A Toolbox for Herpesvirus miRNA Research: Construction of a Complete Set of KSHV miRNA Deletion Mutants
Abstract
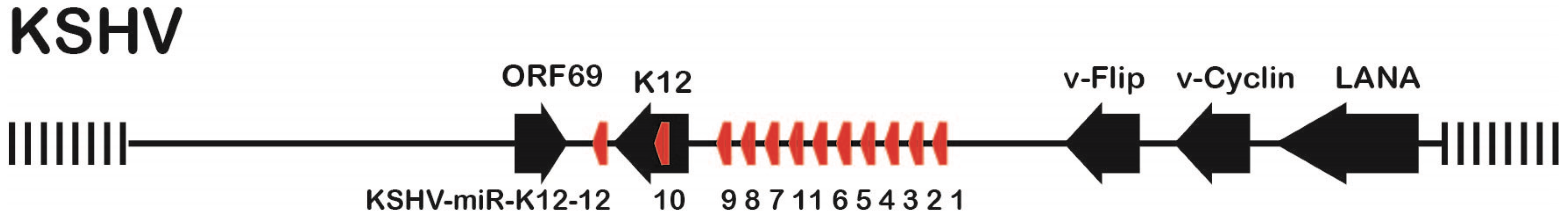
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
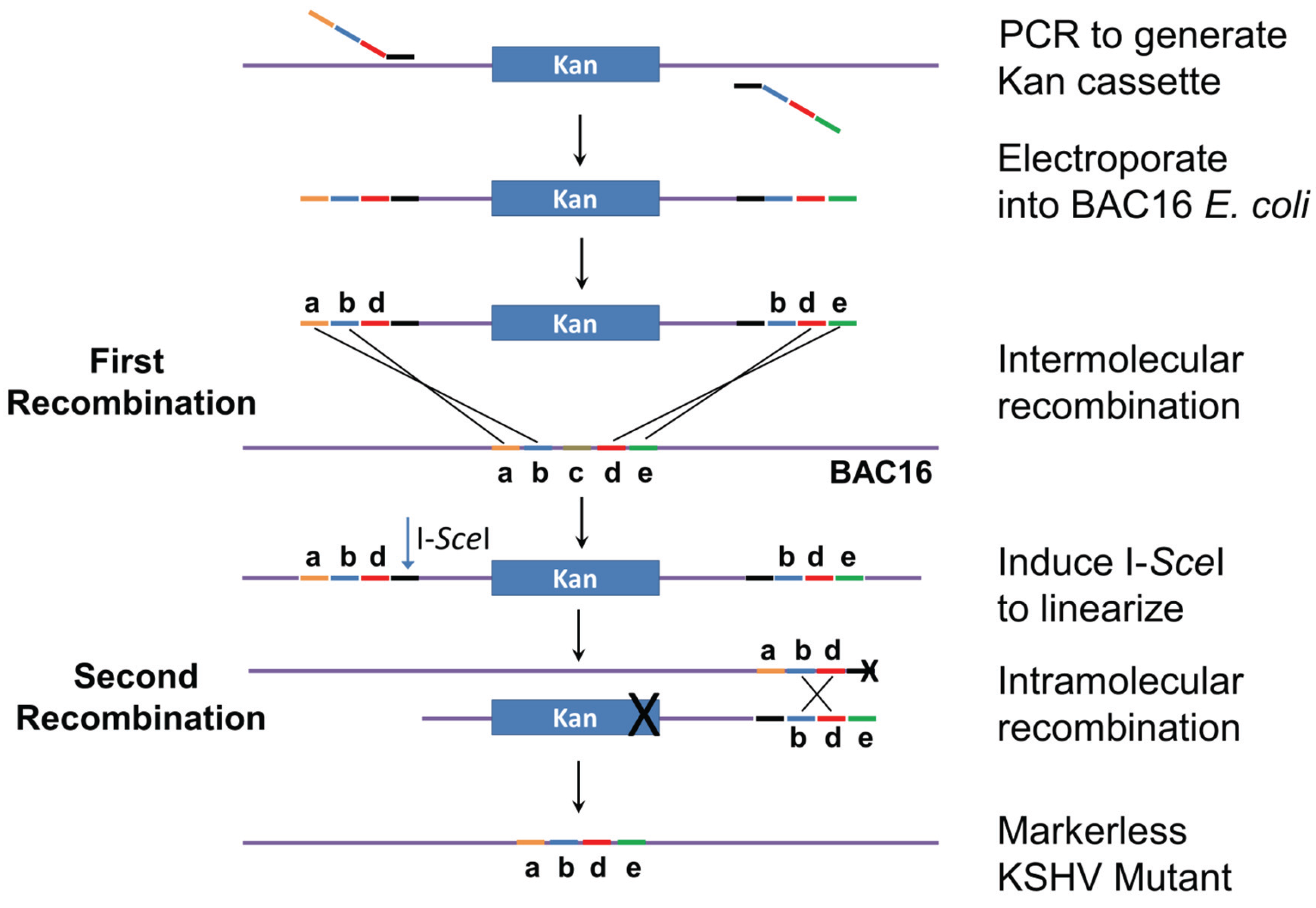
2.2. Mutagenesis Strategy
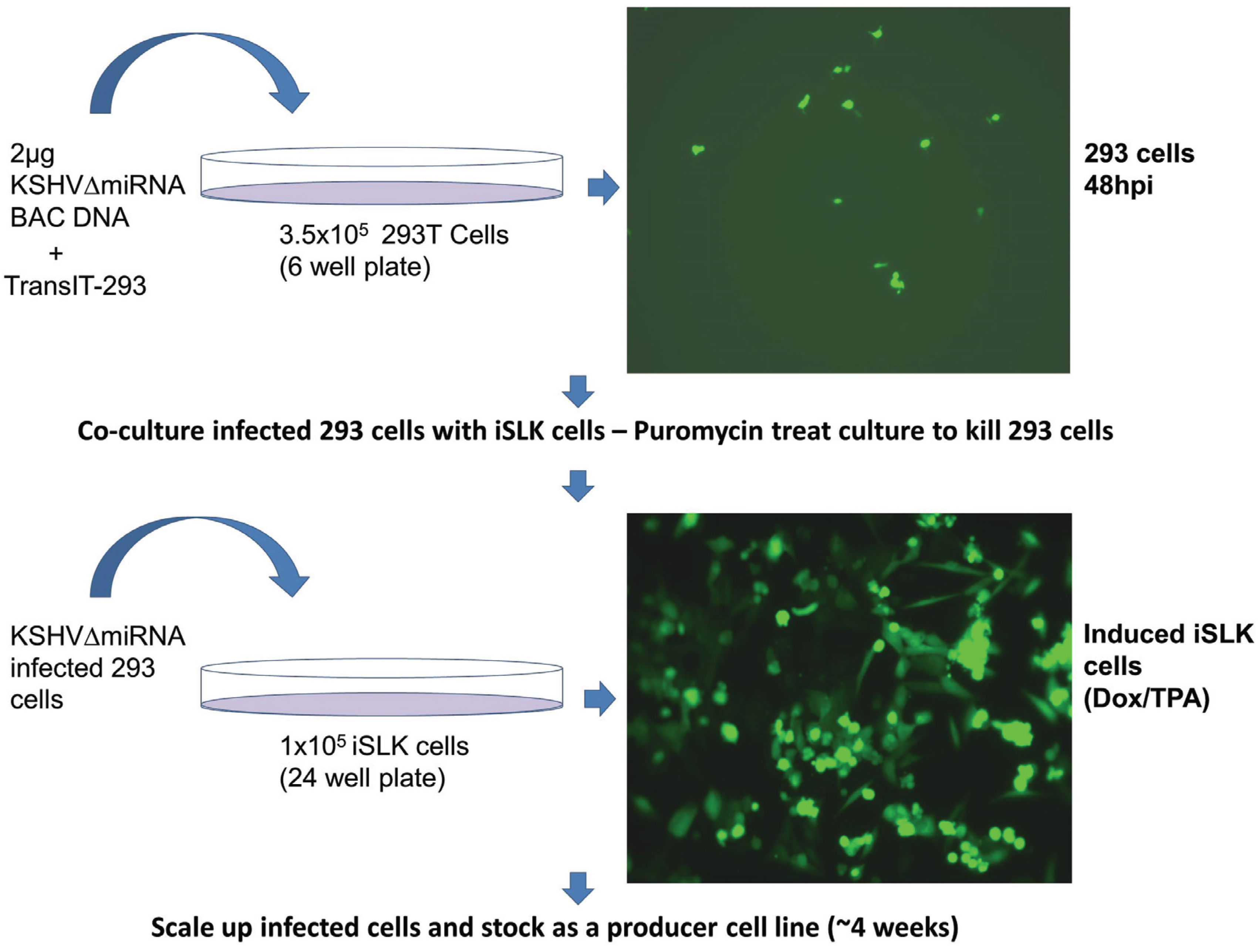
2.3. Co-Culture Infection of iSLK Cells
2.4. Isolation and Quantification of Recombinant Virus from iSLK Cells
2.5. TaqMan Assays
2.6. Illumina Sequencing and Enrichment of Viral Genomes Using SureSelect System
3. Results and Discussion
3.1. Mutation Strategy
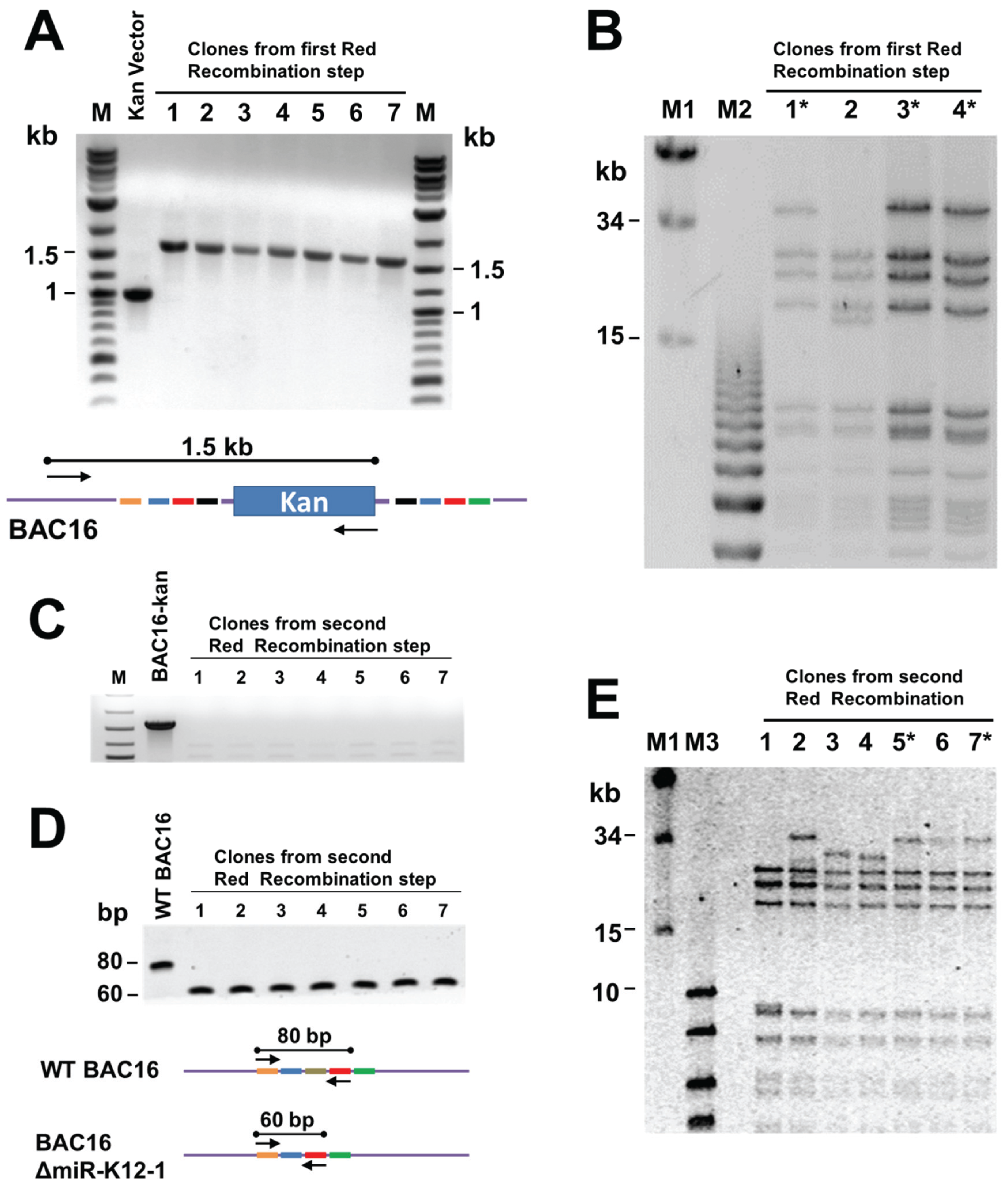
3.2. Generation of Recombinant KSHV Bacmids and Quality Control
3.3. Generation of iSLK Cells Infected with KSHV miRNA Mutants
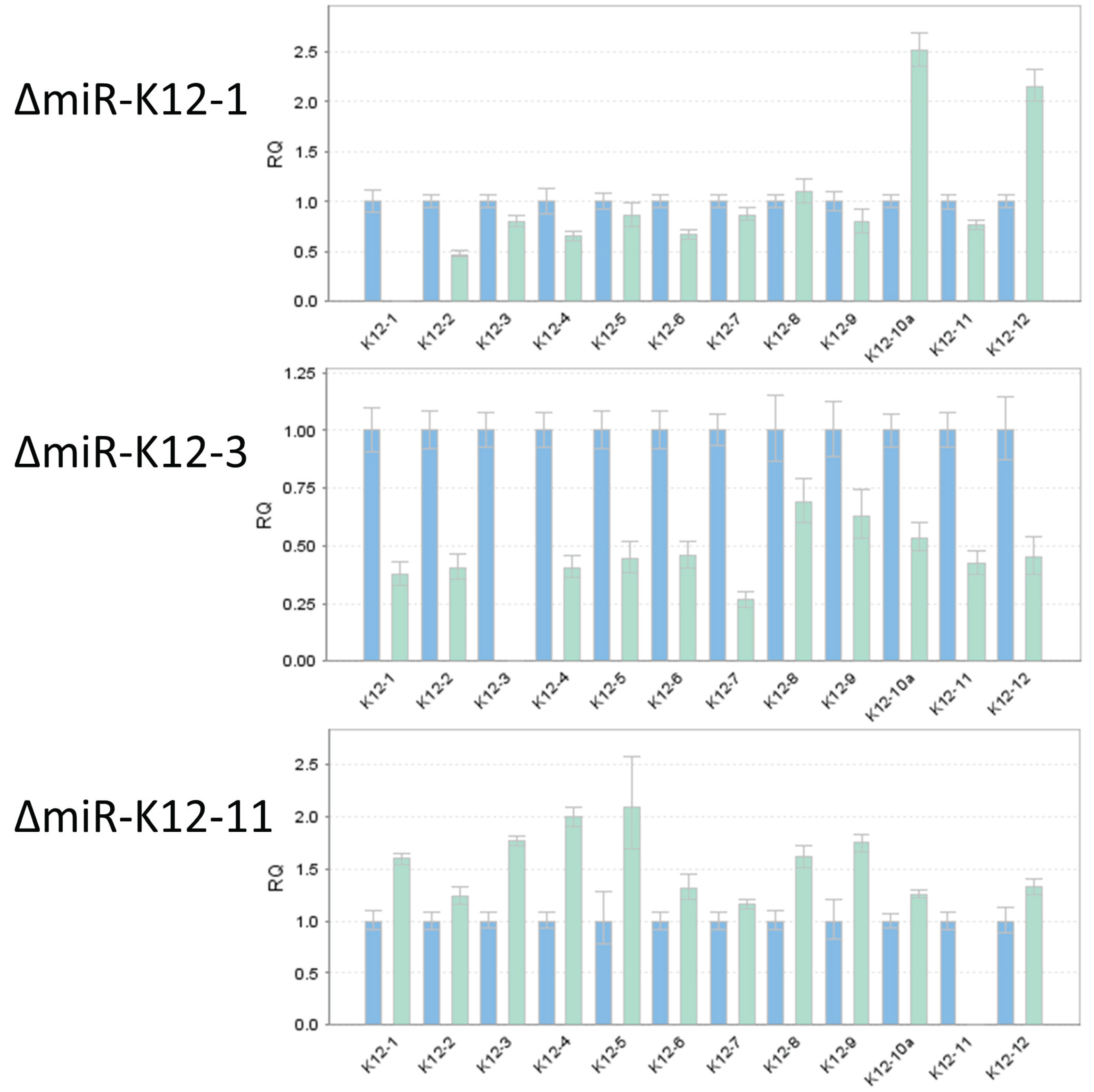
3.4. TaqMan Analysis of miRNA Mutants Confirms Loss of Expression of Individual miRNAs
3.5. Illumina Sequencing Shows No Unintended Changes in DNA Sequences of Individual Mutants
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cesarman, E.; Chang, Y.; Moore, P.S.; Said, J.W.; Knowles, D.M. Kaposi′s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 1995, 332, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Cesarman, E.; Pessin, M.S.; Lee, F.; Culpepper, J.; Knowles, D.M.; Moore, P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi′s sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef] [PubMed]
- Soulier, J.; Grollet, L.; Oksenhendler, E.; Cacoub, P.; Cazals-Hatem, D.; Babinet, P.; d′Agay, M.F.; Clauvel, J.P.; Raphael, M.; Degos, L.; et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman′s disease. Blood 1995, 86, 1276–1280. [Google Scholar] [PubMed]
