Abstract
Chronic inflammation and dysregulated epithelial differentiation, especially of hair follicle keratinocytes, have been suggested as the major pathogenetic pathways of hidradenitis suppurativa/acne inversa (HS). On the other hand, obesity and metabolic syndrome have additionally been considered as an important risk factor. With adalimumab, a drug has already been approved and numerous other compounds are in advanced-stage clinical studies. A systematic review was conducted to detect and corroborate HS pathogenetic mechanisms at the molecular level and identify HS molecular markers. The obtained data were used to confirm studied and off-label administered drugs and to identify additional compounds for drug repurposing. A robust, strongly associated group of HS biomarkers was detected. The triad of HS pathogenesis, namely upregulated inflammation, altered epithelial differentiation and dysregulated metabolism/hormone signaling was confirmed, the molecular association of HS with certain comorbid disorders, such as inflammatory bowel disease, arthritis, type I diabetes mellitus and lipids/atherosclerosis/adipogenesis was verified and common biomarkers were identified. The molecular suitability of compounds in clinical studies was confirmed and 31 potential HS repurposing drugs, among them 10 drugs already launched for other disorders, were detected. This systematic review provides evidence for the importance of molecular studies to advance the knowledge regarding pathogenesis, future treatment and biomarker-supported clinical course follow-up in HS.
1. Introduction
Hidradenitis suppurativa/acne inversa (HS) is a chronic, inflammatory, recurrent, debilitating skin disease of the hair follicle that usually presents after puberty with painful, deep-seated, inflamed lesions in the apocrine gland-bearing areas of the body, most commonly at the axillae, inguinal and anogenital regions [1]. A consistent finding, regardless of disease duration, is follicular hyperkeratosis, leading to follicular rupture, inflammation and possible secondary bacterial colonization. The deep part of the follicle appears to be involved. HS is further associated with an initial lymphohistiocytic inflammation, granulomatous reaction, sinus tract formation and scarring [2].
Current own transcriptome and proteome studies highlighted a panel of immune-related drivers in HS, which induce an innate immunity response in epithelial skin cells in a targeted manner [3]. An inflammatory process coupled to impaired barrier function and bacterial activity were detected at the follicular and epidermal keratinocyte and at a minor grade at the skin-gland level. In addition, the adipose tissue was shown to be involved in HS at a real-world immune histochemical study [4].
Despite the beneficial therapeutic effectiveness of several compounds [5,6], treatment of HS is still challenging, since most patients only respond partially with subsequent recurrences. The large unmet need of new therapies requires the elucidation of disease-driving mechanisms and the recognition of the skin compartment initially involved [7,8]. This need can be covered by the development of novel therapeutic regimens for HS [9,10] or by drug repurposing through drug–gene interaction profiling [11,12].
New technology, including inverse virtual screening [13] and computational drug repurposing screening approaches [14], are widely engaged in identifying existing compounds as potential drugs for various diseases. The interaction level of disease and compound molecular profile patterns defines the probability of therapeutic activity of a certain drug. The aim of this study is to provide a wide and robust application of molecular pharmacology in HS through a systematic review of the relevant literature and identification of key molecular mediators in a real-world setting. Using the latter data, therapeutic agents that are currently available or under development for other indications are identified and potential paths for use in the medical management of HS are proposed.
2. Materials and Methods
2.1. Literature Search
This systematic review was conducted and narrated in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [15] utilizing datasets from publicly available studies, as previously described [11]. A rigorous search of academic databases including PubMed, Web of Science and Ovid databases through August 2021 was conducted. A search strategy predefined and adapted for each aforementioned database included the following keywords: (transcriptome OR proteome OR biomarker(s) OR repurposing OR repositioning OR reprogramming) AND (hidradenitis suppurativa OR acne inversa OR Verneuil’s disease). Additional records were obtained through the Gene Expression Omnibus, National Institutes of Health (Bethesda, MD, USA) [16] and the citation search of the bibliographic records obtained from the academic databases. There were no search filters pertaining to language or publication year.
2.2. Study Selection
First the duplicates among bibliographic records were removed. Titles and abstracts were then scrutinized by two reviewers (V.A.Z. and K.C.Z.) working independently according to predefined inclusion and exclusion criteria. This was followed by scrutiny of full texts of eligible studies. Discrepancies were resolved by discussion with the senior investigator (C.C.Z.). After eligible studies were identified, their bibliographies were screened for studies judged suitable for inclusion. Original investigations of HS molecular signatures and protein studies followed by the identification of molecular mediators were selected for further analysis.
2.3. Data Extraction
Data pertaining to characteristics of publications under study and quantitative data were extracted by two of the reviewers (V.A.Z. and K.C.Z.) working independently using a predetermined customized extraction form. Characteristics of publications included publication year and affiliation of corresponding authors. Molecular characteristics included transcriptome and/or proteome of HS, and drug repurposing/repositioning/reprogramming.
2.4. Data Analysis
Qualitative gene/protein data from the studies were pooled to detect HS signature pathways. Gene nomenclature was verified through the HUGO Gene Nomenclature Committee, European Bioinformatics Institute (Cambridge, UK) public domain [17]. Gene taxonomy was assessed through the biological DataBase network, National Cancer Institute (Frederick, MD, USA) [18]. The molecular pathways were assessed according to the g:Profiler, University of Tartu (Tartu, Estonia) [19], the Kyoto Encyclopedia of Genes and Genomes [KEGG, gene ontology (GO); Kyoto, Japan] [20], the Reactome (REAC), Ontario Institute for Cancer Research (Toronto, ON, Canada), New York University (New York, NY, USA), Oregon Health and Science University (Portland, OR, USA) and the European Molecular Biology Laboratory—European Bioinformatics Institute (Heidelberg, Germany) [21], the WikiPathways (WP) [22] and the Human Phenotype Ontology (HP; The Jackson Laboratory for Genomic Medicine, Farmington, CT, USA) [23] public domains. Random effects were applied throughout the analysis due to expected clinical heterogeneity encountered in different studies supported by g:Profiler [19]. This approach allows heterogeneity in the data to be addressed by considering that differences between studies are random.
2.5. Drug Repurposing Sources
For drug repurposing, the detected overall HS molecular signature was compared with the drugs’ molecular signatures of The Drug Repurposing Hub public domain, Eli and Edy L. Broad Institute, MIT and Harvard University (Cambridge, MA, USA) [24] and the Gene Cards, Weizmann Institute of Science (Rehovot, Israel) [25] public domains.
2.6. Statistics
Statistics were automatically performed by the applied public domains used [19,20,21,22,23].
3. Results
3.1. Study Selection Process
A total of 123 bibliographic records were identified after electronic database searches, 36 through other sources and six through bibliographic record citation search. Among them, 61 records were removed as duplicates, leaving 104 titles and abstracts to be screened. After careful screening and manual search, six records were excluded based on title and abstract and 49 records due to inappropriate design and two records due to overlapping data sets with another record, resulting in 47 studies that were included in the quantitative synthesis [3,4,11,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69] (Figure 1).
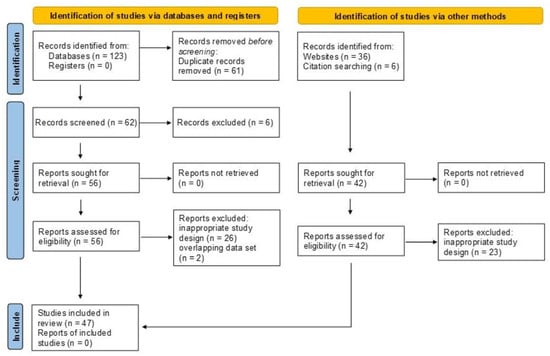
Figure 1.
