrossRef]
- Aliper, A.; Plis, S.; Artemov, A.; Ulloa, A.; Mamoshina, P.; Zhavoronkov, A. Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol. Pharm. 2016, 13, 2524–2530. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, F.; Tang, J.; Nussinov, R.; Cheng, F. Artificial intelligence in COVID-19 drug repurposing. Lancet Dig. Health 2020, 2, e667–e676. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef] [PubMed]
- Udrescu, L.; Sbârcea, L.; Topîrceanu, A.; Iovanovici, A.; Kurunczi, L.; Bogdan, P.; Udrescu, M. Clustering drug-drug interaction networks with energy model layouts: Community analysis and drug repurposing. Sci. Rep. 2016, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Desai, R.J.; Handy, D.E.; Wang, R.; Schneeweiss, S.; Barabasi, A.L.; Loscalzo, J. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Luo, Y.; Zhao, X.; Zhou, J.; Yang, J.; Zhang, Y.; Kuang, W.; Peng, J.; Chen, L.; Zeng, J. A network integration approach for drug-target interaction prediction and computational drug repositioning from heterogeneous information. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- AY, M.; Goh, K.I.; Cusick, M.E.; Barabasi, A.L.; Vidal, M. Drug–target network. Nat. Biotechnol. 2007, 25, 1119–1127. [Google Scholar]
- Ye, H.; Liu, Q.; Wei, J. Construction of drug network based on side effects and its application for drug repositioning. PLoS ONE 2014, 9, e87864. [Google Scholar] [CrossRef]
- Lotfi Shahreza, M.; Ghadiri, N.; Mousavi, S.R.; Varshosaz, J.; Green, J.R. A review of network-based approaches to drug repositioning. Brief. Bioinf. 2018, 19, 878–892. [Google Scholar] [CrossRef]
- de Oliveira, T.B.; Zhao, L.; Faceli, K.; de Carvalho, A.C. Data clustering based on complex network community detection. In Proceedings of the 2008 IEEE Congress on Evolutionary Computation (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–6 June 2008; pp. 2121–2126. [Google Scholar]
- Yang, Z.; Algesheimer, R.; Tessone, C.J. A comparative analysis of community detection algorithms on artificial networks. Sci. Rep. 2016, 6, 1–18. [Google Scholar]
- Udrescu, L.; Bogdan, P.; Chiş, A.; Sîrbu, I.O.; Topîrceanu, A.; Văruţ, R.M.; Udrescu, M. Uncovering New Drug Properties in Target-Based Drug-Drug Similarity Networks. Pharmaceutics 2020, 12, 879. [Google Scholar] [CrossRef] [PubMed]
- Badkas, A.; De Landtsheer, S.; Sauter, T. Topological network measures for drug repositioning. Brief. Bioinf. 2021, 22, bbaa357. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Moraga, R.; Forés-Martos, J.; Suay-García, B.; Duval, J.L.; Falcó, A.; Climent, J. A COVID-19 Drug Repurposing Strategy through Quantitative Homological Similarities Using a Topological Data Analysis-Based Framework. Pharmaceutics 2021, 13, 488. [Google Scholar] [CrossRef] [PubMed]
- Iorio, F.; Bosotti, R.; Scacheri, E.; Belcastro, V.; Mithbaokar, P.; Ferriero, R.; Murino, L.; Tagliaferri, R.; Brunetti-Pierri, N.; Isacchi, A.; et al. Discovery of drug mode of action and drug repositioning from transcriptional responses. Proc. Natl. Acad. Sci. USA 2010, 107, 14621–14626. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Li, W.; Wu, Z.; Wang, X.; Zhang, C.; Li, J.; Liu, G.; Tang, Y. Prediction of polypharmacological profiles of drugs by the integration of chemical, side effect, and therapeutic space. J. Chem. Inf. Model. 2013, 53, 753–762. [Google Scholar] [CrossRef]
- Gottlieb, A.; Stein, G.Y.; Ruppin, E.; Sharan, R. PREDICT: A method for inferring novel drug indications with application to personalized medicine. Mol. Syst. Biol. 2011, 7, 496. [Google Scholar] [CrossRef] [PubMed]