- Speck, S.H.; Ganem, D. Viral latency and its regulation: Lessons from the gamma-herpesviruses. Cell Host Microbe 2010, 8, 100–115. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Zavolan, M.; Grasser, F.A.; Chien, M.; Russo, J.J.; Ju, J.; John, B.; Enright, A.J.; Marks, D.; Sander, C.; et al. Identification of virus-encoded microRNAs. Science 2004, 304, 734–736. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Lu, S.; Zhang, Z.; Gonzalez, C.M.; Damania, B.; Cullen, B.R. Kaposi′s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA 2005, 102, 5570–5575. [Google Scholar] [CrossRef] [PubMed]
- Grundhoff, A.; Sullivan, C.S.; Ganem, D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 2006, 12, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.; Sewer, A.; Lagos-Quintana, M.; Sheridan, R.; Sander, C.; Grasser, F.A.; van Dyk, L.F.; Ho, C.K.; Shuman, S.; Chien, M.; et al. Identification of microRNAs of the herpesvirus family. Nat. Methods 2005, 2, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Samols, M.A.; Hu, J.; Skalsky, R.L.; Renne, R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi′s sarcoma-associated herpesvirus. J. Virol. 2005, 79, 9301–9305. [Google Scholar] [CrossRef] [PubMed]
- Umbach, J.L.; Cullen, B.R. In-depth analysis of Kaposi′s sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J. Virol. 2010, 84, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Skalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010, 64, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Haecker, I.; Yang, Y.; Gao, S.-J.; Renne, R. γ-Herpesvirus-encoded miRNAs and their roles in viral biology and pathogenesis. Curr. Opin. Virol. 