Preferred reporting items for systematic reviews and meta-analyses (PRISMA 2020 [15]) flow diagram.
3.2. Differentially Expressed Genes and Proteins in HS
The comparison of lesional skin vs. non-lesional skin as well as of blood of patients vs. controls at the mRNA and protein levels (cumulatively reported as “targets”) without restrictions revealed 386 differentially expressed genes (DEGs) in HS (Table S1).
3.3. HS Biomarkers
DEGs and differentially expressed proteins in blood and involved skin of HS patients in comparison to controls in at least two relevant articles or two targets were defined as HS biomarkers. Among the 109 detected genes/proteins out of the 386 genes/proteins detected without restrictions, which fulfilled this requirement, 43 DEGs (including the coding genes of detected differentially expressed proteins) have been described in 2/4 targets in two articles, seven in 3/4 targets (CXCL10, IL6, IL17A, IL36A, IL36G, S100A8, S100A9) and none in all four targets (Table 1). Additional 10 DEGs have been described in 2/4 targets, however, in a diversified direction (upregulated/downregulated). Among the 109 HS biomarkers, 65 are druggable.

Table 1.
HS biomarkers resulting from the DEGs after transcriptomic profiling and protein expression studies between lesional HS and non-lesional skin biopsies and blood samples from HS patients and healthy controls, respectively and reported in at least two relevant articles. Bold letters indicate druggable genes. Background: white = similar results reported in one target (biological material) in at least two independent studies; orange = similar results reported in two targets in at least two independent studies; yellow = similar results reported in three targets in at least two independent studies. Gray = diversified result reported in at least two independent studies; + = upregulation; − = downregulation; +/− = diversified dysregulation in different studies; () = lower level of evidence.
3.4. Enrichment Analysis of HS-Associated Genes
The 386 detected HS-associated DEGs and the 109 HS biomarkers were enriched into relevant signaling pathways, which were assessed according to the g:Profiler [19], the KEGG GO, [20], the REAC [21], the WP [22] and the HP [23] public domains in order to identify the major organismal and signal transduction pathways involved in HS. Gene clustering in chromosome 2 and 4 was detected.
Among the 386 HS-associated DEGs, 101 genes were enriched in the cytokine–cytokine (C–C) receptor interaction pathway (−log10 = 2.5 × 10−74), 51 in the JAK-STAT signaling pathway (2.6 × 10−34), 39 in the chemokine signaling pathway (2.7 × 10−18), 32 in the IL-17 signaling pathway (1.8 × 10−22), 31 in the Th17 cell differentiation pathway (2.6 × 10−18), 28 in the Toll-like receptor (TLR) pathway (2.2 × 10−16) and 26 in the inflammatory bowel disease pathway (3.6 × 10−26) (Figure S1).
Furthermore, 45 HS biomarkers were enriched in the C–C receptor interaction pathway (5.6 × 10−43, Figure 2, 19 in the IL-17 signaling pathway (8.8 × 10−19, Figure 3), 19 in the JAK-STAT signaling pathway (6.0 × 10−14, Figure 4), 18 in the inflammatory bowel disease pathway (1.1 × 10−20), 18 in the rheumatoid arthritis pathway (1.2 × 10−17), 13 in the Th17 cell differentiation pathway (1.5 × 10−9), 13 in the lipid and atherosclerosis pathway (1.2 × 10−5), 10 in the TLR pathway (4.3 × 10−6), 9 in C-type leptin receptor signaling pathway (6.1 × 10−5), 8 in the tumor necrosis factor (TNF) signaling pathway (1.1 × 10−3) and 7 in the type I diabetes mellitus pathway (8.5 × 10−6) (Figure 5).
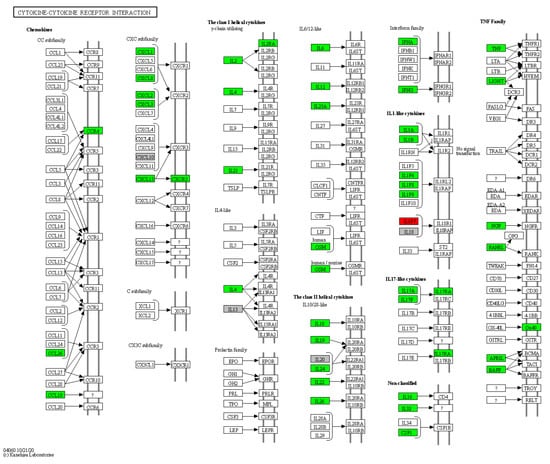
Figure 2.
Hierarchical clustering of HS biomarkers in the KEGG GO C-C receptor interaction pathway. Genes which are positively regulated in HS are shown in green color, those downregulated with red color. Gray color corresponds to genes with a diversified reported regulation.
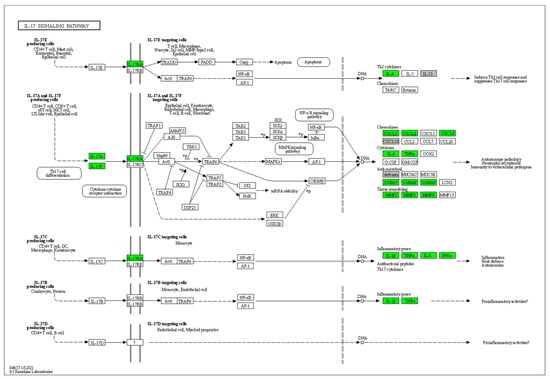
Figure 3.
Hierarchical clustering of HS biomarkers in the KEGG GO IL-17 signaling pathway. Genes which are positively regulated in HS are shown in green color. Gray color corresponds to genes with a diversified reported regulation.
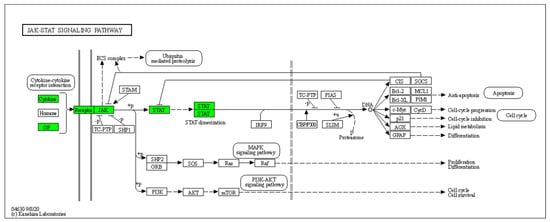
Figure 4.
Hierarchical clustering of HS biomarkers in the KEGG GO JAK-STAT signaling pathway. Genes which are positively regulated in HS are shown in green color. Gray color corresponds to genes with a diversified reported regulation.
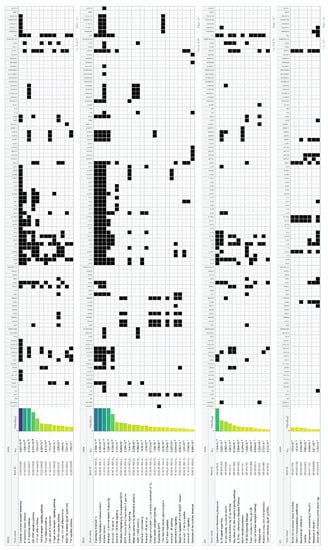
Figure 5.
Enrichment of HS biomarkers resulting from the comparison of transcriptomic profiles and protein expression studies between lesional HS and non-lesional skin biopsies and blood samples from HS patients and healthy controls, respectively, in signaling pathways.
Concerning the individual cytokine signaling, IL-17, IL-4, IL-13, IL-10, IL-20 family, IL-1 family, IL-18, IL-36, IL-2 family, IL-21 and IL-12 family signaling included DEGs in HS (Figure 5).
