A Sequence-Independent Strategy for Amplification and Characterisation of Episomal Badnavirus Sequences Reveals Three Previously Uncharacterised Yam Badnaviruses
Abstract
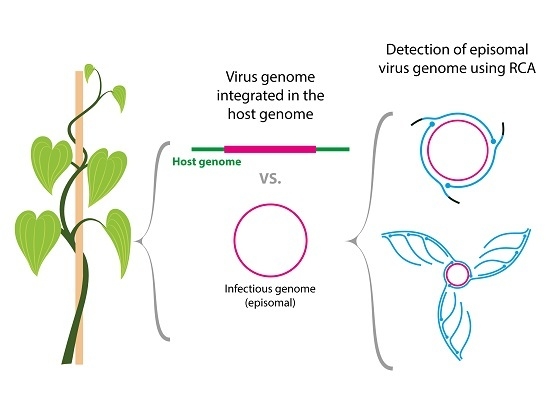
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Total Nucleic Acid Extractions from Yam Leaves and PCR Amplification of Badnavirus Sequences
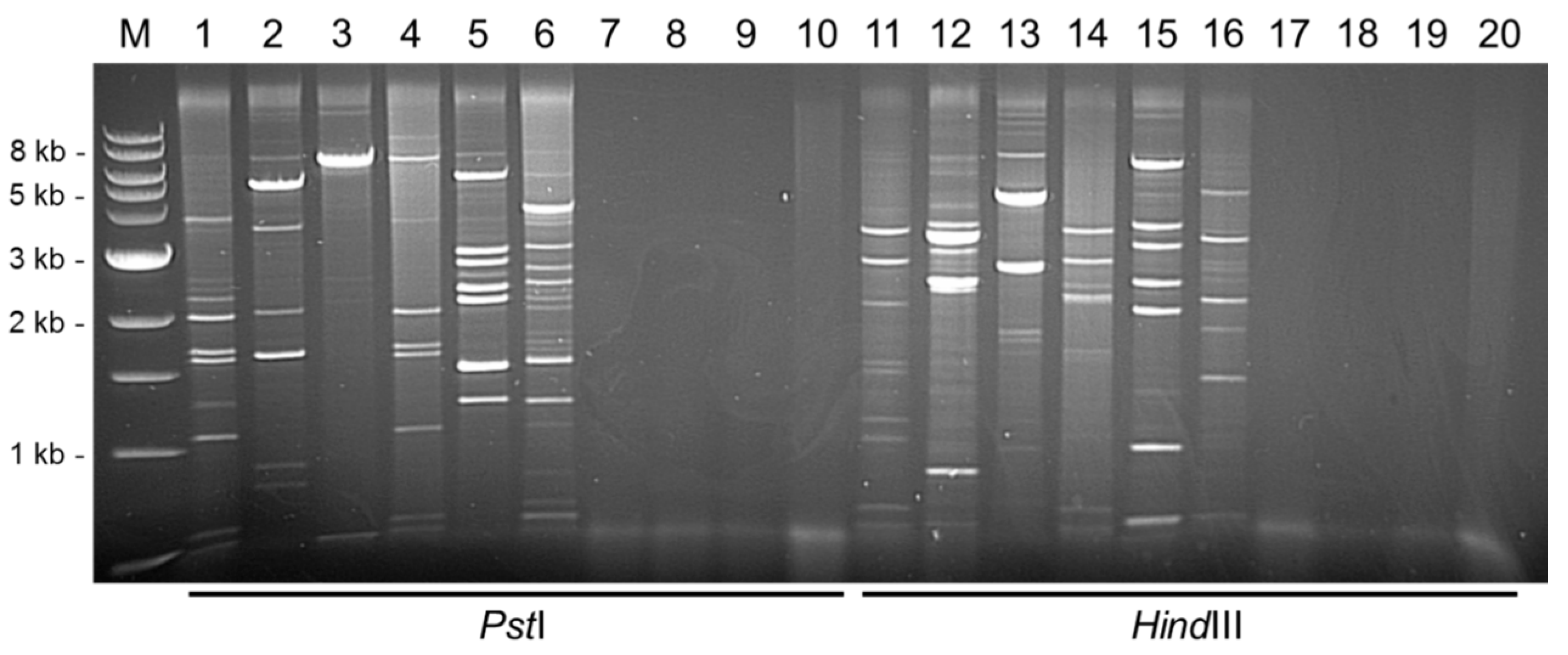
2.3. RCA, Restriction Digestion Analysis, Cloning and Sequencing
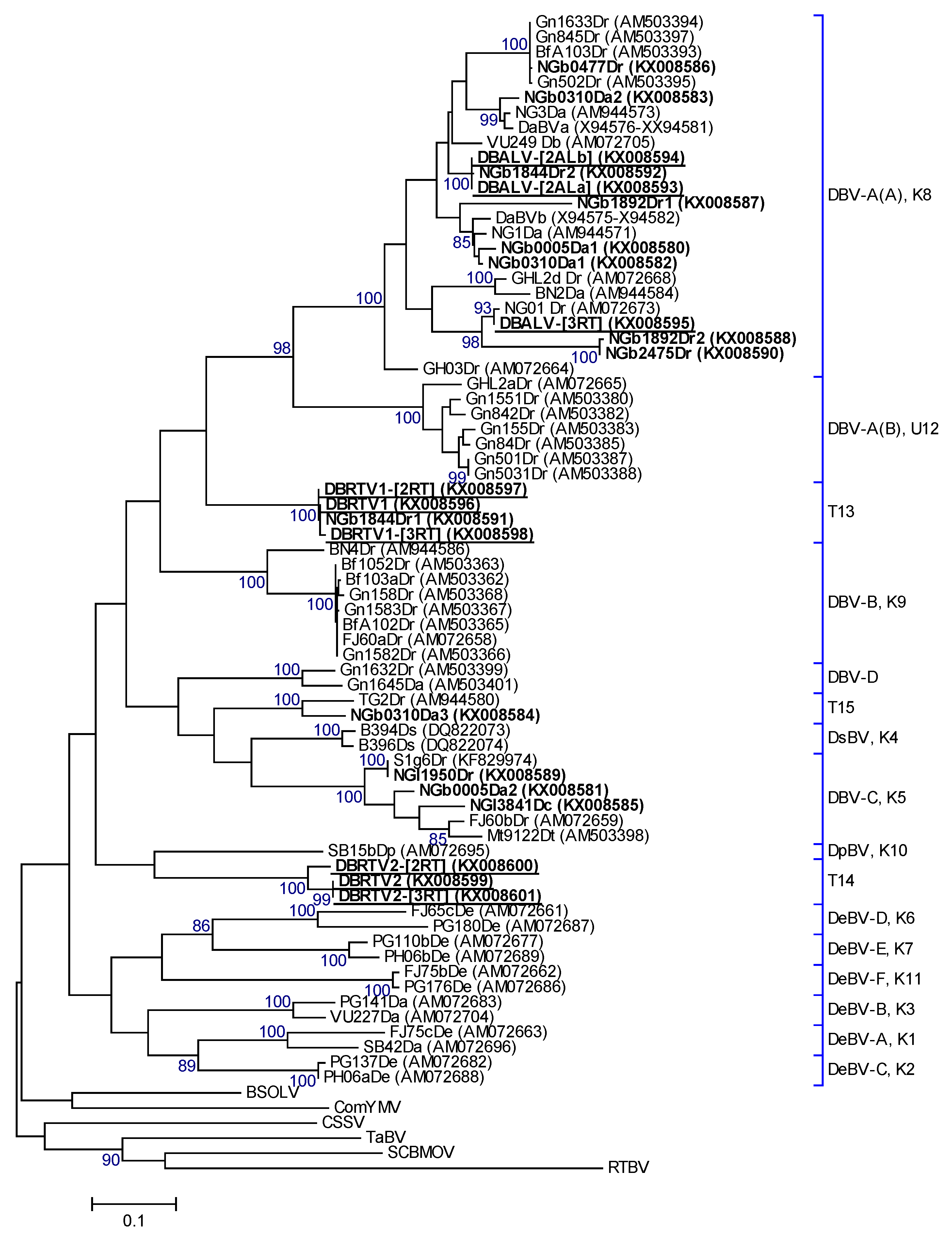
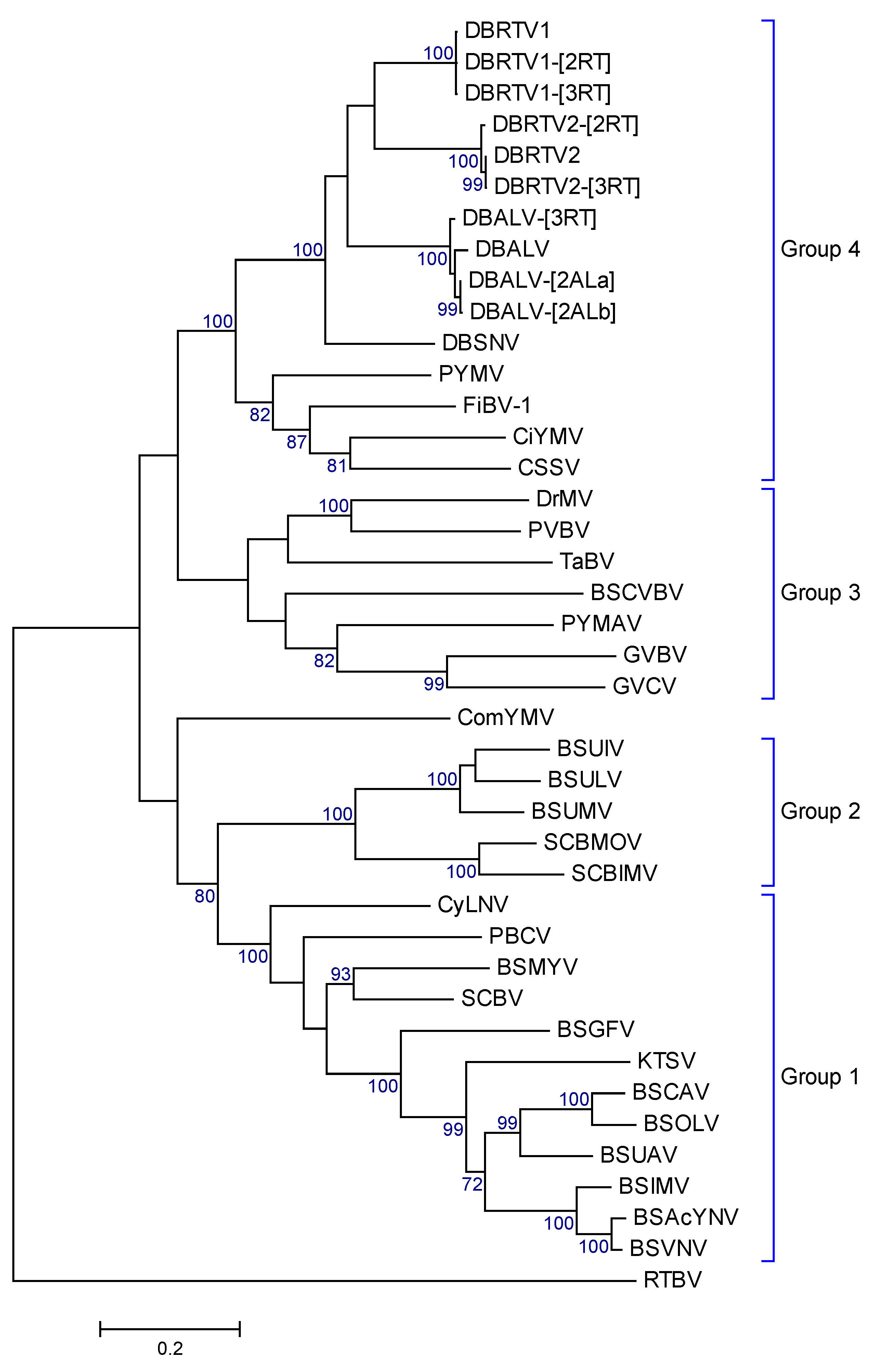
2.4. Sequence Analysis and Phylogeny
3. Results
3.1. Rolling Circle Amplification Combined with Restriction Fragment Length Polymorphism (RCA/RFLP)
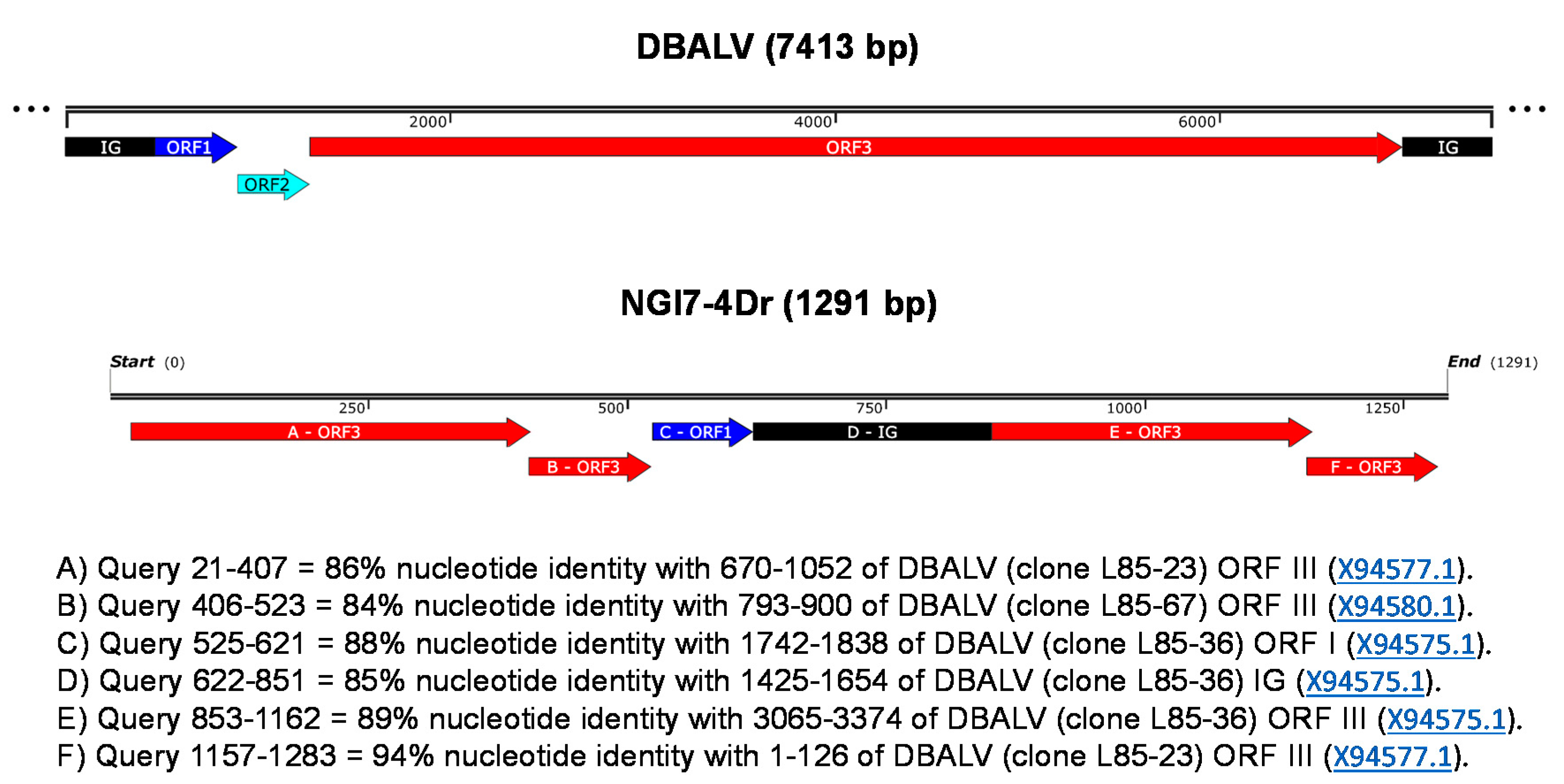
3.2. Analysis of the Partial RT-RNaseH Region from Episomal RCA Sequences
3.3. Complete Genome Characterisations
3.4. Amino Acid Analysis and Phylogenetic Relationships
4. Discussion
4.1. Potential of RCA/RFLP in Yam Badnavirus Diagnostics
4.2. RCA-Captured Badnavirus Diversity
4.3. Full-Length Sequences of Dioscorea Bacilliform Viruses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| aa | amino acid |
| BLAST | basic local alignment search tool |
| BSV | banana streak virus |
| CP | capsid protein |
| CTAB | cetyltrimethylammonium bromide |
| DBALV | Dioscorea bacilliform alata virus |
| DBRTV | Dioscorea bacilliform rotundata (RT) virus |
| DBSNV | Dioscorea bacilliform sansibarensis virus |
| DBV | Dioscorea bacilliform virus |
| DOAJ | Directory of open access journals |
| ds | double stranded |
| eDBVs | endogenous Dioscorea bacilliform viruses |
| EPRV | endogenous pararetrovirus |
| IC-PCR | immunocapture-PCR |
| ICTV | International Committee on Taxonomy of Viruses |
| IG | intergenic region |
| ISEM | immunosorbent electron microscopy |
| kbp | kilo base pairs |
| LD | linear dichroism |
| MAFFT | Multiple Alignment using Fast Fourier Transform |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MP | movement protein |
| NCBI | National Centre for Biotechnology Information |
| ORF | open reading frame |
| PCR | polymerase chain reaction |
| PR | pepsin-like aspartate protease |
| RCA | rolling circle amplification |
| RFLP | restriction fragment length polymorphism |
| RNaseH | ribonuclease H |
| RT | reverse transcriptase |
| RT-PCR | reverse transcription-PCR |
| SDW | sterile distilled deionised water |
| SSA | Sub-Saharan Africa |
| TDa | Dioscorea alata accession |
| TDc | Dioscorea cayenensis accession |
| TDr | Dioscorea rotundata accession |
| TLA | Three letter acronym |