2013, 3, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Gottwein, E.; Corcoran, D.L.; Mukherjee, N.; Skalsky, R.L.; Hafner, M.; Nusbaum, J.D.; Shamulailatpam, P.; Love, C.L.; Dave, S.S.; Tuschl, T.; et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe 2011, 10, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Haecker, I.; Gay, L.A.; Yang, Y.; Hu, J.; Morse, A.M.; McIntyre, L.; Renne, R. Ago-HITS-CLIP expands understanding of Kaposi’s sarcoma-associated herpesvirus miRNA function in primary effusion lymphomas. PLoS Pathog. 2012, 8, e1002884. [Google Scholar] [CrossRef] [PubMed]
- Haecker, I.; Renne, R. HITS-CLIP and PAR-CLIP advance viral miRNA targetome analysis. Crit. Rev. Eukaryot. Gene Expr. 2014, 24, 101–116. [Google Scholar] [CrossRef] [PubMed]
- Riley, K.J.; Rabinowitz, G.S.; Yario, T.A.; Luna, J.M.; Darnell, R.B.; Steitz, J.A. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012, 31, 2207–2221. [Google Scholar] [CrossRef] [PubMed]
- Brulois, K.; Toth, Z.; Wong, L.-Y.; Feng, P.; Gao, S.-J.; Ensser, A.; Jung, J.U. Kaposi’s sarcoma-associated herpesvirus K3 and K5 ubiquitin E3 ligases have stage-specific immune evasion roles during lytic replication. J. Virol. 2014, 88, 9335–9349. [Google Scholar] [CrossRef] [PubMed]
- Brulois, K.F.; Chang, H.; Lee, A.S.; Ensser, A.; Wong, L.Y.; Toth, Z.; Lee, S.H.; Lee, H.R.; Myoung, J.; Ganem, D.; et al. Construction and manipulation of a new Kaposi′s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol. 2012, 86, 9708–9720. [Google Scholar] [CrossRef] [PubMed]