Epithelial differentiation signaling dysregulation in HS was represented by the epidermal growth factor receptor (EGFR), IL-1, IL-1 receptor, formation of the cornified envelope, TLRs and antimicrobial peptides (Figure 5).
Metabolic/obesity-associated dysregulation in HS was detected through type I diabetes mellitus signaling, lipid and atherosclerosis, C-type leptin receptor signaling, estrogen-dependent nuclear events and extranuclear signaling, adipogenesis and resistin signaling (Figure 5).
Interestingly, infection-indicating signaling pathways did not exhibit any major involvement in our study (Figure 5).
At last, the REAC evaluation of globally involved pathways [70] revealed the innate immune system, the cytokine signaling in immune system (major pathways: regulation of IFNG signaling), signal transduction (nuclear receptor, GPCR and leptin pathways) and developmental biology (formation of the cornified envelope pathway) pathways as the mainly HS-associated ones (Figure S2).
The protein-based connectivity map occurring from an assumed gene biomarker translation (103 proteins our of 109 genes) resulted in 2465 interactions compared with the expected 531 interactions (4.64-fold; p < 0.0001), a result that indicates a robust strong protein–protein association in HS (Figure 6). On the other hand, the protein-based connectivity map occurring from the 386 HS-associated DEGs (372 proteins out of 386 genes) resulted in 19,823 interactions compared with the expected 6502 interactions (3.05-fold; p < 0.0001), indicating that the biomarker selection procedure increased the HS/protein association.
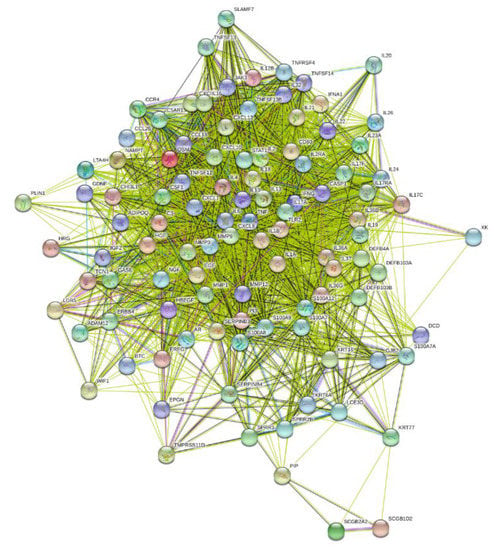
Figure 6.
Biomarker-resulting protein-based connectivity map of HS.
3.5. Enrichment Analysis of HS Druggable Genes
Among the 386 HS-associated DEGs, 105 druggable genes were recognized. With the 11 additional druggable genes described by Zouboulis et al. [12], namely ABAT, ADRA1A, CYP3A4, GRM4, HRH1, OPRD1, OPRM, PRKAB1, PTGS1, PTGS2 and SLC6A4, the overall detected druggable genes in HS are 116.
The 116 druggable genes were enriched in relevant signaling pathways according to the KEGG GO [20] and the Gene Cards [25] public domains to identify the major targeted organismal and signal transduction pathways (Figure S3). Twenty-two druggable genes were enriched in the lipid and atherosclerosis pathway (8.4 × 10−13), 19 in the JAK-STAT signaling pathway (6.2 × 10−12), 17 in the Th17 cell differentiation pathway (5.2 × 10−13), 17 in the IL-17 signaling pathway (6.0 × 10−14), 16 in the inflammatory bowel disease pathway (1.5 × 10−16), 14 in the TLR signaling pathway (6.0 × 10−14), 14 in the C-type leptin receptor signaling pathway (2.4 × 10−9) and 13 in the TNF signaling pathway (8.4 × 10−8).
3.6. Study Drugs and Drug Repurposing for HS
The majority of registered, studied or off-label administered drugs modify HS-associated DEGs. On the other hand, the evaluation of the detected 105 HS-associated druggable genes proposed 452 potentially therapeutic compounds, among them 120 launched drugs, 178 compounds in clinical studies and 154 in preclinical evaluation (Table S2). Among these potentially therapeutic compounds, the 31 drugs, which regulate three or more genes with all of them being HS-associated DEGs or at least four genes with 60% of them been DEGs were classified as probable repurposing drugs for HS (Table 2).

Table 2.
Probable HS repurposing drugs * and molecular profile of drugs registered ** or off-label administered in HS.
4. Discussion
4.1. HS Pathogenesis
Inflammation doubtlessly plays a major role in the pathogenesis of HS [3,7,8]. Proteome studies provide evidence that the innate immunity system and both IL-1 and IL-17 signaling pathways are activated in HS lesions and circulating neutrophils [27,40,45,71,72,73], findings that have been confirmed in our systematic review. In addition, Th17 differentiation of CD4+ lymphocytes is activated in HS [57]. Among others, Kelly et al. [38] provided evidence that CD45+CD4+ T cells are responsible for IL-17 production and CD11c+CD1a-CD14+ dendritic cells are the main producers of IL-1β in lesional HS skin. The IL-17 cytokine family has been linked to the pathogenesis of diverse autoimmune and inflammatory diseases and also plays an essential role in host defense against extracellular microorganisms [2,74]. IL-17 has been shown to increase the expression of skin antimicrobial peptides, including human β-defensin 2, psoriasin (S100A7) and calprotectin (S100A8/9) in keratinocytes and of a number of cytokines attracting neutrophils [75]. Thus, IL-17 may contribute to inflammation by increasing the influx of neutrophils, dendritic cells and memory T cells into the lesions. On the other hand, the involvement of IL-1 signaling pathway is also prominent in HS with upregulation of molecules causing immune cell infiltration and extracellular matrix degradation and could be reversed by application of IL-1 receptor antagonist [40,76]. IL1B signaling pathway-associated genes, such as IL1R1, IL1RN, IFNG, IL6, IL18, IL18R1, IL32, IL33, IL36A, IL36B, IL36G, IL36RN, IL37, TLR2, TLR3, TLR4, S100A7, S100A7A, S100A8, S100A9 and S100A12 were HS-associated DEGs, as detected in our systemic review.
The inflammatory process in HS seems to be coupled with impaired barrier function, altered epidermal cell differentiation, formation of the cornified envelope, TLRs and antimicrobial peptides [3], the latter not being associated with any infection, as clearly shown in the present study. These events have been observed at the follicular and epidermal keratinocytes and at a minor grade at the skin glands [3]. Moreover, we could confirm a dysregulated expression pattern of serpins, small proline-rich proteins and certain keratins, which further support the involvement of the follicular infundibulum in the initiation of the lesions, especially at the anatomic area of communication with the apocrine gland duct and the ductus seboglandularis [3].
Although HS has well-documented associations with the metabolic syndrome, which is characterized by systemic inflammation identified at a molecular level [77], the role of adipose tissue in HS has barely been investigated. Obesity is currently shown to represent the primary risk factor in HS at the molecular level [4,28]. A chronic low-grade subclinical inflammatory response is strongly implicated in the pathogenesis of insulin resistance and metabolic syndrome. The clinically relevant peroxisome proliferator-activated receptor (PPAR) pathway was down-regulated in adipocytes of HS lesions [4]. In agreement with these data, reduced serum levels of adiponectin were currently found in non-diabetic patients with HS [28]. Since adiponectin inhibits the production of TNF-α, IL-6 and chemokines of human macrophages the upregulation of ADIPOQ and PLIN1, shown in this systematic review, might be beneficial in HS treatment. Indeed, thiazolidine derivatives act as PPARγ agonists and effectively increase the adiponectin concentration and adipogenic gene expression [28,78]. Unsaturated fatty acids, eicosanoids and non-steroidal anti-inflammatory drugs function in a similar manner [79]. Further metabolic pathways, e.g., the IGF transport and uptake of IGF-binding proteins pathway, type I diabetes mellitus signaling, lipid and atherosclerosis, C-type leptin receptor signaling, estrogen-dependent nuclear events and extranuclear signaling and RETN signaling, encoding resistin, are dysregulated in HS, as shown in the present review.
