CRISPR/Cas9 Delivery System Engineering for Genome Editing in Therapeutic Applications
Abstract
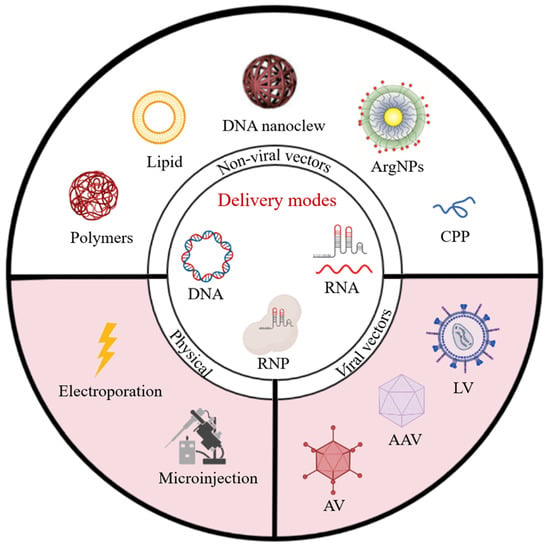
:1. Introduction
2. Clinical CRISPR/Cas9 Delivery Strategies: Physical Import and Viral Vector Transfection
2.1. Clinical Applications of CRISPR-Based Genome Editing
2.2. Physical Import
2.3. Viral Vector Transfection
| Vector Type | Package Limitation | Superiority | Deficiency | References |
|---|---|---|---|---|
| AAV | 4.5 kb | low immunogenicity, serotype-related targeting, stable transgene expression | low packaging capacity | [32] |
| LV | 8 kb | large packaging capacity, low cell cycle tendency | long lasting expression of Cas9 | [33] |
| AV | >8 kb | large packaging capacity, no-integration to host genome | high immunogenicity | [17] |
2.4. The Obstacles of CRISPR/Cas9 Genome Therapy In Vivo
3. Non-Viral Nanovectors for CRISPR/Cas9 Delivery
3.1. CRISPR/Cas9 Plasmid Delivery with Nanovectors for Compression, Nucleus Targeting, and Off-Target Reduction
3.1.1. Delivery Systems for Effective Compression
3.1.2. Delivery Systems for Guiding Plasmids into the Nucleus
3.1.3. Delivery Systems for Reducing the Off-Target Effect by Enhanced Cell Selectivity
3.2. Cas9 mRNA and sgRNA Delivery with Nanovectors for Protection, Compression, and Controllable Release
3.2.1. Delivery Systems for mRNA and sgRNA Encapsulation and Protection
3.2.2. Delivery Systems for RNA Responsive Release
3.3. Cas9 Ribonucleoprotein Delivery with Nanovectors for Package, Controllable Liberation, and Nuclear Localization
3.3.1. Encapsulation and Protection of Cas9 RNP
3.3.2. Bio-Responsive Nanoparticles for Endosomal Escape and Controlled Release

3.3.3. RNP Delivery Designs for Nuclear Localization
4. Concluding Remarks and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [Green Version]
- Mojica, F.J.M.; Diez-Villasenor, C.; Garcia-Martinez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palermo, G.; Chen, J.S.; Ricci, C.G.; Rivalta, I.; Jinek, M.; Batista, V.S.; Doudna, J.A.; McCammon, J.A. Key role of the REC lobe during CRISPR-Cas9 activation by ‘sensing’, ‘regulating’, and ‘locking’ the catalytic HNH domain. Q. Rev. Biophys. 2018, 51, e91. [Google Scholar] [CrossRef] [Green Version]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.B.; Exline, C.M.; DeClercq, J.J.; Llewellyn, G.N.; Hayward, S.B.; Li, P.W.L.; Shivak, D.A.; Surosky, R.T.; Gregory, P.D.; Holmes, M.C.; et al. Homology-driven genome editing in hematopoietic stem and progenitor cells using ZFN mRNA and AAV6 donors. Nat. Biotechnol. 2015, 33, 1256–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.Q.; Fine, E.J.; Wu, X.B.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Platt, R.J.; Chen, S.; Zhou, Y.; Yim, M.J.; Swiech, L.; Kempton, H.R.; Dahlman, J.E.; Parnas, O.; Eisenhaure, T.M.; Jovanovic, M.; et al. CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell 2014, 159, 440–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mali, P.; Aach, J.; Stranges, P.B.; Esvelt, K.M.; Moosburner, M.; Kosuri, S.; Yang, L.H.; Church, G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A.T to G.C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.W.; Wang, H.; Yang, H.; Shi, L.; Katz, Y.; Theunissen, T.W.; Rangarajan, S.; Shivalila, C.S.; Dadon, D.B.; Jaenisch, R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013, 23, 1163–1171. [Google Scholar] [CrossRef]