- Myoung, J.; Ganem, D. Generation of a doxycycline-inducible KSHV producer cell line of endothelial origin: Maintenance of tight latency with efficient reactivation upon induction. J. Virol. Methods 2011, 174, 12–21. [Google Scholar] [CrossRef] [PubMed]
- An, F.Q.; Folarin, H.M.; Compitello, N.; Roth, J.; Gerson, S.L.; McCrae, K.R.; Fakhari, F.D.; Dittmer, D.P.; Renne, R. Long-term-infected telomerase-immortalized endothelial cells: A model for Kaposi’s sarcoma-associated herpesvirus latency in vitro and in vivo. J. Virol. 2006, 80, 4833–4846. [Google Scholar] [CrossRef] [PubMed]
- Tischer, B.K.; Kaufer, B.B. Viral bacterial artificial chromosomes: Generation, mutagenesis, and removal of mini-F sequences. J. Biomed. Biotechnol. 2012. [Google Scholar] [CrossRef] [PubMed]
- Tischer, B.K.; von Einem, J.; Kaufer, B.; Osterrieder, N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 2006, 40, 191–197. [Google Scholar] [PubMed]
- Gandy, S.Z.; Linnstaedt, S.D.; Muralidhar, S.; Cashman, K.A.; Rosenthal, L.J.; Casey, J.L. RNA editing of the human herpesvirus 8 kaposin transcript eliminates its transforming activity and is induced during lytic replication. J. Virol. 2007, 81, 13544–13551. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Yang, Y.; Turner, P.C.; Jain, V.; McIntyre, L.M.; Renne, R. LANA binds to multiple active viral and cellular promoters and associates with the H3K4methyltransferase hSET1 complex. PLoS Pathog. 2014, 10, e1004240. [Google Scholar] [CrossRef] [PubMed]
- Cannon, J.S.; Ciufo, D.; Hawkins, A.L.; Griffin, C.A.; Borowitz, M.J.; Hayward, G.S.; Ambinder, R.F. A new primary effusion lymphoma-derived cell line yields a highly infectious Kaposi′s sarcoma herpesvirus-containing supernatant. J. Virol. 2000, 74, 10187–10193. [Google Scholar] [CrossRef] [PubMed]