| VAP | virion-associated protein |
| Zn knuckle | zinc-finger domain |
References
- Asiedu, R.; Sartie, A. Crops that feed the World 1. Yams. Food Secur. 2010, 2, 305–315. [Google Scholar] [CrossRef]
- Kenyon, L.; Lebas, B.S.M.; Seal, S.E. Yams (Dioscorea spp.) from the South Pacific Islands contain many novel badnaviruses: Implications for international movement of yam germplasm. Arch. Virol. 2008, 153, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Seal, S.; Turaki, A.; Muller, E.; Kumar, P.L.; Kenyon, L.; Filloux, D.; Galzi, S.; Lopez-Montes, A.; Iskra-Caruana, M.L. The prevalence of badnaviruses in West African yams (Dioscorea cayenensis-rotundata) and evidence of endogenous pararetrovirus sequences in their genomes. Virus Res. 2014, 186, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Phillips, S.; Brunt, A.; Hull, R. Analysis of the sequence of Dioscorea alata bacilliform virus; comparison to other members of the badnavirus group. Virus Genes 1999, 18, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, L.; Shoyinka, S.A.; Hughes, J.D.; Odu, B.O. An overview of viruses infecting Dioscorea yams in sub-Saharan Africa. In 1st Symposium of Plant Virology for Sub-Saharan Africa (PVSSA), IITA, Ibadan, Nigeria, January 2001; Hughes, J.D., Odu, B.O., Eds.; ResearchGate Corporation: San Francisco, CA, USA, 2001; pp. 432–439. [Google Scholar]
- Seal, S.; Muller, E. Molecular analysis of a full-length sequence of a new yam badnavirus from Dioscorea sansibarensis. Arch. Virol. 2007, 152, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Eni, A.O.; Hughes, J.D.; Rey, M.E.C. Survey of the incidence and distribution of five viruses infecting yams in the major yam-producing zones in Benin. Ann. Appl. Biol. 2008, 153, 223–232. [Google Scholar] [CrossRef]
- Eni, A.O.; Hughes, J.D.; Asiedu, R.; Rey, M.E.C. Sequence diversity among badnavirus isolates infecting yam (Dioscorea spp.) in Ghana, Togo, Benin and Nigeria. Arch. Virol. 2008, 153, 2263–2272. [Google Scholar] [CrossRef] [PubMed]
- Bousalem, M.; Durand, O.; Scarcelli, N.; Lebas, B.S.M.; Kenyon, L.; Marchand, J.L.; Lefort, F.; Seal, S.E. Dilemmas caused by endogenous pararetroviruses regarding the taxonomy and diagnosis of yam (Dioscorea spp.) badnaviruses: Analyses to support safe germplasm movement. Arch. Virol. 2009, 154, 297–314. [Google Scholar] [CrossRef] [PubMed]
- Eni, A.O.; Hughes, J.D.; Asiedu, R.; Rey, M.E.C. Survey of the incidence and distribution of viruses infecting yam (Dioscorea spp.) in Ghana and Togo. Ann. Appl. Biol. 2010, 156, 243–251. [Google Scholar] [CrossRef]