- Schumann, K.; Lin, S.; Boyer, E.; Simeonov, D.R.; Subramaniam, M.; Gate, R.E.; Haliburton, G.E.; Yee, C.J.; Bluestone, J.A.; Doudna, J.A.; et al. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc. Natl. Acad. Sci. USA 2015, 112, 10437–10442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Ryu, S.-M.; Kim, S.-T.; Baek, G.; Kim, D.; Lim, K.; Chung, E.; Kim, S.; Kim, J.-S. Highly efficient RNA-guided base editing in mouse embryos. Nat. Biotechnol. 2017, 35, 435–437. [Google Scholar] [CrossRef]
- Xu, C.F.; Chen, G.J.; Luo, Y.L.; Zhang, Y.; Zhao, G.; Lu, Z.D.; Czarna, A.; Gu, Z.; Wang, J. Rational designs of in vivo CRISPR-Cas delivery systems. Adv. Drug Deliv. Rev. 2021, 168, 3–29. [Google Scholar] [CrossRef]
- Li, L.; Hu, S.; Chen, X. Non-viral delivery systems for CRISPR/Cas9-based genome editing: Challenges and opportunities. Biomaterials 2018, 171, 207–218. [Google Scholar] [CrossRef]
- Wu, Y.X.; Zhou, H.; Fan, X.Y.; Zhang, Y.; Zhang, M.; Wang, Y.H.; Xie, Z.F.; Bai, M.Z.; Yin, Q.; Liang, D.; et al. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015, 25, 67–79. [Google Scholar] [CrossRef] [Green Version]
- Xu, W. Microinjection and Micromanipulation: A Historical Perspective. In Microinjection; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019; Volume 1874, pp. 1–16. [Google Scholar] [CrossRef]
- Gu, B.; Posfai, E.; Rossant, J. Efficient generation of targeted large insertions by microinjection into two-cell-stage mouse embryos. Nat. Biotechnol. 2018, 36, 632–637. [Google Scholar] [CrossRef]
- Bakhtiar, A.; Chowdhury, E.H. PH-responsive strontium nanoparticles for targeted gene therapy against mammary carcinoma cells. Asian J. Pharm. Sci. 2021, 16, 236–252. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Topkar, V.V.; Zheng, Z.; Joung, J.K. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015, 33, 1293–1298. [Google Scholar] [CrossRef] [PubMed]
- Villiger, L.; Grisch-Chan, H.M.; Lindsay, H.; Ringnalda, F.; Pogliano, C.B.; Allegri, G.; Fingerhut, R.; Haberle, J.; Matos, J.; Robinson, M.D.; et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 2018, 24, 1519–1525. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Wu, Y.Y.; Gemberling, M.P.; Oliver, M.L.; Waller, M.A.; Bohning, J.D.; Robinson-Hamm, J.N.; Bulaklak, K.; Rivera, R.M.C.; Collier, J.H.; et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 2019, 25, 427–432. [Google Scholar] [CrossRef]
- Kruzik, A.; Fetahagic, D.; Hartlieb, B.; Dorn, S.; Koppensteiner, H.; Horling, F.M.; Scheiflinger, F.; Reipert, B.M.; de la Rosa, M. Prevalence of Anti-Adeno-Associated Virus Immune Responses in International Cohorts of Healthy Donors. Mol. Ther.-Methods Clin. Dev. 2019, 14, 126–133. [Google Scholar] [CrossRef] [Green Version]