In conclusion, inflammatory signaling, mainly innate immunity signaling pathways, mostly that of IL-1 and IL-17, epithelial differentiation signaling pathways, primarily of follicular keratinocytes and skin gland duct cells and metabolic signaling pathways, especially that of obesity/adipogenesis, represent pathogenetic HS cascades, whose activity may be targeted by future therapeutic means.
4.2. HS Comorbid Disorders
HS has been associated with a variety of comorbid disorders, such as inflammatory bowel diseases, especially Crohn’s disease, axial spondylarthritis without or with follicular occlusion, triad signs, genetic keratin disorders associated with follicular occlusion, such as pachyonychia congenita, steatocystoma multiplex, Dowling-Degos disease without and with arthritis, as well as other genetic disorders, such as keratitis–ichthyosis–deafness syndrome and Down syndrome [80]. Moreover, HS has been associated with reduced quality of life, metabolic syndrome, sexual dysfunction, working disability, depression and anxiety. Like in psoriasis, HS patients have higher prevalence of cardiovascular disease risk factors and suicide risk [81]. At last, the development of epithelial tumors on chronic HS lesions at the anogenital region may be considered as the consequence of chronic severe inflammatory skin disease. The current work has provided molecular evidence of HS association with inflammatory bowel disease pathway, rheumatoid arthritis pathway, type I diabetes mellitus signaling, lipid and atherosclerosis and adipogenesis signaling.
4.3. Study Drugs and Drug Repurposing for HS
In addition to the only registered drug in HS, namely adalimumab [9,82,83], the majority of studied and off-label administered drugs also regulate differentially expressed genes and their proteins in HS, as shown in the present review [10,65,76,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95]. On the other hand, the 452 HS-associated druggable genes proposed can mostly be classified in receptor ligands, enzyme/protein inhibitors, JAK-STAT inhibitors, PI3K inhibitors, sodium/potassium/calcium channel activators and MMP inhibitors. Additionally, Gentamicin, Ibudilast, Spironolactone, Trastuzumab, Thalidomide, Apremilast, Glucosamine, Interferon-a-2b, Binimetinib and Midostaurin have previously been reported as repurposing drugs for HS [11]. The majority of the 31 probable repurposing drugs shown in Table 2 are JAK inhibitors, with cytokine inhibitors, such as anti-IL-17 compounds, tyrosine kinase receptor inhibitors, TNF inhibitors, cyclooxygenase inhibitors, EGF receptor inhibitors, MMP inhibitors and PPARγ ligands—among others—being represented. Ten of these drugs, which have not yet been administered in HS, are already launched for other indications and 17 are in clinical studies, not including HS.
5. Conclusions
The current review provides robust molecular evidence on the pathogenetic triads of HS, namely upregulated inflammation, dysregulated epithelial cell differentiation and obesity signaling/hormone involvement. In addition, evidence of the negligible role of infectious agents is included. Moreover, HS biomarkers with strong protein–protein connectivity in HS are presented. While adalimumab, the only currently registered drug in HS, and the majority of studied and off-label administered drugs regulate DEGs and their proteins in HS, numerous compounds are eligible for HS repurposing due to their molecular signaling. Among them, 31 compounds are designated probable, following our classification, with 10 of them already being launched for other indications.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/pharmaceutics14010044/s1: Figure S1: Enrichment of HS-associated DEGs in signaling pathways; Figure S2: Global REAC evaluation of possibly involved signaling pathways in HS; Figure S3: Enrichment of druggable HS-associated genes in signaling pathways; Table S1: DEGs resulting from the comparison of transcriptomic profiles and protein expression studies between lesional HS and non-lesional skin biopsies and blood samples from HS patients and healthy controls, respectively; Table S2. Drugs regulating HS-associated DEGs.
Author Contributions
Conceptualization, C.C.Z.; methodology, V.A.Z., K.C.Z. and C.C.Z.; software, V.A.Z. and K.C.Z.; validation, V.A.Z., K.C.Z. and C.C.Z.; formal analysis, V.A.Z. and K.C.Z.; investigation, V.A.Z. and K.C.Z.; resources, V.A.Z., K.C.Z. and C.C.Z.; data curation, V.A.Z. and K.C.Z.; writing—original draft preparation, V.A.Z., K.C.Z. and C.C.Z.; writing—review and editing, C.C.Z.; visualization, V.A.Z.; supervision, C.C.Z.; project administration, C.C.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Ethical review and approval were waived because the article reviews ethically approved published studies involving humans.
Informed Consent Statement
Patient consent was waived because the article reviews published studies involving humans. No patient can be identified.
Data Availability Statement
Data sets related to this article are hosted at the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/ (accessed on 21 November 2021)) data repositories GSE72702, GSE79150, GSE128637, GSE137141, GSE144801, GSE148027, GSE154773, GSE154775, GSE155176, GSE155850 and GSE175990.
Acknowledgments
The Departments of Dermatology, Venereology, Allergology and Immunology, Dessau Medical Center, Dessau, Germany are health care providers of the European Reference Network for Rare and Complex Skin Diseases (ERN Skin—ALLOCATE Skin group).
Conflicts of Interest
V.A.Z. and K.C.Z. declare no conflict of interest. C.C.Z. has received subject-relevant honoraria from AbbVie, Bayer Healthcare, Boehringer-Ingelheim, Idorsia, Incyte, Inflarx, Janssen, Novartis, Regeneron, UCB and Viatris, which were not associated with or have any influence on this study. His departments have received grants from AbbVie, AOTI, AstraZeneca, Celgene, Galderma, Inflarx, NAOS-BIODERMA, Novartis, PPM and UCB for his participation as clinical investigator, which were not associated with this study.