- Galzi, S.; Scutt, R.; Prophète, P.; Roumagnac, P.; Filloux, D. Assessment and characterization of the genetic diversity of viruses infecting cultivated yams (Dioscorea spp.) in Haïti. In Rencontres de Virologie Végétale; Marais Armelle, R.F., Ed.; FRA: Aussois, France, 2013; p. 70. [Google Scholar]
- Lima, J.S.; Lima, G.S.A.; Micheref, S.J. Variabilidade genética de isolados de badnavírus infectando inhame (Dioscorea spp.) no nordeste do Brasil. Trop. Plant Pathol. 2013, 38, 349–353. [Google Scholar] [CrossRef]
- Guimarães, K.M.C.; Silva, S.J.C.; Melo, A.M.; Ramos-Sobrinho, R.; Lima, J.S.; Zerbini, F.M.; Assunção, I.P.; Lima, G.S.A. Genetic variability of badnaviruses infecting yam (Dioscorea spp.) in northeastern Brazil. Trop. Plant Pathol. 2015, 40, 111–118. [Google Scholar] [CrossRef]
- Phillips, S.; Briddon, R.W.; Brunt, A.A.; Hull, R. The partial characterization of a badnavirus infecting the Greater Asiatic or water yam (Dioscorea alata). J. Phytopathol. 1999, 147, 265–269. [Google Scholar] [CrossRef]
- Atiri, G.I.; Winter, S.; Alabi, O.J. Virus and Virus-Like Diseases of Major Crops in Developing Countries; Loebenstein, G., Thottappilly, G., Eds.; Springer: Dordrecht, The Netherlands, 2003; pp. 249–268. [Google Scholar]
- Odu, B.O.; Hughes, J.D.A.; Asiedu, R.; Ng, N.Q.; Shoyinka, S.A.; Oladiran, O.A. Responses of white yam (Dioscorea rotundata) cultivars to inoculation with three viruses. Plant Pathol. 2004, 53, 141–147. [Google Scholar] [CrossRef]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. Family—Caulimoviridae. In Virus Taxonomy; Elsevier: San Diego, CA, USA, 2012; pp. 429–443. [Google Scholar]
- Medberry, S.L.; Lockhart, B.E.L.; Olszewski, N.L. Properties of Commelina yellow mottle virus’s complete DNA sequence, genomic discontinuities and transcript suggest that it is a pararetrovirus. Nucleic Acids Res. 1990, 18, 5505–5513. [Google Scholar] [CrossRef] [PubMed]
- Bouhida, M.; Lockhartz, B.E.L.; Olszewski, N.E. An analysis of the complete sequence of a sugarcane bacilliform virus genome infectious to banana and rice. J. Gen. Virol. 1993, 74, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Harper, G.; Hull, R. Cloning and sequence analysis of Banana streak virus. Virus Genes 1998, 17, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Harrison, B.D.; Roberts, I.M. Association of virus-like particles with internal brown spot of yam (Dioscorea alata). Trop. Agric. 1973, 50, 335–340. [Google Scholar]
