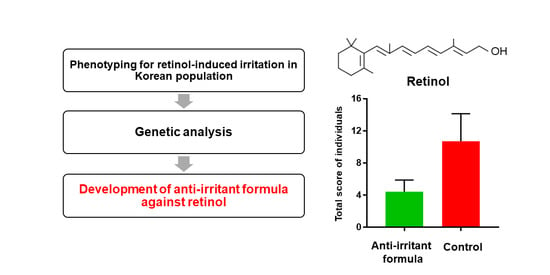
Anti-Irritant Strategy against Retinol Based on the Genetic Analysis of Korean Population: A Genetically Guided Top–Down Approach
Abstract
1. Introduction
2. Materials and Methods
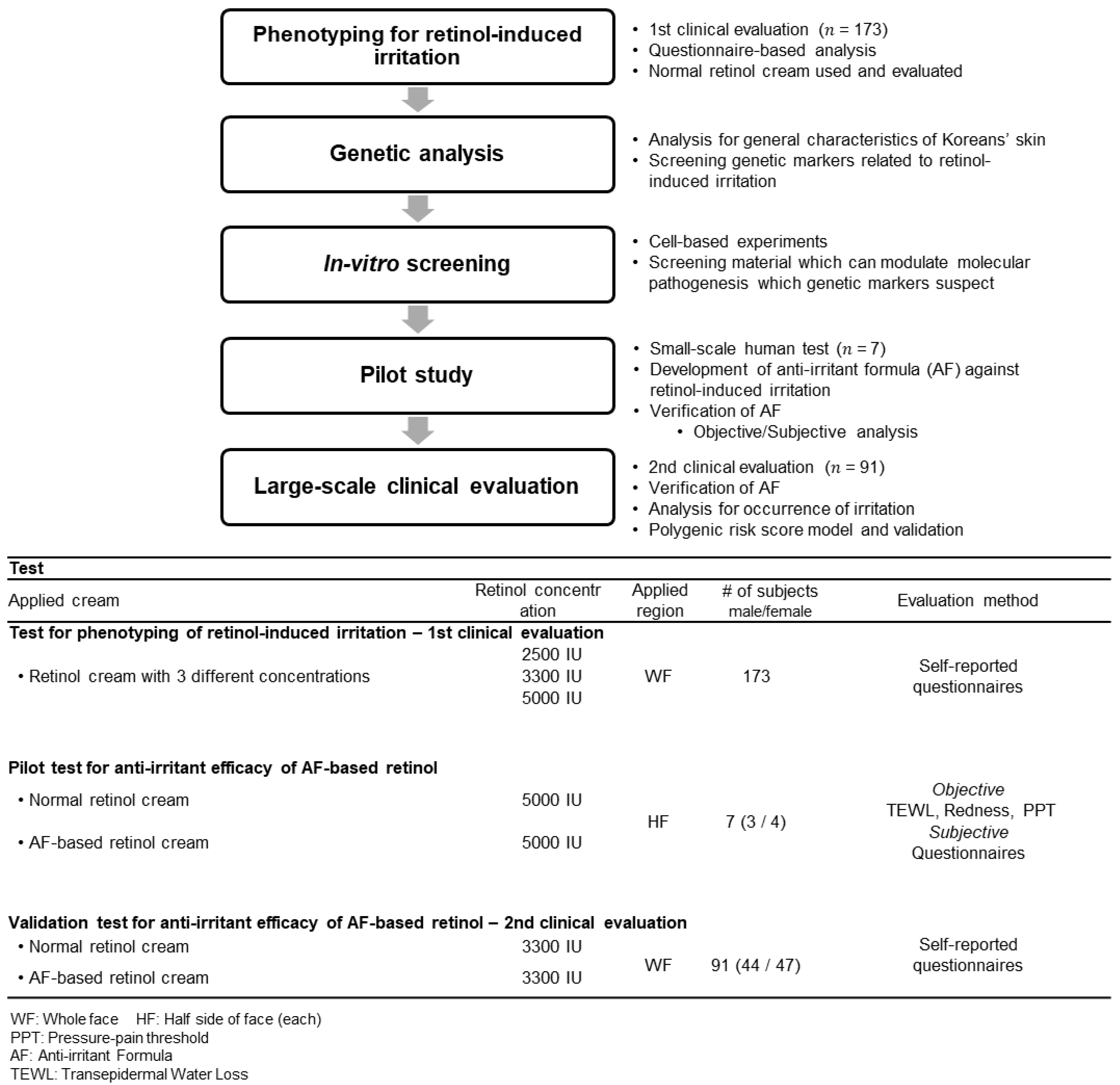
2.1. Study Design
2.2. Phenotyping of Retinol-Induced Irritation
- Using provided retinol cream;
- Providing information through a questionnaire after the experiment;
- Collecting saliva for DNA testing.