- Merienne, N.; Vachey, G.; de Longprez, L.; Meunier, C.; Zimmer, V.; Perriard, G.; Canales, M.; Mathias, A.; Herrgott, L.; Beltraminelli, T.; et al. The Self-Inactivating KamiCas9 System for the Editing of CNS Disease Genes. Cell Rep. 2017, 20, 2980–2991. [Google Scholar] [CrossRef] [Green Version]
- Lombardo, A.; Cesana, D.; Genovese, P.; Di Stefano, B.; Provasi, E.; Colombo, D.F.; Neri, M.; Magnani, Z.; Cantore, A.; Lo Riso, P.; et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat. Methods 2011, 8, 861–869. [Google Scholar] [CrossRef]
- Sanchez-Hernandez, S.; Gutierrez-Guerrero, A.; Martin-Guerra, R.; Cortijo-Gutierrez, M.; Tristan-Manzano, M.; Rodriguez-Perales, S.; Sanchez, L.; Garcia-Perez, J.L.; Chato-Astrain, J.; Fernandez-Valades, R.; et al. The IS2 Element Improves Transcription Efficiency of Integration-Deficient Lentiviral Vector Episomes. Mol. Nucleic Acids 2018, 13, 16–28. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Park, K.H.; Zhao, L.; Xu, J.; El Refaey, M.; Gao, Y.; Zhu, H.; Ma, J.; Han, R. CRISPR-mediated Genome Editing Restores Dystrophin Expression and Function in mdx Mice. Mol. Ther. 2016, 24, 564–569. [Google Scholar] [CrossRef] [Green Version]
- Koo, T.; Yoon, A.R.; Cho, H.Y.; Bae, S.; Yun, C.O.; Kim, J.S. Selective disruption of an oncogenic mutant allele by CRISPR/Cas9 induces efficient tumor regression. Nucleic Acids Res. 2017, 45, 7897–7908. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Zhang, F.; Gao, G.P. CRISPR-Based Therapeutic Genome Editing: Strategies and In Vivo Delivery by AAV Vectors. Cell 2020, 181, 136–150. [Google Scholar] [CrossRef]
- Wilbie, D.; Walther, J.; Mastrobattista, E. Delivery Aspects of CRISPR/Cas for in Vivo Genome Editing. Acc. Chem. Res. 2019, 52, 1555–1564. [Google Scholar] [CrossRef] [Green Version]
- Branden, L.J.; Mohamed, A.J.; Smith, C.I.E. A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat. Biotechnol. 1999, 17, 784–787. [Google Scholar] [CrossRef]
- Chen, F.; Ding, X.; Feng, Y.; Seebeck, T.; Jiang, Y.; Davis, G.D. Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat. Commun. 2017, 8, 14958. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Wang, P.; Feng, Q.; Wang, N.; Chen, Z.; Huang, Y.; Zheng, W.; Jiang, X. Lipid nanoparticle-mediated efficient delivery of CRISPR/Cas9 for tumor therapy. NPG Asia Mater. 2017, 9, e441. [Google Scholar] [CrossRef]
- Bai, J.; Duan, J.L.; Liu, R.; Du, Y.F.; Luo, Q.; Cui, Y.N.; Su, Z.B.; Xu, J.R.; Xie, Y.; Lu, W.L. Engineered targeting tLyp-1 exosomes as gene therapy vectors for efficient delivery of siRNA into lung cancer cells. Asian J. Pharm. Sci. 2020, 15, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Yang, Y.; Oh, S.J.; Hong, Y.; Seo, M.; Jang, M. Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J. Control Release 2017, 266, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wu, J.; Gu, W.; Huang, Y.; Tong, Z.; Huang, L.; Tan, J. Exosome-Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs. Adv. Sci. 2018, 5, 1700611. [Google Scholar] [CrossRef]