- Yakushko, Y.; Hackmann, C.; Gunther, T.; Ruckert, J.; Henke, M.; Koste, L.; Alkharsah, K.; Bohne, J.; Grundhoff, A.; Schulz, T.F.; et al. Kaposi′s sarcoma-associated herpesvirus bacterial artificial chromosome contains a duplication of a long unique-region fragment within the terminal repeat region. J. Virol. 2010, 85, 4612–4617. [Google Scholar] [CrossRef] [PubMed]
- Herndier, B.G.; Werner, A.; Arnstein, P.; Abbey, N.W.; Demartis, F.; Cohen, R.L.; Shuman, M.A.; Levy, J.A. Characterization of a human Kaposi’s sarcoma cell line that induces angiogenic tumors in animals. AIDS 1994, 8, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Sturzl, M.; Gaus, D.; Dirks, W.G.; Ganem, D.; Jochmann, R. Kaposi′s sarcoma-derived cell line SLK is not of endothelial origin, but is a contaminant from a known renal carcinoma cell line. Int. J. Cancer 2013, 132, 1954–1958. [Google Scholar] [CrossRef] [PubMed]
- Contrant, M.; Fender, A.; Chane-Woon-Ming, B.; Randrianjafy, R.; Vivet-Boudou, V.; Richer, D.; Pfeffer, S. Importance of the RNA secondary structure for the relative accumulation of clustered viral microRNAs. Nucleic Acids Res. 2014, 42, 7981–7996. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.J.; Chugh, P.; Wang, L.; Netto, E.M.; Luz, E.; Harrington, W.J.; Dezube, B.J.; Damania, B.; Dittmer, D.P. Pre-micro RNA signatures delineate stages of endothelial cell transformation in Kaposi sarcoma. PLoS Pathog. 2009, 5, e1000389. [Google Scholar] [CrossRef] [PubMed]