2.3. Single Nucleotide Polymorphism (SNP) Microarray
2.4. Assessment of Irritation
2.5. Cell Preparation and In Vitro Experiment
2.6. Statistical Analysis
3. Results and Discussion
3.1. Proposing Target Genes for Screening of Anti-Irritants to Retinol-Induced Irritation
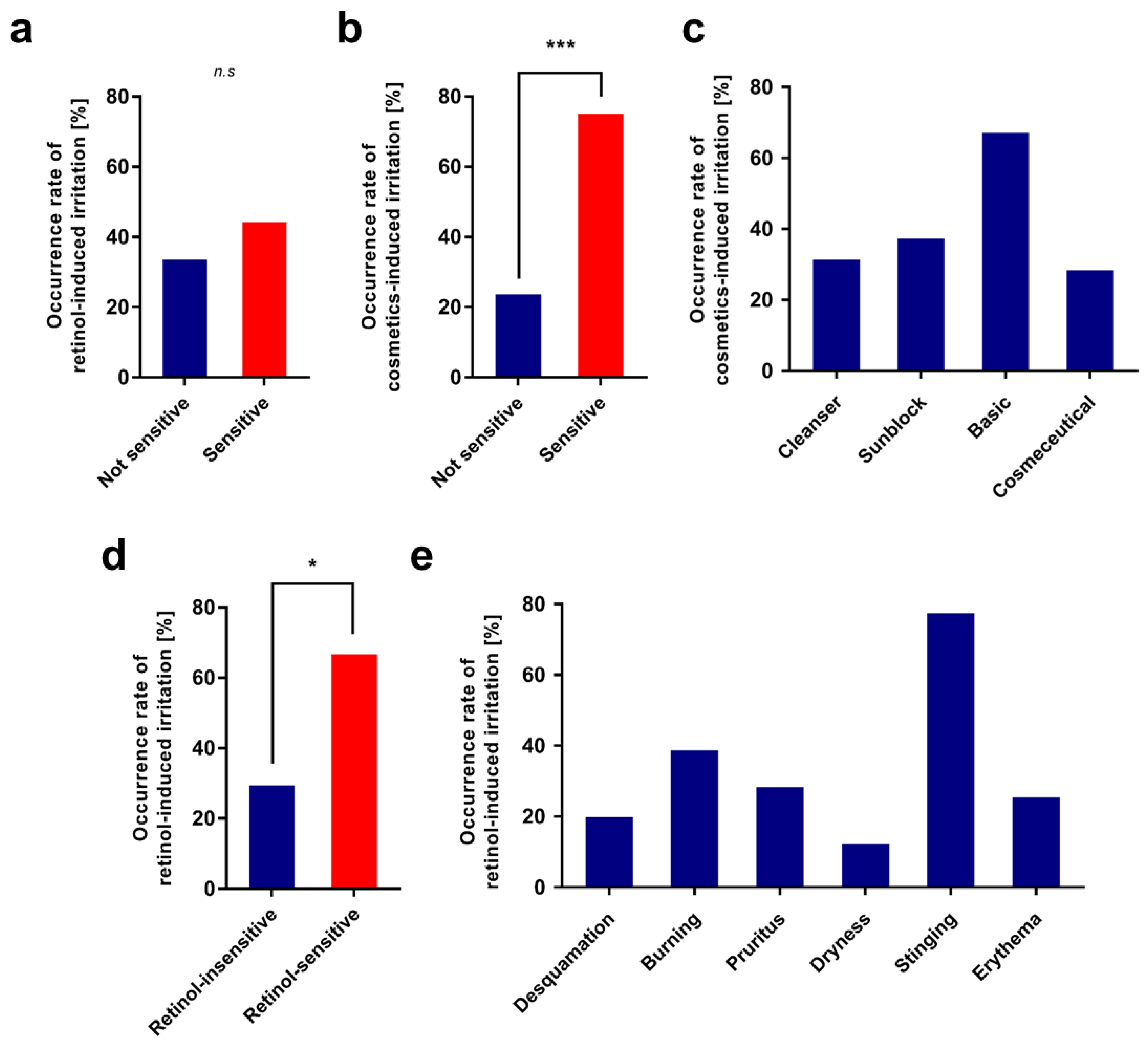
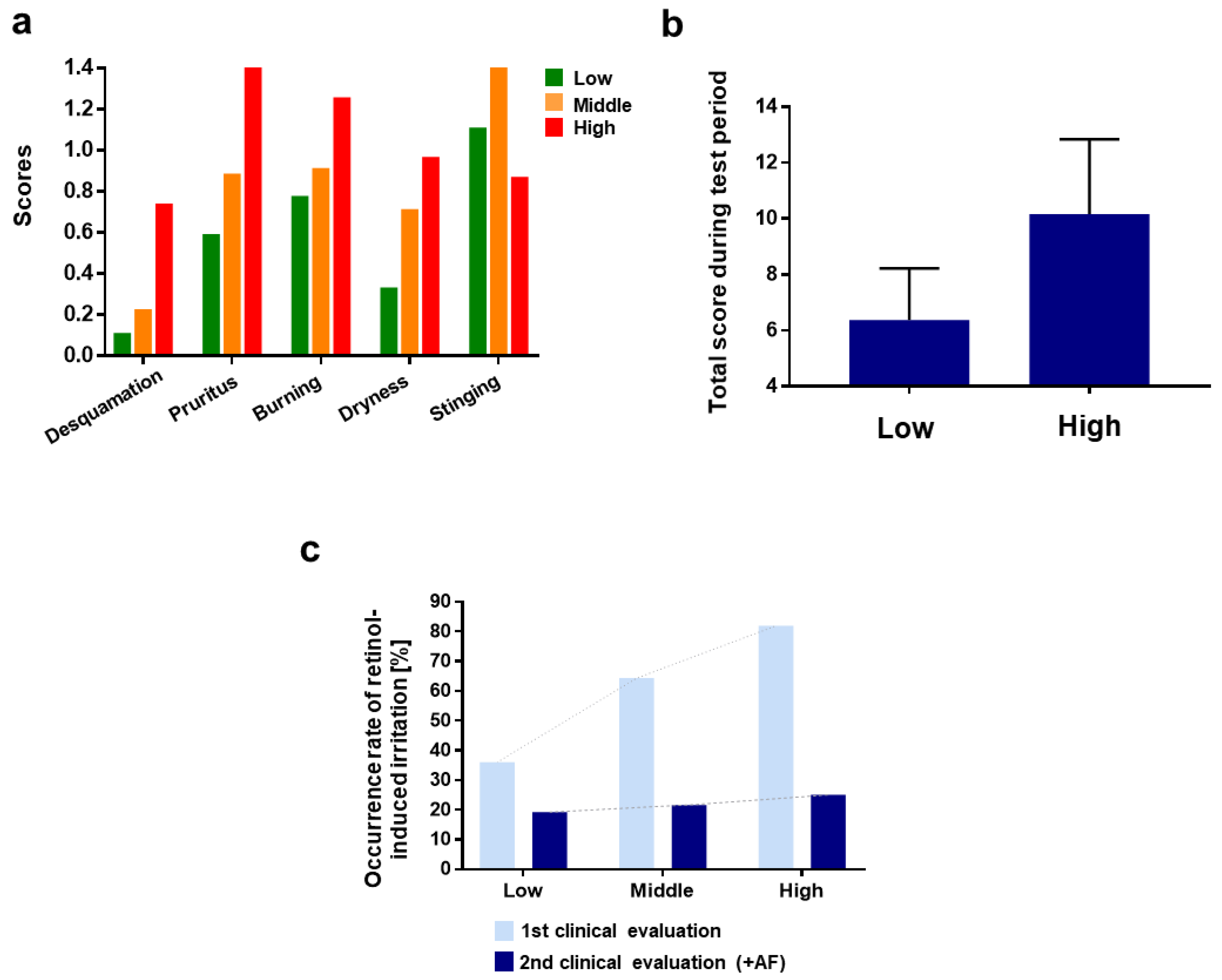
3.1.1. 1st Clinical Evaluation—Topical Application of Retinol and Analysis of Its Irritant Properties
3.1.2. Candidate Gene Analysis for Retinol-Induced Irritation
3.2. Top–Down Approach: Screening Anti-Irritants In Vitro
3.2.1. Tissue Repair-Related Genes, COL6A2, and EGFR
3.2.2. Inflammatory Gene: IL-4R
3.2.3. Neurogenic Inflammation, Adiponectin, and TRPV1
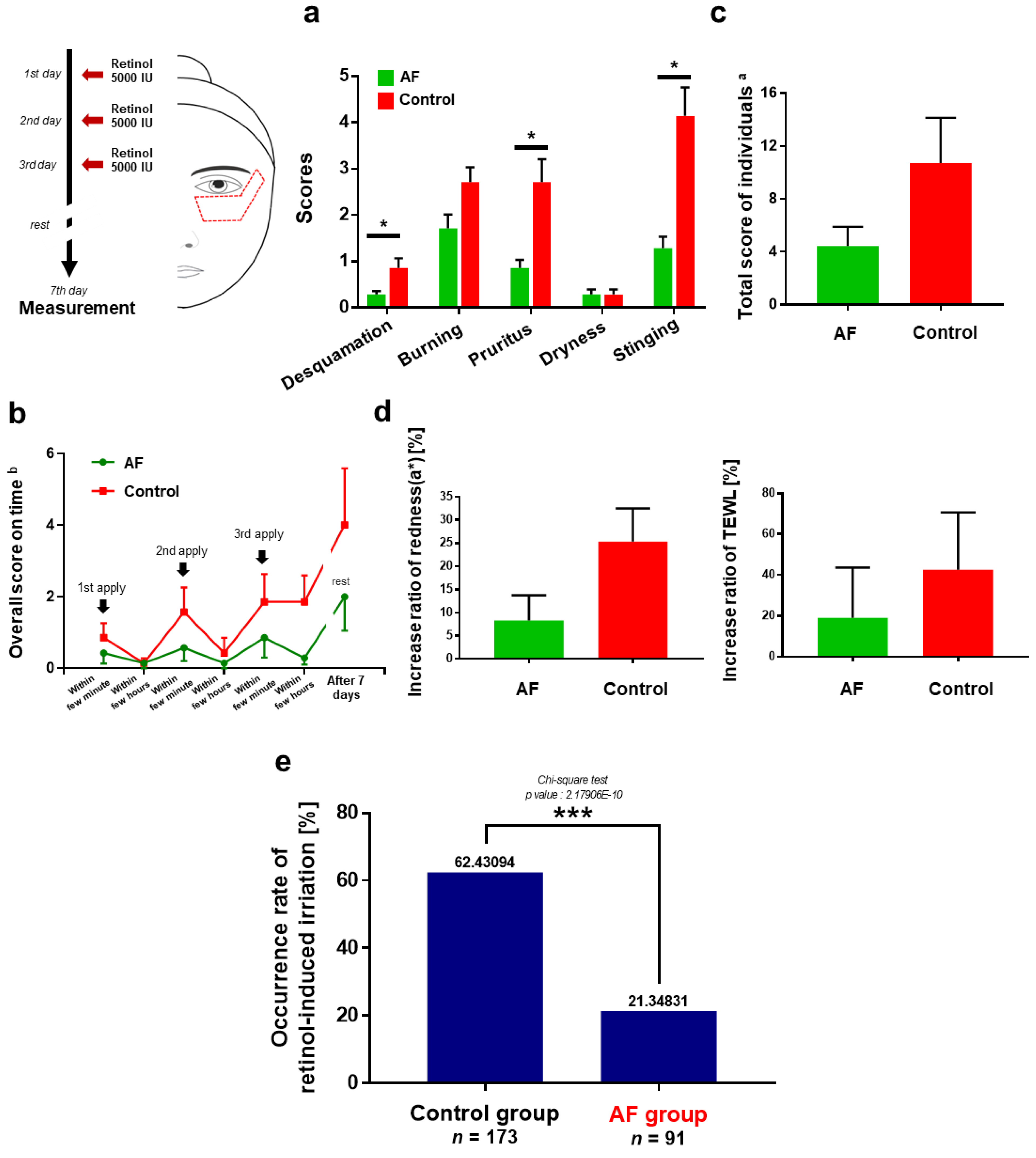
3.3. Anti-Irritation Efficacy of the Formula; Human Tests
3.4. Polygenic Risk Prediction Model for Retinol-Induced Irritation
3.5. Further Consideration; Acute vs. Chronic Irritation?
- As reported earlier, retinol may be converted into retinaldehyde (retinal) and retinoic acid to possess the bioavailability at which RAR or RXR binding is involved.
- Under retinol treatment, fibroblasts or keratinocytes activate the circumjacent cutaneous immune system as quickly as within a few minutes by producing IL-1 or IL-6 through the interaction between RAR and retinoic acid, to which the retinol should be converted.
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chanda, B.; Ditadi, A.; Iscove, N.N.; Keller, G. Retinoic acid signaling is essential for embryonic hematopoietic stem cell development. Cell 2013, 155, 215–227. [Google Scholar] [CrossRef]
- Erkelens, M.N.; Mebius, R.E. Retinoic acid and immune homeostasis: A balancing act. Trends Immunol. 2017, 38, 168–180. [Google Scholar] [CrossRef]
- Tang, X.-H.; Gudas, L.J. Retinoids, retinoic acid receptors, and cancer. Annu. Rev. Pathol. Mech. Dis. 2011, 6, 345–364. [Google Scholar] [CrossRef] [PubMed]
- Povoleri, G.A.; Nova-Lamperti, E.; Scottà, C.; Fanelli, G.; Chen, Y.-C.; Becker, P.D.; Boardman, D.; Costantini, B.; Romano, M.; Pavlidis, P. Human retinoic acid–regulated CD161+ regulatory T cells support wound repair in intestinal mucosa. Nat. Immunol. 2018, 19, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.; Napoli, J.; Enver, T.; Bernardino, L.; Ferreira, L. Advances and challenges in retinoid delivery systems in regenerative and therapeutic medicine. Nat. Commun. 2020, 11, 4265. [Google Scholar] [CrossRef]
- Theodosiou, M.; Laudet, V.; Schubert, M. From carrot to clinic: An overview of the retinoic acid signaling pathway. Cell. Mol. Life Sci. 2010, 67, 1423–1445. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Date, A.; Patravale, V.; Korting, H.C.; Roeder, A.; Weindl, G. Retinoids in the treatment of skin aging: An overview of clinical efficacy and safety. Clin. Interv. Aging 2006, 1, 327–348. [Google Scholar] [CrossRef]