- Li, L.; Song, L.; Liu, X.; Yang, X.; Li, X.; He, T.; Wang, N.; Yang, S.; Yu, C.; Yin, T.; et al. Artificial Virus Delivers CRISPR-Cas9 System for Genome Editing of Cells in Mice. ACS Nano 2017, 11, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhang, L.; Zheng, W.; Cong, L.; Guo, Z.; Xie, Y.; Wang, L.; Tang, R.; Feng, Q.; Hamada, Y.; et al. Thermo-triggered Release of CRISPR-Cas9 System by Lipid-Encapsulated Gold Nanoparticles for Tumor Therapy. Angew. Chem. Int. Ed. Engl. 2018, 57, 1491–1496. [Google Scholar] [CrossRef]
- Liu, B.Y.; He, X.Y.; Xu, C.; Xu, L.; Ai, S.L.; Cheng, S.X.; Zhuo, R.X. A Dual-Targeting Delivery System for Effective Genome Editing and In Situ Detecting Related Protein Expression in Edited Cells. Biomacromolecules 2018, 19, 2957–2968. [Google Scholar] [CrossRef]
- Luo, Y.L.; Xu, C.F.; Li, H.J.; Cao, Z.T.; Liu, J.; Wang, J.L.; Du, X.J.; Yang, X.Z.; Gu, Z.; Wang, J. Macrophage-Specific in Vivo Gene Editing Using Cationic Lipid-Assisted Polymeric Nanoparticles. ACS Nano 2018, 12, 994–1005. [Google Scholar] [CrossRef]
- Shen, J.; Lu, Z.; Wang, J.; Hao, Q.; Ji, W.; Wu, Y.; Peng, H.; Zhao, R.; Yang, J.; Li, Y.; et al. Traceable Nano-Biohybrid Complexes by One-Step Synthesis as CRISPR-Chem Vectors for Neurodegenerative Diseases Synergistic Treatment. Adv. Mater. 2021, 33, e2101993. [Google Scholar] [CrossRef]
- Vaidyanathan, S.; Azizian, K.T.; Haque, A.K.M.A.; Henderson, J.M.; Hendel, A.; Shore, S.; Antony, J.S.; Hogrefe, R.I.; Kormann, M.S.D.; Porteus, M.H.; et al. Uridine Depletion and Chemical Modification Increase Cas9 mRNA Activity and Reduce Immunogenicity without HPLC Purification. Mol. Ther.-Nucleic Acids 2018, 12, 530–542. [Google Scholar] [CrossRef]
- Anderson, B.R.; Muramatsu, H.; Jha, B.K.; Silverman, R.H.; Weissman, D.; Kariko, K. Nucleoside modifications in RNA limit activation of 2’-5’-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res. 2011, 39, 9329–9338. [Google Scholar] [CrossRef] [Green Version]
- Finn, J.D.; Smith, A.R.; Patel, M.C.; Shaw, L.; Youniss, M.R.; van Heteren, J.; Dirstine, T.; Ciullo, C.; Lescarbeau, R.; Seitzer, J.; et al. A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Rep. 2018, 22, 2227–2235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Q.; Wei, T.; Farbiak, L.; Johnson, L.T.; Dilliard, S.A.; Siegwart, D.J. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR-Cas gene editing. Nat. Nanotechnol. 2020, 15, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Farbiak, L.; Cheng, Q.; Wei, T.; Alvarez-Benedicto, E.; Johnson, L.T.; Lee, S.; Siegwart, D.J. All-In-One Dendrimer-Based Lipid Nanoparticles Enable Precise HDR-Mediated Gene Editing In Vivo. Adv. Mater. 2021, 33, e2006619. [Google Scholar] [CrossRef]