- Langhauser, F.; Casas, A.I.; Guney, E.; Menche, J.; Geuss, E.; Kleikers, P.W.; López, M.G.; Barabási, A.L.; Kleinschnitz, C.; Schmidt, H.H.; et al. A diseasome cluster-based drug repurposing of soluble guanylate cyclase activators from smooth muscle relaxation to direct neuroprotection. NPJ Syst. Biol. Appl. 2018, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Goh, K.I.; Choi, I.G. Exploring the human diseasome: The human disease network. Brief. Funct. Gen. 2012, 11, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Goh, K.I.; Cusick, M.E.; Valle, D.; Childs, B.; Vidal, M.; Barabási, A.L. The human disease network. Proc. Natl. Acad. Sci. USA 2007, 104, 8685–8690. [Google Scholar] [CrossRef] [PubMed]
- Lancichinetti, A.; Fortunato, S. Limits of modularity maximization in community detection. Phys. Rev. E 2011, 84, 066122. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucl. Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Guo, A.C.; Lo, E.J.; Marcu, A.; Grant, J.R.; Sajed, T.; Johnson, D.; Li, C.; Sayeeda, Z.; et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucl. Acids Res. 2018, 46, D1074–D1082. [Google Scholar] [CrossRef]
- Merkel, D. Docker: Lightweight linux containers for consistent development and deployment. Linux J. 2014, 2014, 2. [Google Scholar]
- McKinney, W. Pandas: A foundational Python library for data analysis and statistics. Python High Perform. Sci. Comput. 2011, 14, 1–9. [Google Scholar]
- Hagberg, A.; Swart, P.; Chult, D.S. Exploring Network Structure, Dynamics, and Function Using NetworkX; Technical report; Los Alamos National Lab. (LANL): Los Alamos, NM, USA, 2008. [Google Scholar]
- Rossetti, G.; Milli, L.; Cazabet, R. CDLIB: A python library to extract, compare and evaluate communities from complex networks. Appl. Netw. Sci. 2019, 4, 1–26. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the Third international AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Zhou, T.; Ren, J.; Medo, M.; Zhang, Y.C. Bipartite network projection and personal recommendation. Phys. Rev. E 2007, 76, 046115. [Google Scholar] [CrossRef]
- Girvan, M.; Newman, M.E. Community structure in social and biological networks. Proc. Natl. Acad. Sci. USA 2002, 99, 7821–7826. [Google Scholar] [CrossRef] [PubMed]
- Blondel, V.D.; Guillaume, J.L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, 2008, P10008. [Google Scholar] [CrossRef]
- Peng, Y.; Wang, M.; Xu, Y.; Wu, Z.; Wang, J.; Zhang, C.; Liu, G.; Li, W.; Li, J.; Tang, Y. Drug repositioning by prediction of drug’s anatomical therapeutic chemical code via network-based inference approaches. Brief. Bioinf. 2021, 22, 2058–2072. [Google Scholar] [CrossRef]
- Tan, F.; Yang, R.; Xu, X.; Chen, X.; Wang, Y.; Ma, H.; Liu, X.; Wu, X.; Chen, Y.; Liu, L.; et al. Drug repositioning by applying ‘expression profiles’ generated by integrating chemical structure similarity and gene semantic similarity. Mol. BioSyst. 2014, 10, 1126–1138. [Google Scholar] [CrossRef]
- Wang, H.; Kuo, M.; Chou, M.; Hung, P.; Lin, K.; Hsieh, M.; Chang, M. Pyridoxal phosphate is better than pyridoxine for controlling idiopathic intractable epilepsy. Arch. Dis. Child. 2005, 90, 512–515. [Google Scholar] [CrossRef]
- Mills, P.B.; Camuzeaux, S.S.; Footitt, E.J.; Mills, K.A.; Gissen, P.; Fisher, L.; Das, K.B.; Varadkar, S.M.; Zuberi, S.; McWilliam, R.; et al. Epilepsy due to PNPO mutations: Genotype, environment and treatment affect presentation and outcome. Brain 2014, 137, 1350–1360. [Google Scholar] [CrossRef] [PubMed]
- Berthet, N.; Faure, O.; Bakri, A.; Ambroise-Thomas, P.; Grillot, R.; Brugere, J.F. In vitro susceptibility of Aspergillus spp. clinical isolates to albendazole. J. Antimicrob. Chemother. 2003, 51, 1419–1422. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, M.S.; Edlind, T.D.; Lee, C.H.; Dean, R.; Queener, S.F.; Shaw, M.M.; Smith, J.W. Albendazole inhibits Pneumocystis carinii proliferation in inoculated immunosuppressed mice. Antimicrob. Agents Chemother. 1994, 38, 1834–1837. [Google Scholar] [CrossRef]