- Botto, S.; Totonchy, J.E.; Gustin, J.K.; Moses, A.V. Kaposi sarcoma herpesvirus induces HO-1 during de Novo infection of endothelial cells via viral miRNA-dependent and -independent mechanisms. mBio 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Wang, C.; Li, W.; Lu, W.; Bai, Z.; Qin, D.; Yan, Q.; Zhu, J.; Krueger, B.J.; Renne, R.; Gao, S.J.; Lu, C. A KSHV microRNA directly targets G protein-coupled receptor kinase 2 to promote the migration and invasion of Endothelial cells by inducing CXCR2 and activating AKT signaling. PLoS Pathog. 2015, 11, e1005171. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-S.; Yuan, H.; Jeon, H.; Zhu, Y.; Yoo, S.; Shi, S.; Krueger, B.; Renne, R.; Lu, C.; Jung, J.U.; et al. Human mesenchymal stem cells of diverse origins support KSHV persistent infection, and manifest distinct angiogenic, invasive and transforming phenotypes. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.S.; Jain, V.; Krueger, B.; Marshall, V.; Kim, C.H.; Shisler, J.L.; Whitby, D.; Renne, R. Kaposi′s sarcoma-associated herpesvirus (KSHV) induces the oncogenic miR-17–92 cluster and down-regulates TGF-β signaling. PLoS Pathog. 2015, 11, e1005255. [Google Scholar]
- Kieffer-Kwon, P.; Happel, C.; Uldrick, T.S.; Ramalingam, D.; Ziegelbauer, J.M. KSHV microRNAs repress tropomyosin 1 and increase anchorage-independent growth and Endothelial tube formation. PLoS ONE 2015, 10, e0135560. [Google Scholar] [CrossRef] [PubMed]
- Plaisance-Bonstaff, K.; Choi, H.S.; Beals, T.; Krueger, B.J.; Boss, I.W.; Gay, L.A.; Haecker, I.; Hu, J.; Renne, R. KSHV miRNAs decrease expression of lytic genes in latently infected PEL and endothelial cells by targeting host transcription factors. Viruses 2014, 6, 4005–4023. [Google Scholar] [CrossRef] [PubMed]