- Usman, W.M.; Pham, T.C.; Kwok, Y.Y.; Vu, L.T.; Ma, V.; Peng, B.; Chan, Y.S.; Wei, L.; Chin, S.M.; Azad, A.; et al. Efficient RNA drug delivery using red blood cell extracellular vesicles. Nat. Commun. 2018, 9, 2359. [Google Scholar] [CrossRef]
- Li, Z.; Zhou, X.; Wei, M.; Gao, X.; Zhao, L.; Shi, R.; Sun, W.; Duan, Y.; Yang, G.; Yuan, L. In Vitro and In Vivo RNA Inhibition by CD9-HuR Functionalized Exosomes Encapsulated with miRNA or CRISPR/dCas9. Nano Lett. 2019, 19, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, S.; Uchida, S.; Toh, K.; Tockary, T.A.; Dirisala, A.; Hayashi, K.; Fukushima, S.; Kataoka, K. Co-encapsulation of Cas9 mRNA and guide RNA in polyplex micelles enables genome editing in mouse brain. J. Control. Release 2021, 332, 260–268. [Google Scholar] [CrossRef]
- Javanmardi, S.; Tamaddon, A.M.; Aghamaali, M.R.; Ghahramani, L.; Abolmaali, S.S. Redox-sensitive, PEG-shielded carboxymethyl PEI nanogels silencing MicroRNA-21, sensitizes resistant ovarian cancer cells to cisplatin. Asian J. Pharm. Sci. 2020, 15, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chang, J.; Jiang, Y.; Meng, X.; Sun, T.; Mao, L.; Xu, Q.; Wang, M. Fast and Efficient CRISPR/Cas9 Genome Editing In Vivo Enabled by Bioreducible Lipid and Messenger RNA Nanoparticles. Adv. Mater. 2019, 31, e1902575. [Google Scholar] [CrossRef]
- Chen, G.; Abdeen, A.A.; Wang, Y.; Shahi, P.K.; Robertson, S.; Xie, R.; Suzuki, M.; Pattnaik, B.R.; Saha, K.; Gong, S. A biodegradable nanocapsule delivers a Cas9 ribonucleoprotein complex for in vivo genome editing. Nat. Nanotechnol. 2019, 14, 974–980. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wan, T.; Wang, H.; Zhang, S.; Ping, Y.; Cheng, Y. A boronic acid-rich dendrimer with robust and unprecedented efficiency for cytosolic protein delivery and CRISPR-Cas9 gene editing. Sci. Adv. 2019, 5, eaaw8922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, E.Y.; Ryu, J.Y.; Lee, H.A.R.; Hong, S.H.; Park, H.S.; Hong, K.S.; Park, S.G.; Kim, H.P.; Yoon, T.J. Lecithin nano-liposomal particle as a CRISPR/Cas9 complex delivery system for treating type 2 diabetes. J. Nanobiotechnol. 2019, 17, 19. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Ji, W.; Hall, J.M.; Hu, Q.; Wang, C.; Beisel, C.L.; Gu, Z. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew. Chem. Int. Ed. 2015, 54, 12029–12033. [Google Scholar] [CrossRef]
- Lee, K.; Conboy, M.; Park, H.M.; Jiang, F.G.; Kim, H.J.; Dewitt, M.A.; Mackley, V.A.; Chang, K.; Rao, A.; Skinner, C.; et al. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017, 1, 889–901. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.; Lee, K.; Panda, S.; Gonzales-Rojas, R.; Chong, A.; Bugay, V.; Park, H.M.; Brenner, R.; Murthy, N.; Lee, H.Y. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2018, 2, 497–507. [Google Scholar] [CrossRef]
- Yin, J.; Wang, Q.; Hou, S.; Bao, L.; Yao, W.; Gao, X. Potent Protein Delivery into Mammalian Cells via a Supercharged Polypeptide. J. Am. Chem. Soc. 2018, 140, 17234–17240. [Google Scholar] [CrossRef]