- MacGregor, J.L.; Maibach, H.I. The specificity of retinoid-induced irritation and its role in clinical efficacy. Exog. Dermatol. 2002, 1, 68–73. [Google Scholar] [CrossRef]
- Kim, B.-H.; Lee, Y.-S.; Kang, K.-S. The mechanism of retinol-induced irritation and its application to anti-irritant development. Toxicol. Lett. 2003, 146, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Cheong, K.A.; Kim, H.J.; Kim, J.-Y.; Kim, C.-H.; Lim, W.-S.; Noh, M.; Lee, A.-Y. Retinoic acid and hydroquinone induce inverse expression patterns on cornified envelope-associated proteins: Implication in skin irritation. J. Dermatol. Sci. 2014, 76, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Q.; Geng, S. All-trans retinoic acid alters the expression of the tight junction proteins Claudin-1 and-4 and epidermal barrier function-associated genes in the epidermis. Int. J. Mol. Med. 2019, 43, 1789–1805. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.; Cheng, S.-H.; Coop, L.; Xia, Q.; Culp, S.; Tolleson, W.; Wamer, W.; Howard, P. Photoreaction, phototoxicity, and photocarcinogenicity of retinoids. J. Environ. Sci. Health Part C 2003, 21, 165–197. [Google Scholar] [CrossRef] [PubMed]
- Floris, M.; Olla, S.; Schlessinger, D.; Cucca, F. Genetic-driven druggable target identification and validation. Trends Genet. 2018, 34, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.R.; Tipney, H.; Painter, J.L.; Shen, J.; Nicoletti, P.; Shen, Y.; Floratos, A.; Sham, P.C.; Li, M.J.; Wang, J. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015, 47, 856–860. [Google Scholar] [CrossRef]
- Cohen, J.; Pertsemlidis, A.; Kotowski, I.K.; Graham, R.; Garcia, C.K.; Hobbs, H.H. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 2005, 37, 161–165. [Google Scholar] [CrossRef]
- Sabatine, M.S.; Giugliano, R.P.; Wiviott, S.D.; Raal, F.J.; Blom, D.J.; Robinson, J.; Ballantyne, C.M.; Somaratne, R.; Legg, J.; Wasserman, S.M. Efficacy and safety of evolocumab in reducing lipids and cardiovascular events. N. Engl. J. Med. 2015, 372, 1500–1509. [Google Scholar] [CrossRef] [PubMed]
- Kathiresan, S.; Willer, C.J.; Peloso, G.M.; Demissie, S.; Musunuru, K.; Schadt, E.E.; Kaplan, L.; Bennett, D.; Li, Y.; Tanaka, T. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 2009, 41, 56–65. [Google Scholar] [CrossRef]
- Okada, Y.; Wu, D.; Trynka, G.; Raj, T.; Terao, C.; Ikari, K.; Kochi, Y.; Ohmura, K.; Suzuki, A.; Yoshida, S. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 2014, 506, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; He, T.; Fisher, G.J.; Voorhees, J.J.; Quan, T. Molecular basis of retinol anti-ageing properties in naturally aged human skin in vivo. Int. J. Cosmet. Sci. 2017, 39, 56–65. [Google Scholar] [CrossRef]
- Chen, S.; Ostrowski, J.; Whiting, G.; Roalsvig, T.; Hammer, L.; Currier, S.J.; Honeyman, J.; Kwasniewski, B.; Yu, K.-L.; Sterzycki, R. Retinoic acid receptor gamma mediates topical retinoid efficacy and irritation in animal models. J. Investig. Dermatol. 1995, 104, 779–783. [Google Scholar] [CrossRef][Green Version]