- Caruso, A.; Caccuri, F.; Bugatti, A.; Zani, A.; Vanoni, M.; Bonfanti, P.; Cazzaniga, M.E.; Perno, C.F.; Messa, C.; Alberghina, L. Methotrexate inhibits SARS-CoV-2 virus replication “in vitro”. J. Med. Virol. 2021, 93, 1780–1785. [Google Scholar] [CrossRef] [PubMed]
- Beck, S.; Zhu, Z.; Oliveira, M.F.; Smith, D.M.; Rich, J.N.; Bernatchez, J.A.; Siqueira-Neto, J.L. Mechanism of action of methotrexate against Zika virus. Viruses 2019, 11, 338. [Google Scholar] [CrossRef]
- Lembo, D.; Gribaudo, G.; Cavallo, R.; Riera, L.; Angeretti, A.; Hertel, L.; Landolfo, S. Human cytomegalovirus stimulates cellular dihydrofolate reductase activity in quiescent cells. Intervirology 1999, 42, 30–36. [Google Scholar] [CrossRef]
- Jerwood, S.; Cohen, J. Unexpected antimicrobial effect of statins. J. Antimicrob. Chemother. 2008, 61, 362–364. [Google Scholar] [CrossRef] [PubMed]
- Parihar, S.P.; Guler, R.; Brombacher, F. Statins: A viable candidate for host-directed therapy against infectious diseases. Nat. Rev. Immunol. 2019, 19, 104–117. [Google Scholar] [CrossRef]
- Chang, Y.L.; Hsu, Y.J.; Chen, Y.; Wang, Y.W.; Huang, S.M. Theophylline exhibits anti-cancer activity via suppressing SRSF3 in cervical and breast cancer cell lines. Oncotarget 2017, 8, 101461. [Google Scholar] [CrossRef]
- Goldman, A.P.; Williams, C.S.; Sheng, H.; Lamps, L.W.; Williams, V.P.; Pairet, M.; Morrow, J.D.; DuBois, R.N. Meloxicam inhibits the growth of colorectal cancer cells. Carcinogenesis 1998, 19, 2195–2199. [Google Scholar] [CrossRef] [PubMed]
- Tsubouchi, Y.; Mukai, S.; Kawahito, Y.; Yamada, R.; Kohno, M.; Inoue, K.; Sano, H. Meloxicam inhibits the growth of non-small cell lung cancer. Anticancer Res. 2000, 20, 2867–2872. [Google Scholar] [PubMed]
- Naruse, T.; Nishida, Y.; Hosono, K.; Ishiguro, N. Meloxicam inhibits osteosarcoma growth, invasiveness and metastasis by COX-2-dependent and independent routes. Carcinogenesis 2006, 27, 584–592. [Google Scholar] [CrossRef]
- Deeb, K.K.; Trump, D.L.; Johnson, C.S. Vitamin D signalling pathways in cancer: Potential for anticancer therapeutics. Nat. Rev. Cancer 2007, 7, 684–700. [Google Scholar] [CrossRef]
- Chiang, K.C.; Chen, T.C. The anti-cancer actions of vitamin D. In Anti-Cancer Agents in Medicinal Chemistry (Formerly Current Medicinal Chemistry—Anti-Cancer Agents); Bentham Science Publishers: Sharjah, United Arab Emirates, 2013; Volume 13, pp. 126–139. [Google Scholar]
- Weyerhäuser, P.; Kantelhardt, S.R.; Kim, E.L. Re-purposing chloroquine for glioblastoma: Potential merits and confounding variables. Front. Oncol. 2018, 8, 335. [Google Scholar] [CrossRef] [PubMed]
- Verbaanderd, C.; Maes, H.; Schaaf, M.B.; Sukhatme, V.P.; Pantziarka, P.; Sukhatme, V.; Agostinis, P.; Bouche, G. Repurposing Drugs in Oncology (ReDO)—Chloroquine and hydroxychloroquine as anti-cancer agents. Ecancermedicalscience 2017, 11, 781. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. Anticancer autophagy inhibitors attract ‘resurgent’ interest. Nat. Rev. Drug Discov. 2019, 18, 408–410. [Google Scholar] [CrossRef] [PubMed]
- Varisli, L.; Cen, O.; Vlahopoulos, S. Dissecting pharmacological effects of chloroquine in cancer treatment: Interference with inflammatory signaling pathways. Immunology 2020, 159, 257–278. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, H.; Yang, Y.; Chen, Z.S.; Zou, C.; Zhang, J. Chloroquine against malaria, cancers and viral diseases. Drug Discov. Today 2020, 2012–2022. [Google Scholar] [CrossRef]
- Kemp, S.F.; Thrailkill, K.M. Mecasermin rinfabate for severe insulin-like growth factor-I deficiency. Clin. Pract. 2007, 4, 133. [Google Scholar] [CrossRef]
- IPLEXTM (Mecasermin Rinfabate [rDNA Origin] Injection). Available online: https://www.accessdata.fda.gov/drugsatfda_docs/label/2007/021884s001lbl.pdf (accessed on 21 October 2021).