- DiMaio, T.A.; Gutierrez, K.D.; Lagunoff, M. Kaposi′s sarcoma-associated herpesvirus downregulates transforming growth factor β2 to promote enhanced stability of capillary-like tube formation. J. Virol. 2014, 88, 14301–14309. [Google Scholar] [CrossRef] [PubMed]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jain, V.; Plaisance-Bonstaff, K.; Sangani, R.; Lanier, C.; Dolce, A.; Hu, J.; Brulois, K.; Haecker, I.; Turner, P.; Renne, R.; et al. A Toolbox for Herpesvirus miRNA Research: Construction of a Complete Set of KSHV miRNA Deletion Mutants. Viruses 2016, 8, 54. https://doi.org/10.3390/v8020054
Jain V, Plaisance-Bonstaff K, Sangani R, Lanier C, Dolce A, Hu J, Brulois K, Haecker I, Turner P, Renne R, et al. A Toolbox for Herpesvirus miRNA Research: Construction of a Complete Set of KSHV miRNA Deletion Mutants. Viruses. 2016; 8(2):54. https://doi.org/10.3390/v8020054
Chicago/Turabian StyleJain, Vaibhav, Karlie Plaisance-Bonstaff, Rajnikumar Sangani, Curtis Lanier, Alexander Dolce, Jianhong Hu, Kevin Brulois, Irina Haecker, Peter Turner, Rolf Renne, and et al. 2016. "A Toolbox for Herpesvirus miRNA Research: Construction of a Complete Set of KSHV miRNA Deletion Mutants" Viruses 8, no. 2: 54. https://doi.org/10.3390/v8020054
APA StyleJain, V., Plaisance-Bonstaff, K., Sangani, R., Lanier, C., Dolce, A., Hu, J., Brulois, K., Haecker, I., Turner, P., Renne, R., & Krueger, B. (2016). A Toolbox for Herpesvirus miRNA Research: Construction of a Complete Set of KSHV miRNA Deletion Mutants. Viruses, 8(2), 54. https://doi.org/10.3390/v8020054