- Gustafsson, O.; Radler, J.; Roudi, S.; Lehto, T.; Hallbrink, M.; Lehto, T.; Gupta, D.; Andaloussi, S.E.; Nordin, J.Z. Efficient Peptide-Mediated In Vitro Delivery of Cas9 RNP. Pharmaceutics 2021, 13, 878. [Google Scholar] [CrossRef]
- Kang, Y.K.; Lee, J.; Im, S.H.; Lee, J.H.; Jeong, J.; Kim, D.K.; Yang, S.Y.; Jung, K.; Kim, S.G.; Chung, H.J. Cas9 conjugate complex delivering donor DNA for efficient gene editing by homology-directed repair. J. Ind. Eng. Chem. 2021, 102, 241–250. [Google Scholar] [CrossRef]
- Rouet, R.; Thuma, B.A.; Roy, M.D.; Lintner, N.G.; Rubitski, D.M.; Finley, J.E.; Wisniewska, H.M.; Mendonsa, R.; Hirsh, A.; de Onate, L.; et al. Receptor-Mediated Delivery of CRISPR-Cas9 Endonuclease for Cell-Type-Specific Gene Editing. J. Am. Chem. Soc. 2018, 140, 6596–6603. [Google Scholar] [CrossRef]
- Shen, J.; Chen, J.; Ma, J.; Fan, L.; Zhang, X.; Yue, T.; Yan, Y.; Zhang, Y. Enhanced lysosome escape mediated by 1,2-dicarboxylic-cyclohexene anhydride-modified poly-l-lysine dendrimer as a gene delivery system. Asian J. Pharm. Sci. 2020, 15, 759–776. [Google Scholar] [CrossRef]
- Alsaiari, S.K.; Patil, S.; Alyami, M.; Alamoudi, K.O.; Aleisa, F.A.; Merzaban, J.S.; Li, M.; Khashab, N.M. Endosomal Escape and Delivery of CRISPR/Cas9 Genome Editing Machinery Enabled by Nanoscale Zeolitic Imidazolate Framework. J. Am. Chem. Soc. 2018, 140, 143–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Q.; Chen, J.; Yan, J.; Cai, S.; Xiong, H.; Liu, Y.; Peng, D.; Mo, M.; Liu, Z. Tumor microenvironment responsive drug delivery systems. Asian J. Pharm. Sci. 2020, 15, 416–448. [Google Scholar] [CrossRef]
- Guo, J.; Wan, T.; Li, B.; Pan, Q.; Xin, H.; Qiu, Y.; Ping, Y. Rational Design of Poly(disulfide)s as a Universal Platform for Delivery of CRISPR-Cas9 Machineries toward Therapeutic Genome Editing. ACS Cent. Sci. 2021, 7, 990–1000. [Google Scholar] [CrossRef]
- Mout, R.; Ray, M.; Tonga, G.Y.; Lee, Y.-W.; Tay, T.; Sasaki, K.; Rotello, V.M. Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing. ACS Nano 2017, 11, 2452–2458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.M.; Shin, S.C.; Kim, E.E.; Kim, S.H.; Park, K.; Oh, S.J.; Jang, M. Simple In Vivo Gene Editing via Direct Self-Assembly of Cas9 Ribonucleoprotein Complexes for Cancer Treatment. ACS Nano 2018, 12, 7750–7760. [Google Scholar] [CrossRef] [PubMed]








| Target Genes | Condition or Disease | Interventions/ Treatment | Delivery Method | Phase | NCT Identifier |
|---|---|---|---|---|---|
| PD-1 | lymphoma | universal anti-CD19 CAR-T-cells (CB-010) | unknown (ex vivo) | I | NCT04637763 |
| metastatic NSCLC | PD-1 knockout T-cells | electroporation | I | NCT02793856 | |
| hepatocellular carcinoma | PD-1 knockout T-cells | unknown (ex vivo) | I | NCT04417764 | |
| Epstein-Barr virus associated malignancies | PD-1 knockout EBV-CTLs | electroporation | I/II | NCT03044743 | |
| esophageal cancer | PD-1 knockout T-cells | unknown (ex vivo) | II | NCT03081715 | |
| TCR, PD-1 | solid tumor | anti-mesothelin CAR-T-cells | electroporation | I | NCT03545815 |