- INCRELEX, INN: Mecasermin. Scientific Discussion. Available online: https://www.ema.europa.eu/en/documents/scientific-discussion/increlex-epar-scientific-discussion_en.pdf (accessed on 21 October 2021).
- Miyake, M.; Kirisako, T.; Kokubo, T.; Miura, Y.; Morishita, K.; Okamura, H.; Tsuda, A. Randomised controlled trial of the effects of L-ornithine on stress markers and sleep quality in healthy workers. Nutr. J. 2014, 13, 1–8. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, M.; Katsyv, I.; Irie, H.; Zhang, B. EMUDRA: Ensemble of multiple drug repositioning approaches to improve prediction accuracy. Bioinformatics 2018, 34, 3151–3159. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, W.; Yan, C.; Luo, J.; Zhang, G. Predicting Drug-Disease Association Based on Ensemble Strategy. Front. Genet. 2021, 12, 548. [Google Scholar] [CrossRef] [PubMed]
- Ghorbanali, Z.; Zare-Mirakabad, F.; Mohammadpour, B. DRP-VEM: Drug repositioning prediction using voting ensemble. arXiv 2021, arXiv:2110.01403. [Google Scholar]
- Jarada, T.N.; Rokne, J.G.; Alhajj, R. A review of computational drug repositioning: Strategies, approaches, opportunities, challenges, and directions. J. Cheminf. 2020, 12, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Sagi, O.; Rokach, L. Ensemble learning: A survey. Wiley Interdiscip. Rev. Data Min. Knowl. Discov. 2018, 8, e1249. [Google Scholar] [CrossRef]
- Lihu, A.; Holban, Ş. A review of ensemble methods for de novo motif discovery in ChIP-Seq data. Brief. Bioinf. 2015, 16, 964–973. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xue, H.; Li, J.; Xie, H.; Wang, Y. Review of drug repositioning approaches and resources. Int. J. Biol. Sci. 2018, 14, 1232. [Google Scholar] [CrossRef]