- Yin, S.; Luo, J.; Qian, A.; Du, J.; Yang, Q.; Zhou, S.; Yu, W.; Du, G.; Clark, R.B.; Walters, E.T. Retinoids activate the irritant receptor TRPV1 and produce sensory hypersensitivity. J. Clin. Investig. 2013, 123, 3941–3951. [Google Scholar] [CrossRef]
- Zheng, Y.; Che, D.; Peng, B.; Hao, Y.; Zhang, X.; He, L.; Geng, S. All-trans-retinoic acid activated mast cells via Mas-related G-protein-coupled receptor-X2 in retinoid dermatitis. Contact Dermat. 2019, 81, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Frosch, P.J.; Kligman, A.M. The soap chamber test: A new method for assessing the irritancy of soaps. J. Am. Acad. Dermatol. 1979, 1, 35–41. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, s13742-015-0047-8. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Janssens, A.C.J. Validity of polygenic risk scores: Are we measuring what we think we are? Hum. Mol. Genet. 2019, 28, R143–R150. [Google Scholar] [CrossRef]
- Mondul, A.M.; Yu, K.; Wheeler, W.; Zhang, H.; Weinstein, S.J.; Major, J.M.; Cornelis, M.C.; Männistö, S.; Hazra, A.; Hsing, A.W. Genome-wide association study of circulating retinol levels. Hum. Mol. Genet. 2011, 20, 4724–4731. [Google Scholar] [CrossRef]
- Borel, P.; Desmarchelier, C. Genetic variations associated with vitamin A status and vitamin A bioavailability. Nutrients 2017, 9, 246. [Google Scholar] [CrossRef]
- Cottrez, F.; Boitel, E.; Auriault, C.; Aeby, P.; Groux, H. Genes specifically modulated in sensitized skins allow the detection of sensitizers in a reconstructed human skin model. Development of the SENS-IS assay. Toxicol. Vitr. 2015, 29, 787–802. [Google Scholar] [CrossRef] [PubMed]
- Theocharidis, G.; Drymoussi, Z.; Kao, A.P.; Barber, A.H.; Lee, D.A.; Braun, K.M.; Connelly, J.T. Type VI collagen regulates dermal matrix assembly and fibroblast motility. J. Investig. Dermatol. 2016, 136, 74–83. [Google Scholar] [CrossRef]
- Naugle, J.E.; Olson, E.R.; Zhang, X.; Mase, S.E.; Pilati, C.F.; Maron, M.B.; Folkesson, H.G.; Horne, W.I.; Doane, K.J.; Meszaros, J.G. Type VI collagen induces cardiac myofibroblast differentiation: Implications for postinfarction remodeling. Am. J. Physiol.-Heart Circ. Physiol. 2006, 290, H323–H330. [Google Scholar] [CrossRef]
- Oono, T.; Specks, U.; Eckes, B.; Majewski, S.; Hunzelmann, N.; Timpl, R.; Krieg, T. Expression of type VI collagen mRNA during wound healing. J. Investig. Dermatol. 1993, 100, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Peltonen, J.; Hsiao, L.L.; Jaakkola, S.; Sollberg, S.; Aumailley, M.; Timpl, R.; Chu, M.-L.; Uitto, J. Activation of collagen gene expression in keloids: Co-localization of type I and VI collagen and transforming growth factor-β1 mRNA. J. Investig. Dermatol. 1991, 97, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Lettmann, S.; Bloch, W.; Maaß, T.; Niehoff, A.; Schulz, J.-N.; Eckes, B.; Eming, S.A.; Bonaldo, P.; Paulsson, M.; Wagener, R. Col6a1 null mice as a model to study skin phenotypes in patients with collagen VI related myopathies: Expression of classical and novel collagen VI variants during wound healing. PLoS ONE 2014, 9, e105686. [Google Scholar] [CrossRef]