References
- Zouboulis, C.C.; Del Marmol, V.; Mrowietz, U.; Prens, E.P.; Tzellos, T.; Jemec, G.B.E. Hidradenitis suppurativa/acne inversa: Criteria for diagnosis, severity assessment, classification and disease evaluation. Dermatology 2015, 231, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Del Duca, E.; Morelli, P.; Bennardo, L.; Di Raimondo, C.; Nisticò, S.P. Cytokine pathways and investigational target therapies in hidradenitis suppurativa. Int. J. Mol. Sci. 2020, 21, 8436. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Nogueira da Costa, A.; Makrantonaki, E.; Hou, X.X.; Almansouri, D.; Dudley, J.T.; Edwards, H.; Readhead, B.; Balthasar, O.; Jemec, G.B.E.; et al. Alterations in innate immunity and epithelial cell differentiation are the molecular pillars of hidradenitis suppurativa. J. Eur. Acad. Dermatol. Venereol. 2020, 34, 846–861. [Google Scholar] [CrossRef] [PubMed]
- Kaleta, K.P.; Nikolakis, G.; Hossini, A.M.; Balthasar, O.; Almansouri, D.; Vaiopoulos, A.; Knolle, J.; Boguslawska, A.; Wojas-Pelc, A.; Zouboulis, C.C. Metabolic disorders/obesity is a primary risk factor in hidradenitis suppurativa: An immunohistochemical real-world approach. Dermatology 2021. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Desai, N.; Emtestam, L.; Hunger, R.E.; Ioannides, D.; Juhász, I.; Lapins, J.; Matusiak, L.; Prens, E.P.; Revuz, J.; et al. European S1 guideline for the treatment of hidradenitis suppurativa/acne inversa. J Eur. Acad. Dermatol. Venereol. 2015, 29, 619–644. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Bechara, F.G.; Dickinson-Blok, J.L.; Gulliver, W.; Horváth, B.; Hughes, R.; Kimball, A.B.; Kirby, B.; Martorell, A.; Podda, M.; et al. Hidradenitis suppurativa/acne inversa: A practical framework for treatment optimization—Systematic review and recommendations from the HS ALLIANCE working group. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 19–31. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Benhadou, F.; Byrd, A.; Chandran, N.; Giamarellos-Bourboulis, E.; Fabbrocini, G.; Frew, J.; Fujita, H.; González-López, M.A.; Guillem, P.; et al. What causes hidradenitis suppurativa? 15 years after. Exp. Dermatol. 2020, 29, 1154–1170. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Frew, J.W.; Giamarellos-Bourboulis, E.J.; Jemec, G.B.E.; del Marmol, V.; Marzano, A.V.; Nikolakis, G.; Sayed, C.J.; Tzellos, T.; Wolk, K.; et al. Target molecules for future hidradenitis suppurativa treatment. Exp. Dermatol. 2021, 30 (Suppl. 1), 8–17. [Google Scholar] [CrossRef]
- Kimball, A.B.; Okun, M.M.; Williams, D.A.; Gottlieb, A.B.; Papp, K.A.; Zouboulis, C.C.; Armstrong, A.W.; Kerdel, F.; Gold, M.H.; Forman, S.B.; et al. Two phase 3 trials of adalimumab treatment of hidradenitis suppurativa. N. Engl. J. Med. 2016, 375, 422–434. [Google Scholar] [CrossRef]
- Glatt, S.; Jemec, G.B.; Forman, S.; Sayed, C.; Schmieder, G.; Weisman, J.; Rolleri, R.; Seegobin, S.; Baeten, D.; Ionescu, L.; et al. Bimekizumab in moderate-to-severe hidradenitis suppurativa: A phase 2, double-blind, placebo-controlled randomized clinical trial. JAMA Dermatol. 2021, 157, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Zouboulis, C.C.; Nogueira da Costa, A. Drug repurposing through drug-gene interaction profiles for hidradenitis suppurativa/acne inversa treatment. J Eur Acad Dermatol Venereol. 2021, 35, e251–e254. [Google Scholar] [CrossRef]
- Zouboulis, C.C.; Readhead, B.; Dudley, J.T. An additional drug repurposing study for hidradenitis suppurativa/acne inversa. Br. J. Dermatol. 2021, 184, 748–750. [Google Scholar] [CrossRef]
- Giordano, A.; Forte, G.; Massimo, L.; Riccio, R.; Bifulco, G.; Di Micco, S. Discovery of new erbB4 inhibitors: Repositioning an orphan chemical library by inverse virtual screening. Eur. J. Med. Chem. 2018, 152, 253–263. [Google Scholar] [CrossRef]
- Yu, J.L.; Dai, Q.Q.; Li, G.B. Deep learning in target prediction and drug repositioning: Recent advances and challenges. Drug Discov. Today 2021. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. Bmj 2021, 372, n71. [Google Scholar] [CrossRef]
- Gene Expression Omnibus. Available online: https://www.ncbi.nlm.nih.gov/geo/ (accessed on 15 November 2021).
- HUGO Gene Nomenclature Committee. Available online: https://www.genenames.org/tools/multi-symbol-checker/ (accessed on 15 November 2021).
- Biological DataBase network. Available online: https://biodbnet-abcc.ncifcrf.gov/db/db2db.php (accessed on 15 November 2021).
- g:Profiler. Available online: https://biit.cs.ut.ee/gprofiler/gost (accessed on 15 November 2021).
- Kyoto Encyclopedia of Genes and Genomes. Available online: https://www.genome.jp/kegg/ (accessed on 15 November 2021).
- Reactome. Available online: https://reactome.org/ (accessed on 15 November 2021).
- WikiPathways. Available online: https://www.wikipathways.org/ (accessed on 15 November 2021).
- Human Phenotype Ontology. Available online: https://hpo.jax.org/app/ (accessed on 15 November 2021).
- The Drug Repurposing Hub. Available online: https://s3.amazonaws.com/data.clue.io/repurposing/downloads/repurposing_drugs_20200324.txt (accessed on 15 November 2021).
- Gene Cards, The Human Gene Database. Available online: https://www.genecards.org/ (accessed on 15 November 2021).
- Lowe, M.M.; Naik, H.B.; Clancy, S.; Pauli, M.; Smith, K.M.; Bi, Y.; Dunstan, R.; Gudjonsson, J.E.; Paul, M.; Harris, H.; et al. Immunopathogenesis of hidradenitis suppurativa and response to anti-TNF-alpha therapy. JCI Insight 2020, 5, e139932. [Google Scholar] [CrossRef]
- Hoffman, L.K.; Tomalin, L.E.; Schultz, G.; Howell, M.D.; Anandasabapathy, D.; Alavi, A.; Suárez-Fariñas, M.; Lowes, M.A. Integrating the skin and blood transcriptomes and serum proteome in hidradenitis suppurativa reveals complement dysregulation and a plasma cell signature. PLoS ONE 2018, 13, e0203672. [Google Scholar] [CrossRef] [Green Version]
- González-López, M.A.; Vilanova, I.; Ocejo-Viñals, G.; Arlegui, R.; Navarro, I.; Guiral, S.; Mata, C.; Pérez-Paredes, M.G.; Portilla, V.; Corrales, A.; et al. Circulating levels of adiponectin, leptin, resistin and visfatin in non-diabetics patients with hidradenitis suppurativa. Arch. Dermatol. Res. 2020, 312, 595–600. [Google Scholar] [CrossRef]