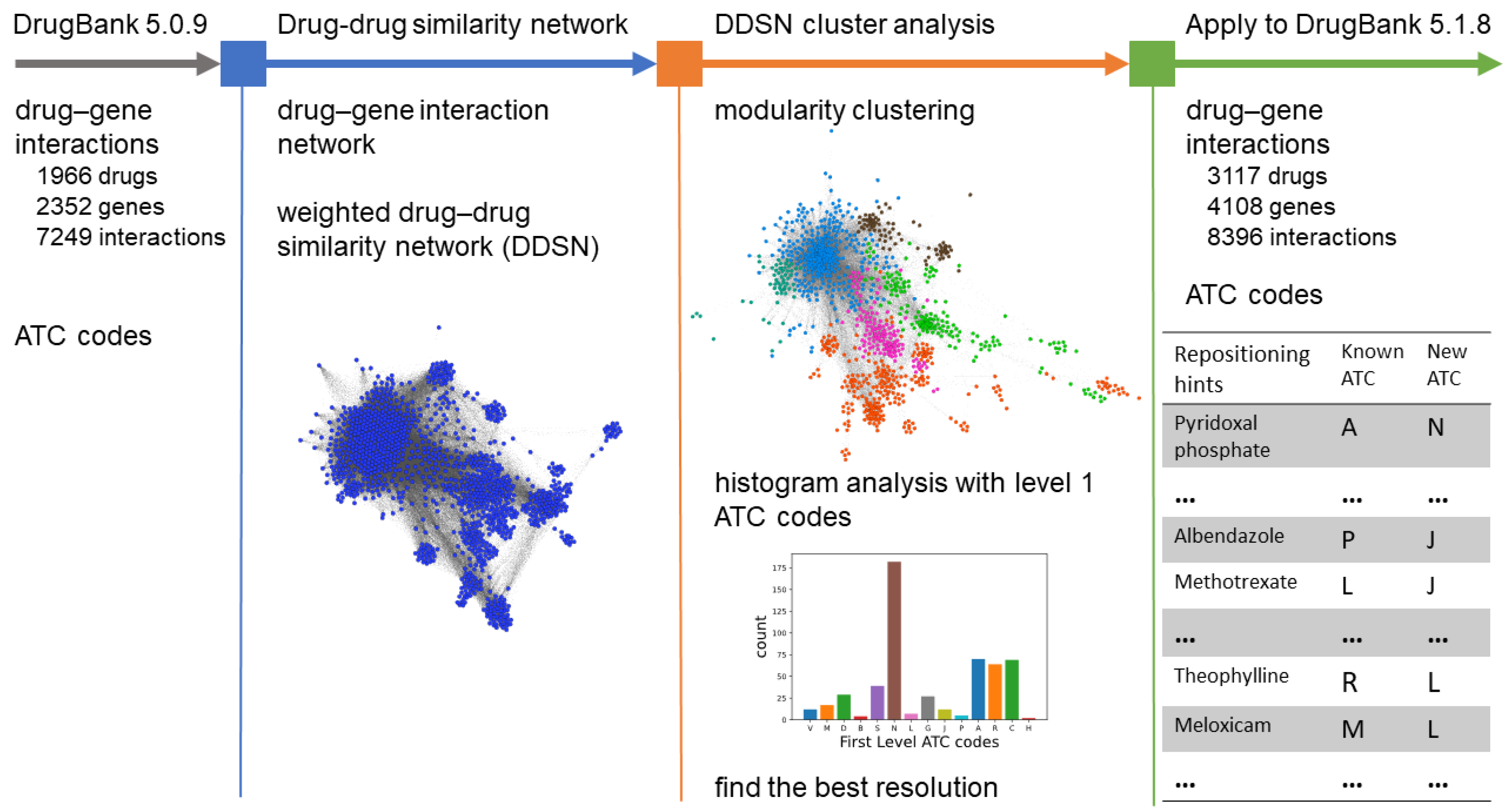

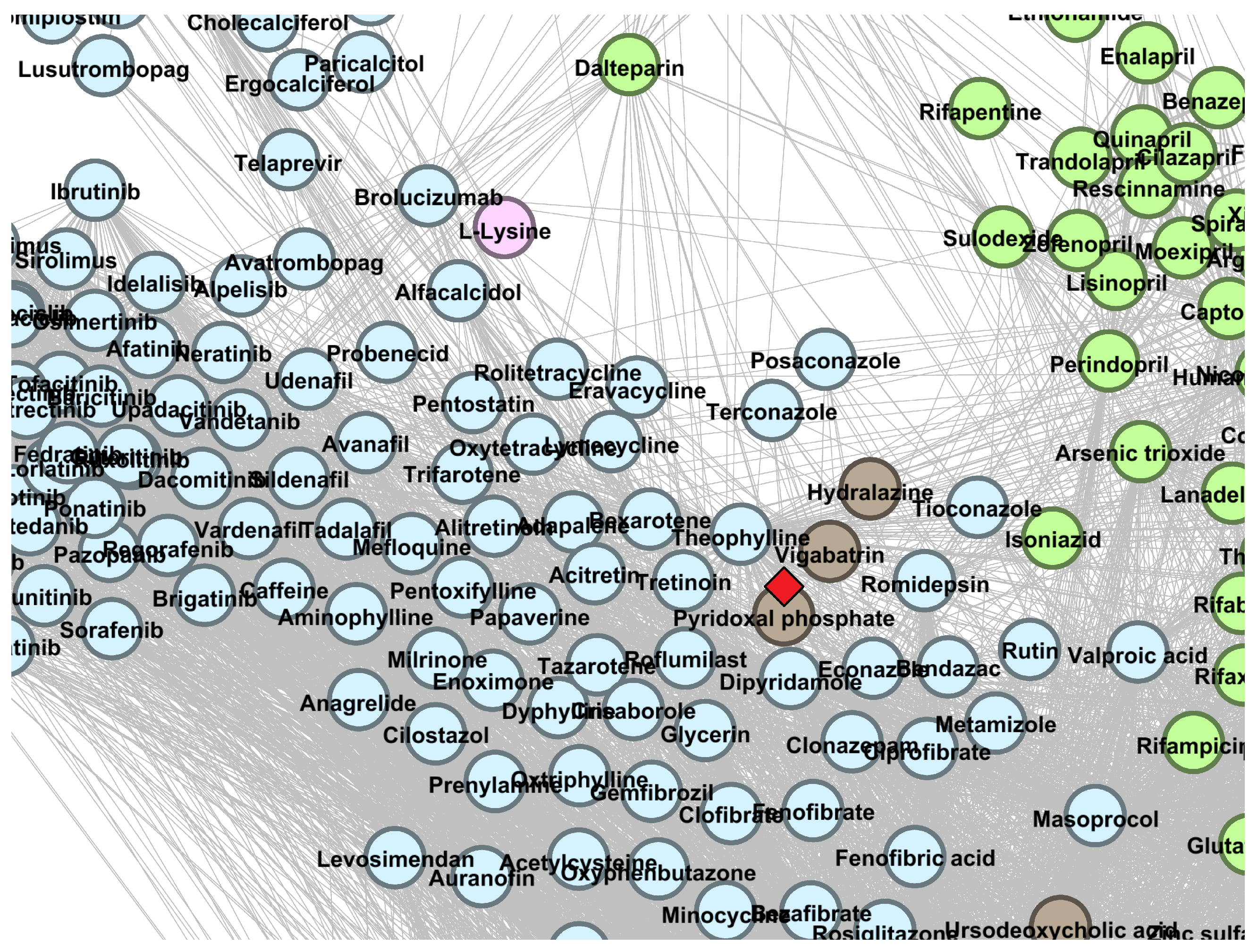

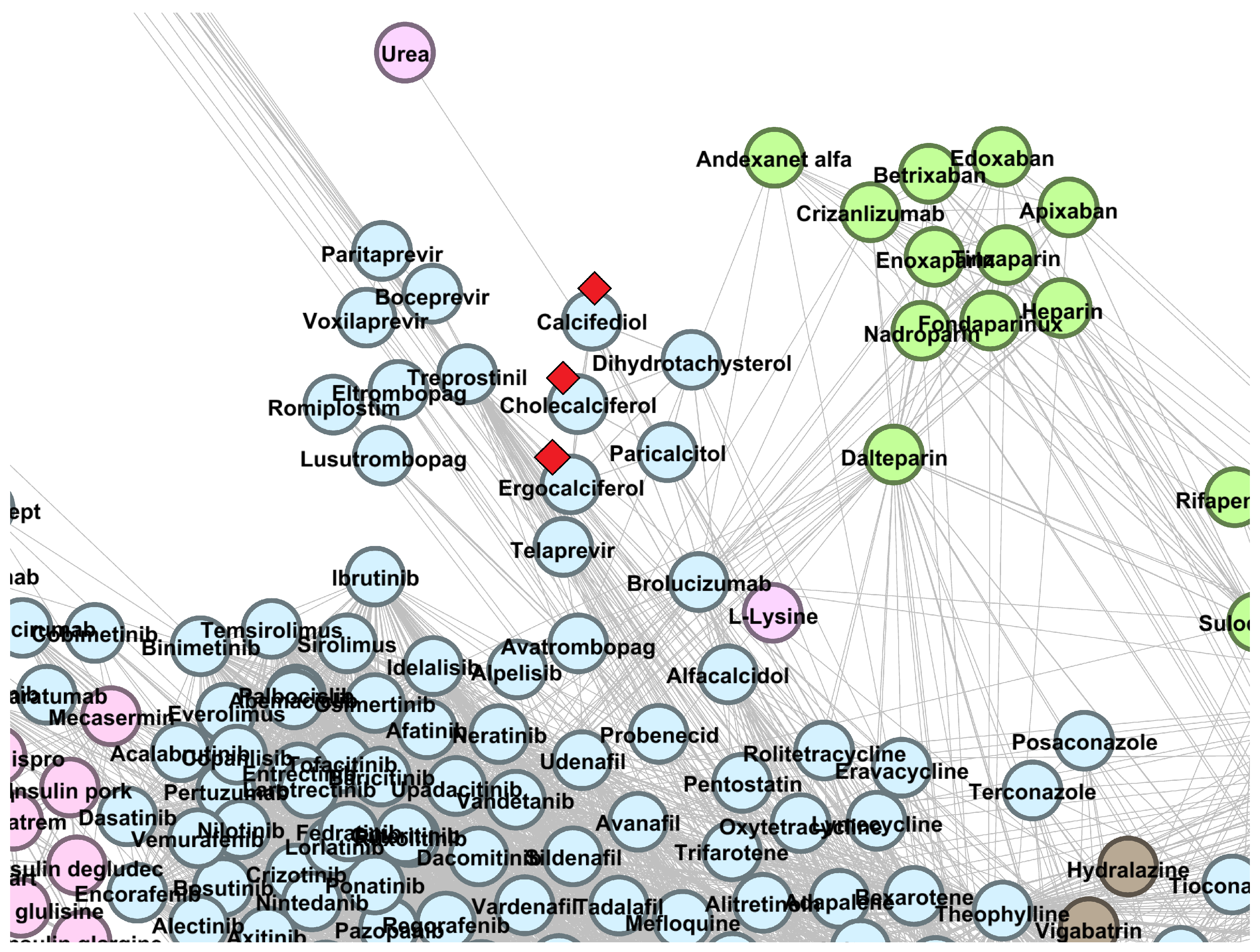

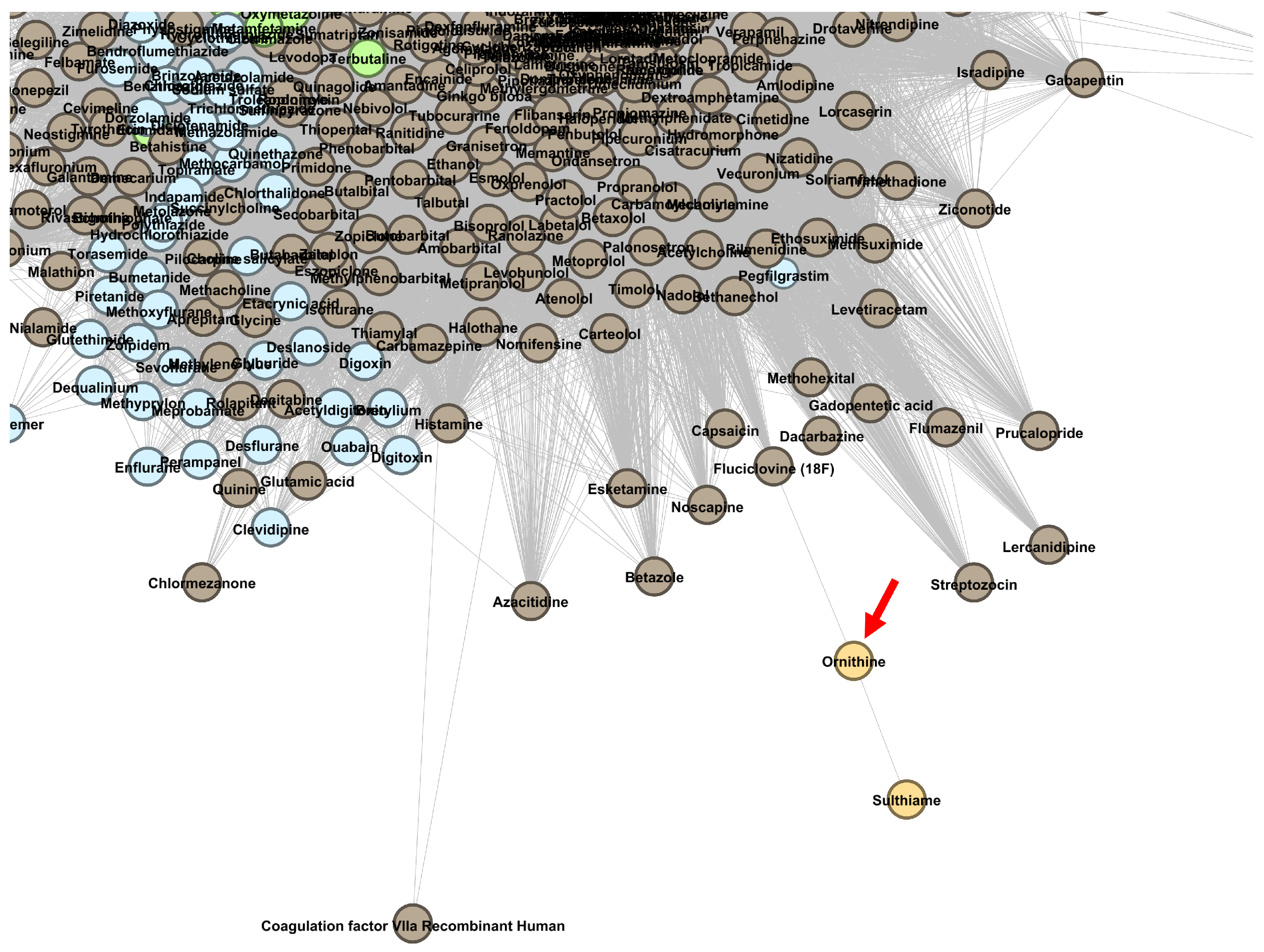
| Drug | Cluster | Current Level 1 ATC | Predicted Level 1 ATC | References |
|---|---|---|---|---|
| Pyridoxal phosphate | A | H | [44,45] | |
| Albendazole | P | J | [46,47] | |
| Methotrexate | L | J | [48,49,50] | |
| C | J | [51,52] | ||
| Theophylline | R | L | [14,53] | |
| Meloxicam | M | L | [54,55,56] | |
| M, A | L | [57,58] | ||
| Chloroquine | P | L | [59,60,61,62,63] | |
| H | A | [64,65,66] | ||
| Ornithine | A | N | [67] |
| Drug Name | Gene Name | Interaction Type |
|---|---|---|
| Alteplase | PLG | activator |
| Hydromorphone | OPRK1 | agonist |
| Varenicline | CHRNB2 | partial agonist |
| Prazosin | ADRA1B | antagonist |
| Ascorbic acid | EGLN1 | chaperone |