- Tran, Q.T.; Kennedy, L.H.; Leon Carrion, S.; Bodreddigari, S.; Goodwin, S.B.; Sutter, C.H.; Sutter, T.R. EGFR regulation of epidermal barrier function. Physiol. Genom. 2012, 44, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Rübsam, M.; Mertz, A.F.; Kubo, A.; Marg, S.; Jüngst, C.; Goranci-Buzhala, G.; Schauss, A.C.; Horsley, V.; Dufresne, E.R.; Moser, M. E-cadherin integrates mechanotransduction and EGFR signaling to control junctional tissue polarization and tight junction positioning. Nat. Commun. 2017, 8, 1250. [Google Scholar] [CrossRef]
- Franzke, C.-W.; Cobzaru, C.; Triantafyllopoulou, A.; Löffek, S.; Horiuchi, K.; Threadgill, D.W.; Kurz, T.; van Rooijen, N.; Bruckner-Tuderman, L.; Blobel, C.P. Epidermal ADAM17 maintains the skin barrier by regulating EGFR ligand–dependent terminal keratinocyte differentiation. J. Exp. Med. 2012, 209, 1105–1119. [Google Scholar] [CrossRef]
- Cheng, X.; Jin, J.; Hu, L.; Shen, D.; Dong, X.-p.; Samie, M.A.; Knoff, J.; Eisinger, B.; Liu, M.-l.; Huang, S.M. TRP channel regulates EGFR signaling in hair morphogenesis and skin barrier formation. Cell 2010, 141, 331–343. [Google Scholar] [CrossRef]
- Sachin, P.; Gadani, S.; Cronk, J.; Norris, G.; Kipnis, J. Interleukin-4: A cytokine to remember. J. Immunol. 2012, 189, 4213–4421. [Google Scholar]
- Bitton, A.; Avlas, S.; Reichman, H.; Itan, M.; Karo-Atar, D.; Azouz, N.P.; Rozenberg, P.; Diesendruck, Y.; Nahary, L.; Rothenberg, M.E. A key role for IL-13 signaling via the type 2 IL-4 receptor in experimental atopic dermatitis. Sci. Immunol. 2020, 5, eaaw2938. [Google Scholar] [CrossRef]
- Hurdayal, R.; Brombacher, F. Interleukin-4 receptor alpha: From innate to adaptive immunity in murine models of cutaneous leishmaniasis. Front. Immunol. 2017, 8, 1354. [Google Scholar] [CrossRef]
- Lee, J.E.; Chang, J.Y.; Lee, S.E.; Kim, M.Y.; Lee, J.S.; Lee, M.G.; Kim, S.-C. Epidermal hyperplasia and elevated HB-EGF are more prominent in retinoid dermatitis compared with irritant contact dermatitis induced by benzalkonium chloride. Ann. Dermatol. 2010, 22, 290–299. [Google Scholar] [CrossRef]
- Stücker, M.; Hoffmann, M.; Altmeyer, P. Instrumental evaluation of retinoid-induced skin irritation. Ski. Res. Technol. 2002, 8, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Behrens, D.T.; Villone, D.; Koch, M.; Brunner, G.; Sorokin, L.; Robenek, H.; Bruckner-Tuderman, L.; Bruckner, P.; Hansen, U. The epidermal basement membrane is a composite of separate laminin-or collagen IV-containing networks connected by aggregated perlecan, but not by nidogens. J. Biol. Chem. 2012, 287, 18700–18709. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Pan, W.; Wallin, B.; Kivlin, R.; Lu, S.; Cao, C.; Bi, Z.; Wan, Y. Nicotinamide attenuates aquaporin 3 overexpression induced by retinoic acid through inhibition of EGFR/ERK in cultured human skin keratinocytes. Int. J. Mol. Med. 2008, 22, 229–236. [Google Scholar] [PubMed]