- Hessam, S.; Sand, M.; Skrygan, M.; Bechara, F.G. The microRNA effector RNA-induced silencing complex in hidradenitis suppurativa: A significant dysregulation within active inflammatory lesions. Arch. Dermatol. Res. 2017, 309, 557–565. [Google Scholar] [CrossRef]
- Rumberger, B.E.; Boarder, E.L.; Owens, S.L.; Howell, M.D. Transcriptomic analysis of hidradenitis suppurativa skin suggests roles for multiple inflammatory pathways in disease pathogenesis. Inflamm. Res. 2020, 69, 967–973. [Google Scholar] [CrossRef]
- Penno, C.A.; Jäger, P.; Laguerre, C.; Hasler, F.; Hofmann, A.; Gass, S.K.; Wettstein-Ling, B.; Schaefer, D.J.; Avrameas, A.; Raulf, F.; et al. Lipidomics profiling of hidradenitis suppurativa skin lesions reveals lipoxygenase pathway dysregulation and accumulation of proinflammatory leukotriene B4. J. Investig. Dermatol. 2020, 140, 2421–2432. [Google Scholar] [CrossRef]
- Coates, M.; Mariottoni, P.; Corcoran, D.L.; Kirshner, H.F.; Jaleel, T.; Brown, D.A.; Brooks, S.R.; Murray, J.; Morasso, M.I.; MacLeod, A.S. The skin transcriptome in hidradenitis suppurativa uncovers an antimicrobial and sweat gland gene signature which has distinct overlap with wounded skin. PLoS ONE 2019, 14, e0216249. [Google Scholar] [CrossRef] [Green Version]
- Shanmugam, V.K.; Jones, D.; McNish, S.; Bendall, M.L.; Crandall, K.A. Transcriptome patterns in hidradenitis suppurativa: Support for the role of antimicrobial peptides and interferon pathways in disease pathogenesis. Clin. Exp. Dermatol. 2019, 44, 882–892. [Google Scholar] [CrossRef]
- Blok, J.L.; Li, K.; Brodmerkel, C.; Jonkman, M.F.; Horváth, B. Gene expression profiling of skin and blood in hidradenitis suppurativa. Br. J. Dermatol. 2016, 174, 1392–1394. [Google Scholar] [CrossRef]
- Buimer, M.G.; Wobbes, T.; Klinkenbijl, J.H.; Reijnen, M.M.; Blokx, W.A. Immunohistochemical analysis of steroid hormone receptors in hidradenitis suppurativa. Am. J. Dermatopathol. 2015, 37, 129–132. [Google Scholar] [CrossRef]
- Vossen, A.; van der Zee, H.H.; Davelaar, N.; Mus, A.M.C.; van Doorn, M.B.A.; Prens, E.P. Apremilast for moderate hidradenitis suppurativa: No significant change in lesional skin inflammatory biomarkers. J. Eur. Acad. Dermatol. Venereol. 2019, 33, 761–765. [Google Scholar] [CrossRef]
- Kanni, T.; Zenker, O.; Habel, M.; Riedemann, N.; Giamarellos-Bourboulis, E.J. Complement activation in hidradenitis suppurativa: A new pathway of pathogenesis? Br. J. Dermatol. 2018, 179, 413–419. [Google Scholar] [CrossRef] [Green Version]
- Kelly, G.; Hughes, R.; McGarry, T.; van den Born, M.; Adamzik, K.; Fitzgerald, R.; Lawlor, C.; Tobin, A.M.; Sweeney, C.M.; Kirby, B. Dysregulated cytokine expression in lesional and nonlesional skin in hidradenitis suppurativa. Br. J. Dermatol. 2015, 173, 1431–1439. [Google Scholar] [CrossRef]
- Vossen, A.; Stubbs, A.; van Doorn, M.; van Straalen, K.; van der Zee, H.; Prens, E. Profiling of the transcriptome in hidradenitis suppurativa: A case-control sample. J. Investig. Dermatol. 2016, 136, S193. [Google Scholar] [CrossRef]
- Witte-Händel, E.; Wolk, K.; Tsaousi, A.; Irmer, M.L.; Mößner, R.; Shomroni, O.; Lingner, T.; Witte, K.; Kunkel, D.; Salinas, G.; et al. The IL-1 pathway is hyperactive in hidradenitis suppurativa and contributes to skin infiltration and destruction. J. Investig. Dermatol. 2019, 139, 1294–1305. [Google Scholar] [CrossRef]
- Vossen, A.; van der Zee, H.H.; Tsoi, L.C.; Xing, X.; Devalaraja, M.; Gudjonsson, J.E.; Prens, E.P. Novel cytokine and chemokine markers of hidradenitis suppurativa reflect chronic inflammation and itch. Allergy 2019, 74, 631–634. [Google Scholar] [CrossRef]
- Garcet, S.; Frew, J.W.; Navrazhina, K.; Krueger, J. Hidradenitis suppurativa RNA-seq skin transcriptome overlaps with psoriasis vulgaris and reveals a marked upregulation of multiple targetable cytokines. J. Investig. Dermatol. 2020, 140, S7. [Google Scholar] [CrossRef]
- Byrd, A.S.; Kerns, M.L.; Williams, D.W.; Zarif, J.C.; Rosenberg, A.Z.; Delsante, M.; Liu, H.; Dillen, C.A.; Maynard, J.P.; Caffrey, J.A.; et al. Collagen deposition in chronic hidradenitis suppurativa: Potential role for CD163(+) macrophages. Br. J. Dermatol. 2018, 179, 792–794. [Google Scholar] [CrossRef]
- Hotz, C.; Boniotto, M.; Guguin, A.; Surenaud, M.; Jean-Louis, F.; Tisserand, P.; Ortonne, N.; Hersant, B.; Bosc, R.; Poli, F.; et al. Intrinsic defect in keratinocyte function leads to inflammation in hidradenitis suppurativa. J. Investig. Dermatol. 2016, 136, 1768–1780. [Google Scholar] [CrossRef] [Green Version]
- Gudjonsson, J.E.; Tsoi, L.C.; Ma, F.; Billi, A.C.; van Straalen, K.R.; Vossen, A.R.J.V.; van der Zee, H.H.; Harms, P.W.; Wasikowski, R.; Yee, C.M.; et al. Contribution of plasma cells and B cells to hidradenitis suppurativa pathogenesis. JCI Insight 2020, 5, e139930. [Google Scholar] [CrossRef] [PubMed]
- Wolk, K.; Warszawska, K.; Hoeflich, C.; Witte, E.; Schneider-Burrus, S.; Witte, K.; Kunz, S.; Buss, A.; Roewert, H.J.; Krause, M.; et al. Deficiency of IL-22 contributes to a chronic inflammatory disease: Pathogenetic mechanisms in acne inversa. J. Immunol. 2011, 186, 1228–1239. [Google Scholar] [CrossRef] [Green Version]
- Nelson, A.M.; Cong, Z.; Gettle, S.L.; Longenecker, A.L.; Kidacki, M.; Kirby, J.S.; Adams, D.R.; Stairs, D.B.; Danby, F.W. E-cadherin and p120ctn protein expression are lost in hidradenitis suppurativa lesions. Exp. Dermatol. 2019, 28, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Dany, M.; Elston, D. Gene expression of sphingolipid metabolism pathways is altered in hidradenitis suppurativa. J. Am. Acad. Dermatol. 2017, 77, 268–273. [Google Scholar] [CrossRef]
- Matusiak, L.; Salomon, J.; Nowicka-Suszko, D.; Bieniek, A.; Szepietowski, J.C. Chitinase-3-like protein 1 (YKL-40): Novel biomarker of hidradenitis suppurativa disease activity? Acta Dermatovenereol. 2015, 95, 736–737. [Google Scholar] [CrossRef]
- Salomon, J.; Piotrowska, A.; Matusiak, L.; Dziegiel, P.; Szepietowski, J.C. Chitinase-3-like protein 1 (YKL-40) Is expressed in lesional skin in hidradenitis suppurativa. Vivo 2019, 33, 141–143. [Google Scholar] [CrossRef] [Green Version]