| TCR, B2M | B-cell leukemia B-cell lymphoma | CAR-T-cells targeting CD19 | electroporation | I/II | NCT03166878 |
| TCR, MHC I | renal cell carcinoma | universal anti-CD70 CAR-T-cells (CTX130) | electroporation | I | NCT04438083 |
| T-cell lymphoma | universal anti-CD70 CAR-T-cells (CTX130) | electroporation | I | NCT04502446 | |
| TRAC, β2M | multiple myeloma | universal anti-BCMA CAR-T-cells (CTX120) | electroporation | I | NCT04244656 |
| B-cell malignancy lymphoma | universal anti-CD19 CAR-T-cells (CTX110) | unknown (ex vivo) | I | NCT04035434 | |
| TRAC, CD52 | lymphoblastic leukemia | CAR-T-cells targeting CD19 | unknown (ex vivo) | I | NCT04557436 |
| CD5 | relapsed/refractory hematopoietic malignancies | anti-CD5 CAR-T-cells (CT125A) | unknown (ex vivo) | I | NCT04767308 |
| CD7 | high risk T-cell malignancies | CD7-specific CAR-T-cells | unknown (ex vivo) | I | NCT03690011 |
| CD19, CD20 or CD22 | B-cell leukemia B-cell lymphoma | universal dual specificity CAR-T-cells | electroporation | I/II | NCT03398967 |
| GISH | gastrointestinal neoplasms | tumor infiltrating lymphocytes | unknown (ex vivo) | I | NCT04426669 |
| HPK1 | leukemia lymphocytic | CD19-specific CAR-T-cells | lentivirus and electroporation | I | NCT04037566 |
| BCL11A | β-thalassemia | CD34+ HSPCs (CTX001) | electroporation | I/II | NCT03655678 |
| sickle cell disease | CD34+ HSPCs (CTX001) | electroporation | I/II | NCT03745287 | |
| β-thalassemia | CD34+ HSPCs (ET-01) | electroporation | I | NCT04925206 | |
| β-globin | sickle cell disease | CD34+ HSPCs (GPH101) | unknown (ex vivo) | I/II | NCT04819841 |
| HbF | sickle cell disease | HPSCs | unknown (ex vivo) | I/II | NCT04774536 |
| TGF-β receptor II | advanced biliary tract cancer | CAR-EGFR T-cells | unknown (ex vivo) | I | NCT04976218 |
| CCR5 | HIV | modified CD34+ HSPCs | unknown (ex vivo) | I | NCT03164135 |
| CEP290 | retinal disease | photoreceptor cells | adeno-associated virus-5 (in vivo) | I/II | NCT03872479 |
| E6, E7 | human papillomavirus-related malignant neoplasm | cervical epithelium | local gel administration (in vivo) | I | NCT03057912 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, H.; Zhang, F.; Ding, Y. CRISPR/Cas9 Delivery System Engineering for Genome Editing in Therapeutic Applications. Pharmaceutics 2021, 13, 1649. https://doi.org/10.3390/pharmaceutics13101649
Cheng H, Zhang F, Ding Y. CRISPR/Cas9 Delivery System Engineering for Genome Editing in Therapeutic Applications. Pharmaceutics. 2021; 13(10):1649. https://doi.org/10.3390/pharmaceutics13101649
Chicago/Turabian StyleCheng, Hao, Feng Zhang, and Yang Ding. 2021. "CRISPR/Cas9 Delivery System Engineering for Genome Editing in Therapeutic Applications" Pharmaceutics 13, no. 10: 1649. https://doi.org/10.3390/pharmaceutics13101649
APA StyleCheng, H., Zhang, F., & Ding, Y. (2021). CRISPR/Cas9 Delivery System Engineering for Genome Editing in Therapeutic Applications. Pharmaceutics, 13(10), 1649. https://doi.org/10.3390/pharmaceutics13101649