- Mantell, S.H.; Haque, S.Q. Incidence of internal brown spot disease in white Lisbon yams (Dioscorea alata) during storage. Exp. Agric. 1978, 14, 167–172. [Google Scholar] [CrossRef]
- Asala, S.; Alegbejo, M.D.; Kashina, B.; Banwo, O.O.; Asiedu, R. Distribution and incidence of viruses infecting yam (Dioscorea spp.) in Nigeria. Glob. J. Biotechnol. Biosci. 2012, 1, 163–167. [Google Scholar]
- Toualy, M.N.Y.; Diallo, H.A.; Akinbade, S.A.; Séka, K.; Kumar, P.L. Distribution, incidence and severity of viral diseases of yam (Dioscorea spp.) in Côte d’Ivoire. Afr. J. Biotechnol. 2014, 13, 465–470. [Google Scholar] [CrossRef]
- Lockhart, B.E.L. Purification and serology of a bacilliform virus associated with Banana streak disease. Phytopathology 1986, 80, 995–999. [Google Scholar] [CrossRef]
- Geering, A.D.; McMichael, L.A.; Dietzgen, R.G.; Thomas, J.E. Genetic diversity among Banana streak virus isolates from Australia. Phytopathology 2000, 90, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Harper, G.; Hart, D.; Moult, S.; Hull, R. Banana streak virus is very diverse in Uganda. Virus Res. 2004, 100, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Harper, G.; Hart, D.; Moult, S.; Hull, R.; Geering, A.; Thomas, J. The diversity of Banana streak virus isolates in Uganda. Arch. Virol. 2005, 150, 2407–2420. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.; Dupuy, V.; Blondin, L.; Bauffe, F.; Daugrois, J.H.; Nathalie, L.; Iskra-Caruana, M.L. High molecular variability of sugarcane bacilliform viruses in Guadeloupe implying the existence of at least three new species. Virus Res. 2011, 160, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Umber, M.; Filloux, D.; Muller, E.; Laboureau, N.; Galzi, S.; Roumagnac, P.; Iskra-Caruana, M.L.; Pavis, C.; Teycheney, P.Y.; Seal, S.E. The genome of African yam (Dioscorea cayenensis-rotundata complex) hosts endogenous sequences from four distinct badnavirus species. Mol. Plant Pathol. 2014, 15, 790–801. [Google Scholar] [CrossRef] [PubMed]
- Harper, G.; Dahal, G.; Thottappilly, G.; Hull, R. Detection of episomal banana streak badnavirus by IC-PCR. J. Virol. Methods 1999, 79, 1–8. [Google Scholar] [CrossRef]
- Le Provost, G.; Iskra-Caruana, M.L.; Acina, I.; Teycheney, P.Y. Improved detection of episomal Banana streak viruses by multiplex immunocapture PCR. J. Virol. Methods 2006, 137, 7–13. [Google Scholar] [CrossRef] [PubMed]
- James, A.P.; Geijskes, R.J.; Dale, J.L.; Harding, R.M. Development of a novel rolling-circle amplification technique to detect Banana streak virus that also discriminates between integrated and episomal virus sequences. Plant Dis. 2011, 95, 57–62. [Google Scholar] [CrossRef]