- Wolk, K.; Brembach, T.C.; Simaite, D.; Bartnik, E.; Cucinotta, S.; Pokrywka, A.; Irmer, M.L.; Triebus, J.; Witte-Handel, E.; Salinas, G.; et al. Activity and components of the granulocyte colony-stimulating factor pathway in hidradenitis suppurativa. Br. J. Dermatol. 2021, 185, 164–176. [Google Scholar] [CrossRef]
- Hofmann, S.C.; Saborowski, V.; Lange, S.; Kern, W.V.; Bruckner-Tuderman, L.; Rieg, S. Expression of innate defense antimicrobial peptides in hidradenitis suppurativa. J. Am. Acad. Dermatol. 2012, 66, 966–974. [Google Scholar] [CrossRef]
- Dréno, B.; Khammari, A.; Brocard, A.; Moyse, D.; Blouin, E.; Guillet, G.; Leonard, F.; Knol, A.C. Hidradenitis suppurativa: The role of deficient cutaneous innate immunity. Arch. Dermatol. 2012, 148, 182–186. [Google Scholar] [CrossRef] [Green Version]
- Argyropoulou, M.; Grundhuber, M.; Kanni, T.; Tzanetakou, V.; Micha, S.; Stergianou, D.; Swiniarski, S.; Giamarellos-Bourboulis, E.J. A composite biomarker score for the diagnosis of hidradenitis suppurativa. Exp. Dermatol. 2019, 28, 18. [Google Scholar] [CrossRef] [Green Version]
- Hessam, S.; Sand, M.; Skrygan, M.; Gambichler, T.; Bechara, F.G. Inflammation induced changes in the expression levels of components of the microRNA maturation machinery Drosha, Dicer, Drosha co-factor DGRC8 and Exportin-5 in inflammatory lesions of hidradenitis suppurativa patients. J. Dermatol. Sci. 2016, 82, 166–174. [Google Scholar] [CrossRef]
- Van der Zee, H.H.; de Ruiter, L.; van den Broecke, D.G.; Dik, W.A.; Laman, J.D.; Prens, E.P. Elevated levels of tumour necrosis factor (TNF)-α, interleukin (IL)-1β and IL-10 in hidradenitis suppurativa skin: A rationale for targeting TNF-α and IL-1β. Br. J. Dermatol. 2011, 164, 1292–1298. [Google Scholar] [CrossRef]
- Wieland, C.W.; Vogl, T.; Ordelman, A.; Vloedgraven, H.G.; Verwoolde, L.H.; Rensen, J.M.; Roth, J.; Boer, J.; Hessels, J. Myeloid marker S100A8/A9 and lymphocyte marker, soluble interleukin 2 receptor: Biomarkers of hidradenitis suppurativa disease activity? Br. J. Dermatol. 2013, 168, 1252–1258. [Google Scholar] [CrossRef]
- Montaudié, H.; Seitz-Polski, B.; Cornille, A.; Benzaken, S.; Lacour, J.-P.; Passeron, T. Interleukin 6 and high-sensitivity C-reactive protein are potential predictive markers of response to infliximab in hidradenitis suppurativa. J. Am. Acad. Dermatol. 2017, 76, 156–158. [Google Scholar] [CrossRef] [Green Version]
- Matusiak, L.; Szczech, J.; Bieniek, A.; Nowicka-Suszko, D.; Szepietowski, J.C. Increased interleukin (IL)-17 serum levels in patients with hidradenitis suppurativa: Implications for treatment with anti-IL-17 agents. J. Am. Acad. Dermatol. 2017, 76, 670–675. [Google Scholar] [CrossRef]
- Schlapbach, C.; Hanni, T.; Yawalkar, N.; Hunger, R.E. Expression of the IL-23/Th17 pathway in lesions of hidradenitis suppurativa. J. Am. Acad. Dermatol. 2011, 65, 790–798. [Google Scholar] [CrossRef]
- Thomi, R.; Kakeda, M.; Yawalkar, N.; Schlapbach, C.; Hunger, R.E. Increased expression of the interleukin-36 cytokines in lesions of hidradenitis suppurativa. J. Eur. Acad. Dermatol. Venereol. 2017, 31, 2091–2096. [Google Scholar] [CrossRef]
- Hayran, Y.; Allı, N.; Yücel, Ç.; Akdoğan, N.; Turhan, T. Serum IL-36alpha, IL-36beta, and IL-36gamma levels in patients with hidradenitis suppurativa: Association with disease characteristics, smoking, obesity, and metabolic syndrome. Arch. Dermatol. Res. 2020, 312, 187–196. [Google Scholar] [CrossRef]
- Akdogan, N.; Alli, N.; Uysal, P.I.; Topcuoglu, C.; Candar, T.; Turhan, T. Visfatin and insulin levels and cigarette smoking are independent risk factors for hidradenitis suppurativa: A case-control study. Arch. Dermatol. Res. 2018, 310, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Wolk, K.; Witte, E.; Tsaousi, A.; Witte, K.; Volk, H.; Sterry, W.; Wenzel, J.; Schneider-Burrus, S.; Sabat, R. Lipocalin-2 as a novel biomarker in acne inversa. J. Investig. Dermatol. 2016, 136, S235. [Google Scholar] [CrossRef]
- Blok, J.L.; Li, K.; Brodmerkel, C.; Horvátovich, P.; Jonkman, M.F.; Horváth, B. Ustekinumab in hidradenitis suppurativa: Clin-ical results and a search for potential biomarkers in serum. Br. J. Dermatol. 2016, 174, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Tsaousi, A.; Witte, E.; Witte, K.; Rowert-Huber, H.J.; Volk, H.D.; Sterry, W.; Wolk, K.; Schneider-Burrus, S.; Sabat, R. MMP8 is increased in lesions and blood of acne inversa patients: A potential link to skin destruction and metabolic alterations. Mediators Inflamm. 2016, 2016, 4097574. [Google Scholar] [CrossRef]
- Wang, B.; Yang, W.; Wen, W.; Sun, J.; Su, B.; Liu, B.; Ma, D.; Lv, D.; Wen, Y.; Qu, T.; et al. Gamma-secretase gene mutations in familial acne inversa. Science 2010, 330, 1065. [Google Scholar] [CrossRef]
- Hunger, R.E.; Surovy, A.M.; Hassan, A.S.; Braathen, L.R.; Yawalkar, N. Toll-like receptor 2 is highly expressed in lesions of acne inversa and colocalizes with C-type lectin receptor. Br. J. Dermatol. 2008, 158, 691–697. [Google Scholar] [CrossRef]
- Sartorius, K.; Emtestam, L.; Lapins, J.; Johansson, O. Cutaneous PGP 9.5 distribution patterns in hidradenitis suppurativa. Arch. Dermatol. Res. 2010, 302, 461–468. [Google Scholar] [CrossRef]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Forner, O.; Marin-Garcia, P.; Arnau, V.; D’Eustachio, P.; Stein, L.; Hermjakob, H. Reactome pathway analysis: A high-performance in-memory approach. BMC Bioinform. 2017, 18, 142. [Google Scholar] [CrossRef] [Green Version]
- Frew, J.W.; Hawkes, J.E.; Krueger, J.G. A systematic review and critical evaluation of immunohistochemical associations in hidradenitis suppurativa. F1000Research 2018, 7, 1923. [Google Scholar] [CrossRef] [PubMed]
- Vossen, A.R.J.V.; van der Zee, H.H.; Prens, E.P. Hidradenitis suppurativa: A systematic review integrating inflammatory pathways into a cohesive pathogenic model. Front. Immunol. 2018, 9, 2965. [Google Scholar] [CrossRef] [Green Version]