- Jung, Y.; Kim, J.-C.; Park, N.-J.; Bong, S.-K.; Lee, S.; Jegal, H.; Jin, L.T.; Kim, S.M.; Kim, Y.K.; Kim, S.-N. Eupatilin, an activator of PPARα, inhibits the development of oxazolone-induced atopic dermatitis symptoms in Balb/c mice. Biochem. Biophys. Res. Commun. 2018, 496, 508–514. [Google Scholar] [CrossRef]
- Eichner, R.; Kahn, M.; Capetola, R.J.; Gendimenico, G.J.; Mezick, J.A. Effects of topical retinoids on cytoskeletal proteins: Implications for retinoid effects on epidermal differentiation. J. Investig. Dermatol. 1992, 98, 154–161. [Google Scholar] [CrossRef]
- Abdel-Aziz, H.; Wadie, W.; Abdallah, D.M.; Lentzen, G.; Khayyal, M.T. Novel effects of ectoine, a bacteria-derived natural tetrahydropyrimidine, in experimental colitis. Phytomedicine 2013, 20, 585–591. [Google Scholar] [CrossRef]
- Unfried, K.; Kroker, M.; Autengruber, A.; Gotić, M.; Sydlik, U. The compatible solute ectoine reduces the exacerbating effect of environmental model particles on the immune response of the airways. J. Allergy 2014, 2014. [Google Scholar] [CrossRef]
- Bethlehem, L.; van Echten-Deckert, G. Ectoines as novel anti-inflammatory and tissue protective lead compounds with special focus on inflammatory bowel disease and lung inflammation. Pharmacol. Res. 2020, 164, 105389. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Kim, Y.K.; Kim, K.H.; Park, S.J.; Kim, S.J.; Chung, J.H. A novel role for the TRPV1 channel in UV-induced matrix metalloproteinase (MMP)-1 expression in HaCaT cells. J. Cell. Physiol. 2009, 219, 766–775. [Google Scholar] [CrossRef]
- Lee, Y.M.; Kim, Y.K.; Chung, J.H. Increased expression of TRPV1 channel in intrinsically aged and photoaged human skin in vivo. Exp. Dermatol. 2009, 18, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S. Role of Nitric Oxide in the TRPV1-mediated Melanogenesis. Yakhak Hoeji 2020, 64, 387–393. [Google Scholar] [CrossRef]
- Denda, M.; Sokabe, T.; Fukumi-Tominaga, T.; Tominaga, M. Effects of skin surface temperature on epidermal permeability barrier homeostasis. J. Investig. Dermatol. 2007, 127, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Lee, D.H.; Kim, Y.K.; Eun, H.C.; Chung, J.H. Adiponectin deficiency contributes to sensitivity in human skin. J. Investig. Dermatol. 2015, 135, 2331. [Google Scholar] [CrossRef]
- Effendy, I.; Kwangsukstith, C.; Lee, J.; Maibach, H. Functional changes in human stratum corneum induced by topical glycolic acid: Comparison with all-trans retinoic acid. Acta Derm.-Venereol. 1995, 75, 455–458. [Google Scholar]
- Wong, B.J.; Hollowed, C.G. Current concepts of active vasodilation in human skin. Temperature 2017, 4, 41–59. [Google Scholar] [CrossRef]