- Laney, A.G.; Hassan, M.; Tzanetakis, I.E. An integrated badnavirus is prevalent in fig germplasm. Phytopathology 2012, 102, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Jaufeerally-Fakim, Y.; Khorugdharry, A.; Harper, G. Genetic variants of Banana streak virus in Mauritius. Virus Res. 2006, 115, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Péréfarres, F.; le Provost, G.; Acina, I.; Lockhart, B.E.L.; Allah Dghim, A.; Iskra-Caruana, M.L.; Candresse, T.; Teycheney, P.Y. Detection, incidence and diversity of banana streak viruses, banana mild mosaic virus and banana virus X in guadeloupe. Acta Hortic. 2009, 828, 205–212. [Google Scholar] [CrossRef]
- Harper, G.; Hart, D.; Moult, S.; Hull, R. Detection of Banana streak virus in field samples of bananas from Uganda. Ann. Appl. Biol. 2002, 141, 247–257. [Google Scholar] [CrossRef]
- Rector, A.; Tachezy, R.; van Ranst, M. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J. Virol. 2004, 78, 4993–4998. [Google Scholar] [CrossRef] [PubMed]
- James, A.P.; Geijskes, R.J.; Dale, J.L.; Harding, R.M. Molecular characterisation of six badnavirus species associated with leaf streak disease of banana in East Africa. Ann. Appl. Biol. 2011, 158, 346–353. [Google Scholar] [CrossRef]
- Paprotka, T.; Boiteux, L.S.; Fonseca, M.E.N.; Resende, R.O.; Jeske, H.; Faria, J.C.; Ribeiro, S.G. Genomic diversity of sweet potato geminiviruses in a Brazilian germplasm bank. Virus Res. 2010, 149, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Homs, M.; Kober, S.; Kepp, G.; Jeske, H. Mitochondrial plasmids of sugar beet amplified via rolling circle method detected during curtovirus screening. Virus Res. 2008, 136, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Wambulwa, M.C. Rolling circle amplification is more sensitive than PCR and serology-based methods in detection of Banana streak virus in musa germplasm. Am. J. Plant Sci. 2012, 3, 1581–1587. [Google Scholar] [CrossRef]
- Mumford, R.A.; Seal, S.E. Rapid single-tube immunocapture RT-PCR for the detection of two yam potyviruses. J. Virol. Methods 1997, 69, 73–79. [Google Scholar] [CrossRef]
- Yang, I.C.; Hafner, G.J.; Revill, P.A.; Dale, J.L.; Harding, R.M. Sequence diversity of South Pacific isolates of Taro bacilliform virus and the development of a PCR-based diagnostic test. Arch. Virol. 2003, 148, 1957–1968. [Google Scholar] [CrossRef] [PubMed]