| Pyridoxal phosphate | GAD1 | cofactor |
| Vardenafil | PDE6G | allosteric modulator |
| Trastuzumab | ERBB2 | antibody |
| Nusinersen | SMN2 | antisense oligonucleotide |
| Methysergide | HTR1F | binder |
| Tiapride | DRD2 | blocker |
| Carvedilol | KCNJ4 | inhibitor |
| Clobetasol propionate | ANXA1 | inducer |
| Clofazimine | PPARG | modulator |
| Cerliponase alfa | IGF2R | ligand |
| Filgrastim | CSF3R | stimulator |
| Dalteparin | SERPINC1 | potentiator |
| Vitamin A | RDH13 | substrate |
| Nedocromil | CYSLTR1 | suppressor |
| Belimumab | TNFSF13B | neutralizer |
| Esmirtazapine | HRH1 | inverse agonist |
| Procainamide | DNMT1 | other |
| Haloperidol | HTR2A | other/unknown |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Groza, V.; Udrescu, M.; Bozdog, A.; Udrescu, L. Drug Repurposing Using Modularity Clustering in Drug-Drug Similarity Networks Based on Drug–Gene Interactions. Pharmaceutics 2021, 13, 2117. https://doi.org/10.3390/pharmaceutics13122117
Groza V, Udrescu M, Bozdog A, Udrescu L. Drug Repurposing Using Modularity Clustering in Drug-Drug Similarity Networks Based on Drug–Gene Interactions. Pharmaceutics. 2021; 13(12):2117. https://doi.org/10.3390/pharmaceutics13122117
Chicago/Turabian StyleGroza, Vlad, Mihai Udrescu, Alexandru Bozdog, and Lucreţia Udrescu. 2021. "Drug Repurposing Using Modularity Clustering in Drug-Drug Similarity Networks Based on Drug–Gene Interactions" Pharmaceutics 13, no. 12: 2117. https://doi.org/10.3390/pharmaceutics13122117
APA StyleGroza, V., Udrescu, M., Bozdog, A., & Udrescu, L. (2021). Drug Repurposing Using Modularity Clustering in Drug-Drug Similarity Networks Based on Drug–Gene Interactions. Pharmaceutics, 13(12), 2117. https://doi.org/10.3390/pharmaceutics13122117