- Ale, S.; Laugier, J.P.; Maibach, H. Differential irritant skin responses to tandem application of topical retinoic acid and sodium lauryl sulphate: II. Effect of time between first and second exposure. Br. J. Dermatol. 1997, 137, 226–233. [Google Scholar] [CrossRef]
- Patruno, C.; Napolitano, M.; Balato, N.; Ayala, F.; Megna, M.; Patrì, A.; Cirillo, T.; Balato, A. Psoriasis and skin pain: Instrumental and biological evaluations. Acta Derm.-Venereol. 2015, 95, 432–438. [Google Scholar] [CrossRef]
- Andrew, D.; Greenspan, J.D. Mechanical and heat sensitization of cutaneous nociceptors after peripheral inflammation in the rat. J. Neurophysiol. 1999, 82, 2649–2656. [Google Scholar] [CrossRef]
- Ro, J.Y.; Lee, J.-S.; Zhang, Y. Activation of TRPV1 and TRPA1 leads to muscle nociception and mechanical hyperalgesia. Pain 2009, 144, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Arendt-Nielsen, L.; Harris, S.; Whiteside, G.T.; Hummel, M.; Knappenberger, T.; O’Keefe, S.; Kapil, R.; Kyle, D. A randomized, double-blind, positive-controlled, 3-way cross-over human experimental pain study of a TRPV1 antagonist (V116517) in healthy volunteers and comparison with preclinical profile. Pain 2016, 157, 2057–2067. [Google Scholar] [CrossRef] [PubMed]
- Effendy, I.; Weltfriend, S.; Kwangsukstith, C.; Singh, P.; Maibach, H. Effects of all-trans retinoic acid and sodium lauryl sulphate on the permeability of human skin in vitro. Br. J. Dermatol. 1996, 135, 428–432. [Google Scholar] [CrossRef]
- Marill, J.; Idres, N.; Capron, C.C.; Nguyen, E.; Chabot, G.G. Retinoic acid metabolism and mechanism of action: A review. Current drug metabolism 2003, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Bailly, J.; Crettaz, M.; Schifflers, M.; Marty, J. In vitro metabolism by human skin and fibroblasts of retinol, retinal and retinoic acid. Exp. Dermatol. 1998, 7, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Antille, C.; Tran, C.; Sorg, O.; Saurat, J.-H. Penetration and metabolism of topical retinoids in ex vivo organ-cultured full-thickness human skin explants. Ski. Pharmacol. Physiol. 2004, 17, 124–128. [Google Scholar] [CrossRef] [PubMed]

| SNP | Gene | Chr:Position | Allele 1 | Position | MAF | Effect Size 2 | p-Value |
|---|---|---|---|---|---|---|---|
| rs6550923 | RARB | 3:24924495 | A > G | Intron | 0.368 | 0.52 | 0.0267 |
| rs59964722 | RARB | 3:24950518 | A > G | Intron | 0.108 | 0.27 | 0.0109 |
| rs10510553 | RARB | 3:24966768 | A > G | Intron | 0.221 | 2.27 | 0.0159 |
| rs4280597 | RARB | 3:25053482 | A > G | Intron | 0.255 | 0.51 | 0.0315 |
| rs1021701 | RARB | 3:25058757 | A > G | Intron | 0.255 | 0.51 | 0.0315 |
| rs1604003 | RARB | 3:25068617 | C > A | Intron | 0.123 | 0.36 | 0.0213 |
| rs6804842 | RARB | 3:25106437 | G > A | Intron | 0.304 | 2.15 | 0.0180 |
| rs321526 | RARB | 3:25240188 | C > A | Intron | 0.245 | 2.08 | 0.0378 |
| rs73042351 | RARB | 3:25260568 | A > G | Intron | 0.044 | 4.64 | 0.0420 |