- Silva, G.; Bömer, M.; Nkere, C.; Lava Kumar, P.; Seal, S.E. Rapid and specific detection of Yam mosaic virus by reverse-transcription recombinase polymerase amplification. J. Virol. Methods 2015, 222, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Vincze, T.; Posfai, J.; Roberts, R.J. NEBcutter: A program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003, 31, 3688–3691. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Mariac, C.; Scarcelli, N.; Pouzadou, J.; Barnaud, A.; Billot, C.; Faye, A.; Kougbeadjo, A.; Maillol, V.; Martin, G.; Sabot, F.; et al. Cost-effective enrichment hybridization capture of chloroplast genomes at deep multiplexing levels for population genetics and phylogeography studies. Mol. Ecol. Resour. 2014, 14, 1103–1113. [Google Scholar] [CrossRef] [PubMed]
- Kalischuk, M.L.; Fusaro, A.F.; Waterhouse, P.M.; Pappu, H.R.; Kawchuk, L.M. Complete genomic sequence of a Rubus yellow net virus isolate and detection of genome-wide pararetrovirus-derived small RNAs. Virus Res. 2013, 178, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Borah, B.K.; Johnson, A.M.A.; Sai Gopal, D.V.R.; Dasgupta, I. Sequencing and computational analysis of complete genome sequences of Citrus yellow mosaic badna virus from acid lime and pummelo. Virus Genes 2009, 39, 137–140. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.C.; Hafner, G.J.; Dale, J.L.; Harding, R.M. Genomic characterisation of taro bacilliform virus. Arch. Virol. 2003, 148, 937–949. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Mock, R.; Kinard, G.; Li, R. Molecular analysis of the complete genomic sequences of four isolates of Gooseberry vein banding associated virus. Virus Genes 2011, 43, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Hagen, L.S.; Jacquemond, M.; Lepingle, A.; Lot, H.; Tepfer, M. Nucleotide sequence and genomic organization of cacao swollen shoot virus. Virology 1993, 196, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, X.; Wu, X.; Wang, A.; Wu, X. Characterization of complete genome and small RNA profile of pagoda yellow mosaic associated virus, a novel badnavirus in China. Virus Res. 2014, 188, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Johne, R.; Müller, H.; Rector, A.; van Ranst, M.; Stevens, H. Rolling-circle amplification of viral DNA genomes using phi29 polymerase. Trends Microbiol. 2009, 17, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Haible, D.; Kober, S.; Jeske, H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods 2006, 135, 9–16. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira Ferreira, P.D.T.; Lemos, T.O.; Nagata, T.; Inoue-Nagata, A.K. One-step cloning approach for construction of agroinfectious begomovirus clones. J. Virol. Methods 2008, 147, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-Y.; Lai, Y.