- Jenei, A.; Dajnoki, Z.; Medgyesi, B.; Gáspár, K.; Béke, G.; Kinyó, Á.; Méhes, G.; Hendrik, Z.; Dinya, T.; Törőcsik, D.; et al. Apocrine gland-rich skin has a non-inflammatory IL-17 related immune milieu, which turns to inflammatory IL-17 mediated disease in hidradenitis suppurativa. J. Investig. Dermatol. 2019, 139, 964–968. [Google Scholar] [CrossRef] [Green Version]
- Bartlett, H.S.; Million, R.P. Targeting the IL-17-T(H)17 pathway. Nat. Rev. Drug Discov. 2015, 14, 11–12. [Google Scholar] [CrossRef]
- Archer, N.K.; Adappa, N.D.; Palmer, J.N.; Cohen, N.A.; Harro, J.M.; Lee, S.K.; Miller, L.S.; Shirtliff, M.E. Interleukin-17A (IL-17A) and IL-17F are critical for antimicrobial peptide production and clearance of Staphylococcus aureus nasal colonization. Infect. Immun. 2016, 84, 3575–3583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzanetakou, V.; Kanni, T.; Giatrakou, S.; Katoulis, A.; Papadavid, E.; Netea, M.G.; Dinarello, C.A.; van der Meer, J.W.M.; Rigopoulos, D.; Giamarellos-Bourboulis, E.J. Safety and efficacy of anakinra in severe hidradenitis suppurativa: A randomized clinical trial. JAMA Dermatol. 2016, 152, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Mintoff, D.; Benhadou, F.; Pace, N.P.; Frew, J.W. Metabolic syndrome and hidradenitis suppurativa: Epidemiological, molecular, and therapeutic aspects. Int. J. Dermatol. 2021. [Google Scholar] [CrossRef]
- Salvator, H.; Grassin-Delyle, S.; Brollo, M.; Couderc, L.J.; Abrial, C.; Victoni, T.; Naline, E.; Devillier, P. Adiponectin inhibits the production of TNF-alpha, IL-6 and chemokines by human lung macrophages. Front. Pharmacol. 2021, 12, 718929. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, Q.; Yan, Y.; Sun, B.; Wang, Y.; Tang, L.; Wang, E.; Yu, J.; Corpuz Nogoy, K.M.; Li, X.; et al. Effect of ciglitazone on adipogenic transdifferentiation of bovine skeletal muscle satellite cells. J. Anim. Sci. Technol. 2021, 63, 934–953. [Google Scholar] [CrossRef]
- Fimmel, S.; Zouboulis, C.C. Comorbidities of hidradenitis suppurativa (acne inversa). Derm.-Endocrinol. 2010, 2, 9–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzellos, T.; Zouboulis, C.C. Review of comorbidities of hidradenitis suppurativa: Implications for daily clinical practice. Dermatol. Ther. 2020, 10, 63–71. [Google Scholar] [CrossRef] [Green Version]
- Zouboulis, C.C.; Okun, M.M.; Prens, E.P.; Gniadecki, R.; Foley, P.A.; Lynde, C.; Weisman, J.; Gu, Y.; Williams, D.A.; Jemec, G.B.E. Long-term adalimumab efficacy in patients with moderate-to-severe hidradenitis suppurativa/acne inversa: 3-year results of a phase 3 open-label extention study. J. Am. Acad. Dermatol. 2019, 80, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Zouboulis, C.C. Adalimumab for the treatment of hidradenitis suppurativa/acne inversa. Expert Rev. Clin. Immunol. 2016, 12, 1015–1026. [Google Scholar] [CrossRef]
- Matusiak, L.; Bieniek, A.; Szepietowski, J.C. Acitretin treatment for hidradenitis suppurativa: A prospective series of 17 patients. Br. J. Dermatol. 2014, 171, 170–174. [Google Scholar] [CrossRef]
- Frew, J.W.; Navrazhina, K.; Grand, D.; Sullivan-Whalen, M.; Gilleaudeau, P.; Garcet, S.; Ungar, J.; Krueger, J.G. The effect of subcutaneous brodalumab on clinical disease activity in hidradenitis suppurativa: An open-label cohort study. J. Am. Acad. Dermatol. 2020, 83, 1341–1348. [Google Scholar] [CrossRef]
- Mortimer, P.S.; Dawber, R.P.; Gales, M.A.; Moore, R.A. A double-blind controlled cross-over trial of cyproterone acetate in females with hidradenitis suppurativa. Br. J. Dermatol. 1986, 115, 263–268. [Google Scholar] [CrossRef]
- Nikolakis, G.; Kyrgidis, A.; Zouboulis, C.C. Is there a role for antiandrogen therapy for hidradenitis suppurativa? A systematic review of published data. Am. J. Clin. Dermatol. 2019, 20, 503–513. [Google Scholar] [CrossRef]
- Van Straalen, K.R.; Tzellos, T.; Guillem, P.; Benhadou, F.; Cuenca-Barrales, C.; Daxhelet, M.; Daoud, M.; Efthymiou, O.; Giamarellos-Bourboulis, E.J.; Jemec, G.B.E.; et al. The efficacy and tolerability of tetracyclines and clindamycin plus rifampicin for the treatment of hidradenitis suppurativa: Results of a prospective European cohort study. J. Am. Acad. Dermatol. 2021, 85, 369–378. [Google Scholar] [CrossRef]
- Lee, R.A.; Dommasch, E.; Treat, J.; Sciacca-Kirby, J.; Chachkin, S.; Williams, J.; Shin, D.B.; Leyden, J.J.; Vittorio, C.; Gelfand, J.M. A prospective clinical trial of open-label etanercept for the treatment of hidradenitis suppurativa. J. Am. Acad. Dermatol. 2009, 60, 565–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, D.R.; Yankura, J.A.; Fogelberg, A.C.; Anderson, B.E. Treatment of hidradenitis suppurativa with etanercept injection. Arch. Dermatol. 2010, 146, 501–504. [Google Scholar] [CrossRef] [PubMed]
- Melendez-Gonzalez, M.D.M.; Hamad, J.; Sayed, C. Golimumab for the treatment of hidradenitis suppurativa in patients with previous TNF-alpha treatment failure. J. Investig. Dermatol. 2021, 141, 2975–2979. [Google Scholar] [CrossRef]
- Grant, A.; Gonzalez, T.; Montgomery, M.O.; Cardenas, V.; Kerdel, F.A. Infliximab therapy for patients with moderate to severe hidradenitis suppurativa: A randomized, double-blind, placebo-controlled crossover trial. J. Am. Acad. Dermatol. 2010, 62, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Verdolini, R.; Clayton, N.; Smith, A.; Alwash, N.; Mannello, B. Metformin for the treatment of hidradenitis suppurativa: A little help along the way. J. Eur. Acad. Dermatol. Venereol. 2013, 27, 1101–1108. [Google Scholar] [CrossRef]
- Prussick, L.; Rothstein, B.; Joshipura, D.; Saraiya, A.; Turkowski, Y.; Abdat, R.; Alomran, A.; Zancanaro, P.; Kachuk, C.; Dumont, N.; et al. Open-label, investigator-initiated, single-site exploratory trial evaluating secukinumab, an anti-interleukin-17A monoclonal antibody, for patients with moderate-to-severe hidradenitis suppurativa. Br. J. Dermatol. 2019, 181, 609–611. [Google Scholar] [CrossRef]
- Giamarellos-Bourboulis, E.J.; Argyropoulou, M.; Kanni, T.; Spyridopoulos, T.; Otto, I.; Zenker, O.; Guo, R.; Riedemann, N.C. Clinical efficacy of complement C5a inhibition by IFX-1 in hidradenitis suppurativa: An open-label single-arm trial in patients not eligible for adalimumab. Br. J. Dermatol. 2020, 183, 176–178. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).