| rs73151296 | RARB | 3:25305813 | A > G | Intron | 0.059 | 4.11 | 0.0302 |
| rs1286733 | RARB | 3:25613372 | A > G | Intron | 0.289 | 0.44 | 0.0107 |
| rs3773439 | RARB | 3:25617775 | A > G | Intron | 0.382 | 0.46 | 0.0089 |
| rs1129055 | CD86 | 3:121838319 | A > G | Exon | 0.373 | 1.79 | 0.0452 |
| rs3117040 | RXRB | 6:33164735 | A > C | Intron | 0.039 | 0.15 | 0.0344 |
| rs6970262 | EGFR | 7:55259763 | G > A | Intron | 0.103 | 2.91 | 0.0330 |
| rs2740762 | EGFR | 7:55261342 | C > A | Intron | 0.064 | 4.68 | 0.0152 |
| rs2293348 | EGFR | 7:55266757 | G > A | Intron | 0.083 | 0.20 | 0.0073 |
| rs996076 | CD44 | 11:35210798 | A > G | Intron | 0.123 | 2.43 | 0.0480 |
| rs2295756 | CD44 | 11:35241229 | G > A | Intron | 0.353 | 1.77 | 0.0499 |
| rs10128586 | CD44 | 11:35245907 | A > G | Intron | 0.221 | 0.50 | 0.0490 |
| rs17099562 | MMP10 | 11:102648527 | G > A | Intron | 0.177 | 0.42 | 0.0290 |
| rs5744247 | IL18 | 11:112026156 | G > C | Intron | 0.348 | 0.49 | 0.0136 |
| rs187238 | IL18 | 11: 112034988 | C > G | Intergenic | 0.123 | 2.43 | 0.0480 |
| rs1110470 | IL4R | 16:27336427 | G > A | Intron | 0.275 | 2.32 | 0.0039 |
| rs3024530 | IL4R | 16:27350687 | G > A | Intron | 0.417 | 2.34 | 0.0028 |
| rs12454712 | BCL2 | 18:60845884 | A > G | Intron | 0.480 | 0.58 | 0.0362 |
| rs8098848 | BCL2 | 18:60857793 | A > G | Intron | 0.446 | 0.45 | 0.0086 |
| rs2062010 | BCL2 | 18:60873082 | G > A | Intron | 0.431 | 0.53 | 0.0308 |
| rs17841945 | BCL2 | 18:60881555 | G > A | Intron | 0.078 | 3.06 | 0.0464 |
| rs117668143 | COL6A2 | 21:47551909 | G > A | Exon | 0.049 | 12.79 | 0.0020 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, S.; Kim, K.; Jun, S.-H.; Lee, S.; Kim, J.; Shin, J.-G.; Kim, Y.; Kim, M.; Park, S.-G.; Kang, N.-G. Anti-Irritant Strategy against Retinol Based on the Genetic Analysis of Korean Population: A Genetically Guided Top–Down Approach. Pharmaceutics 2021, 13, 2006. https://doi.org/10.3390/pharmaceutics13122006
Kang S, Kim K, Jun S-H, Lee S, Kim J, Shin J-G, Kim Y, Kim M, Park S-G, Kang N-G. Anti-Irritant Strategy against Retinol Based on the Genetic Analysis of Korean Population: A Genetically Guided Top–Down Approach. Pharmaceutics. 2021; 13(12):2006. https://doi.org/10.3390/pharmaceutics13122006
Chicago/Turabian StyleKang, Seongsu, Kyunghoe Kim, Seung-Hyun Jun, Seonju Lee, Juhyun Kim, Joong-Gon Shin, Yunkwan Kim, Mina Kim, Sun-Gyoo Park, and Nae-Gyu Kang. 2021. "Anti-Irritant Strategy against Retinol Based on the Genetic Analysis of Korean Population: A Genetically Guided Top–Down Approach" Pharmaceutics 13, no. 12: 2006. https://doi.org/10.3390/pharmaceutics13122006
APA StyleKang, S., Kim, K., Jun, S.-H., Lee, S., Kim, J., Shin, J.-G., Kim, Y., Kim, M., Park, S.-G., & Kang, N.-G. (2021). Anti-Irritant Strategy against Retinol Based on the Genetic Analysis of Korean Population: A Genetically Guided Top–Down Approach. Pharmaceutics, 13(12), 2006. https://doi.org/10.3390/pharmaceutics13122006