-C.; Lin, N.-S.; Hsu, Y.-H.; Tsai, H.-T.; Liao, J.-Y.; Hu, C.-C. A simplified method of constructing infectious clones of begomovirus employing limited restriction enzyme digestion of products of rolling circle amplification. J. Virol. Methods 2008, 147, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Chabannes, M.; Baurens, F.C.; Duroy, P.O.; Bocs, S.; Vernerey, M.S.; Rodier-Goud, M.; Barbe, V.; Gayral, P.; Iskra-Caruana, M.L. Three infectious viral species lying in wait in the banana genome. J. Virol. 2013, 87, 8624–8637. [Google Scholar] [CrossRef] [PubMed]
- Burkill, H.M. The Useful Plants of West Tropical Africa, 2nd ed.; Royal Botanic Gardens: Kew, UK, 1985. [Google Scholar]
- Cheng, C.P.; Lockhart, B.E.; Olszewski, N.E. The ORF I and II proteins of Commelina yellow mottle virus are virion-associated. Virology 1996, 223, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Hohn, T.; Fuetterer, J. The proteins and functions of plant pararetroviruses: Knowns and unknowns. CRC Crit. Rev. Plant Sci. 1997, 16, 133–161. [Google Scholar] [CrossRef]
- Jacquot, E.; Hagen, L.S.; Jacquemond, M.; Yot, P. The open reading frame 2 product of cacao swollen shoot badnavirus is a nucleic acid-binding protein. Virology 1996, 225, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, D.; Burri, L.; Kajava, A.V.; Mougeot, J.L.; Hess, D.; Lustig, A.; Kleemann, G.; Hohn, T. The open reading frame III product of cauliflower mosaic virus forms a tetramer through a N-terminal coiled-coil. J. Biol. Chem. 1998, 273, 29015–29021. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, D.; Stavolone, L.; Meier, E.; Guerra-Peraza, O.; Herzog, E.; Hohn, T. The product of ORF III in cauliflower mosaic virus interacts with the viral coat protein through its C-terminal proline rich domain. Virus Genes 2001, 22, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Stavolone, L.; Herzog, E.; Leclerc, D.; Hohn, T. Tetramerization is a conserved feature of the virion-associated protein in plant pararetroviruses. J. Virol. 2001, 75, 7739–7743. [Google Scholar] [CrossRef] [PubMed]
- IITA, 2012. New Project to Invest US$12 Million to Boost Yam Productivity in Ghana and Nigeria 02/04/2012 Press Release-IITA. Available online: http://www.iita.org/2012-press-releases/-/asset_publisher/CxA7/content/new-project-to-invest-us-12-million-to-boost-yam-productivity-in-ghana-and-nigeria?redirect=/2012-press-releases#.V3pp_E1TFaQ (accessed on 4 June 2016).
- Fredericks, D.N.; Relman, D.A. Sequence-based identification of microbial pathogens: A reconsideration of Koch’s postulates. Clin. Microbiol. Rev. 1996, 9, 18–33. [Google Scholar]


| Plant Accession a | RCA Sequence | Accession | Size (bp) | NCBI Nearest Match | Identity (%) | Species Group e |
|---|---|---|---|---|---|---|
| Adaka (TDr) | DBRTV2-[2RT] d | KX008578 | 7462 | S1un5Dr (KF830000) | 72 | T14 |
| TDa 00/00005 | NGb0005Da1 b | KX008580 | 528 | NG1Da (AM944571) | 96 | K08 |
| NGb0005Da2 b | KX008581 | 528 | S1g6Dr (KF829974) | 92 | K05 | |
| TDa 85/00250 | DBALV-[2ALa] | KX008571 | 7544 | VU249Db (AM072705) | 94 | K08 |
| DBALV-[2ALb] | KX008572 | 7544 | VU249Db (AM072705) | 94 | K08 | |
| TDa 95/00310 | NGb0310Da1 b | KX008582 | 528 | NG1Da (AM944571) | 98 | K08 |
| NGb0310Da2 b | KX008583 | 528 | NG3Da (AM944573) | 97 | K08 | |
| NGb0310Da3 b | KX008584 | 528 | TG2Dr (AM944580) | 90 | T15 | |
| TDc 3841A | NGl3841Dc b | KX008585 | 528 | FJ60bDr (AM072659) | 91 | K05 |
| TDr 04/00219 × TDr 97/00777 | NGb0477Dr b | KX008586 | 528 | BfA103Dc (AM503393) | 99 | K08 |
| TDr 1892 | DBRTV2 d | KX008577 | 7438 | FJ60bDr (AM072659) | 72 | T14 |
| NGb1892Dr1 b | KX008587 | 528 | NG1Da (AM944571) | 88 | K08 | |
| NGb1892Dr2 b | KX008588 | 528 | NG01Dr (AM072673) | 86 | K08 | |
| TDr 1892B | DBRTV1-[2RT] c | KX008575 | 7707 | S2f8Dr (KF829993) | 77 | T13 |
| DBRTV2-[3RT] d | KX008579 | 7438 | FJ60bDr (AM072659) | 72 | T14 | |
| DBALV-[3RT] c | KX008573 | 7609 | NG01Dr (AM072673) | 99 | K08 | |
| TDr 1950B | NGl1950Dr b | KX008589 | 528 | S1g6Dr (KF829974) | 100 | K05 |
| TDr 89/02475 | DBRTV1 | KX008574 | 7702 | S2f8Dr (KF829993) | 76 | T13 |
| NGb2475Dr b | KX008590 | 528 | NG01Dr (AM072673) | 86 | K08 | |
| TDr 89/02475A | DBRTV1-[3RT] d | KX008576 | 7708 | S2f8Dr (KF829993) | 76 | T13 |
| TDr 95/18544 | NGb1844Dr1 b | KX008591 | 528 | S2f8Dr (KF829993) | 76 | T13 |
| NGb1844Dr2 b | KX008592 | 528 | VU249Db (AM072705) | 94 | K08 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bömer, M.; Turaki, A.A.; Silva, G.; Kumar, P.L.; Seal, S.E. A Sequence-Independent Strategy for Amplification and Characterisation of Episomal Badnavirus Sequences Reveals Three Previously Uncharacterised Yam Badnaviruses. Viruses 2016, 8, 188. https://doi.org/10.3390/v8070188
Bömer M, Turaki AA, Silva G, Kumar PL, Seal SE. A Sequence-Independent Strategy for Amplification and Characterisation of Episomal Badnavirus Sequences Reveals Three Previously Uncharacterised Yam Badnaviruses. Viruses. 2016; 8(7):188. https://doi.org/10.3390/v8070188
Chicago/Turabian StyleBömer, Moritz, Aliyu A. Turaki, Gonçalo Silva, P. Lava Kumar, and Susan E. Seal. 2016. "A Sequence-Independent Strategy for Amplification and Characterisation of Episomal Badnavirus Sequences Reveals Three Previously Uncharacterised Yam Badnaviruses" Viruses 8, no. 7: 188. https://doi.org/10.3390/v8070188
APA StyleBömer, M., Turaki, A. A., Silva, G., Kumar, P. L., & Seal, S. E. (2016). A Sequence-Independent Strategy for Amplification and Characterisation of Episomal Badnavirus Sequences Reveals Three Previously Uncharacterised Yam Badnaviruses. Viruses, 8(7), 188. https://doi.org/10.3390/v8070188