Origins of the Endogenous and Infectious Laboratory Mouse Gammaretroviruses
Abstract
:1. Introduction
| Type | No. of ERV Copies in C57BL | Ability of XRVs to Infect Mouse and Other Mammalian Cells | Receptor | |||||
|---|---|---|---|---|---|---|---|---|
| Mouse | Mink, Human | Bat, Dog | ||||||
| Laboratory strains | M. m. domesticus | M. m. castaneus | M. m. musculus | |||||
| Ecotropic | 1 | + | + | + | + | - | - | CAT-1 |
| Polytropic | >30 | + | + | - | - | + | - | XPR1 |
| Xenotropic | >20 | - | + | + | + | + | + | XPR1 |
2. Mouse Gammaretrovirus Genome
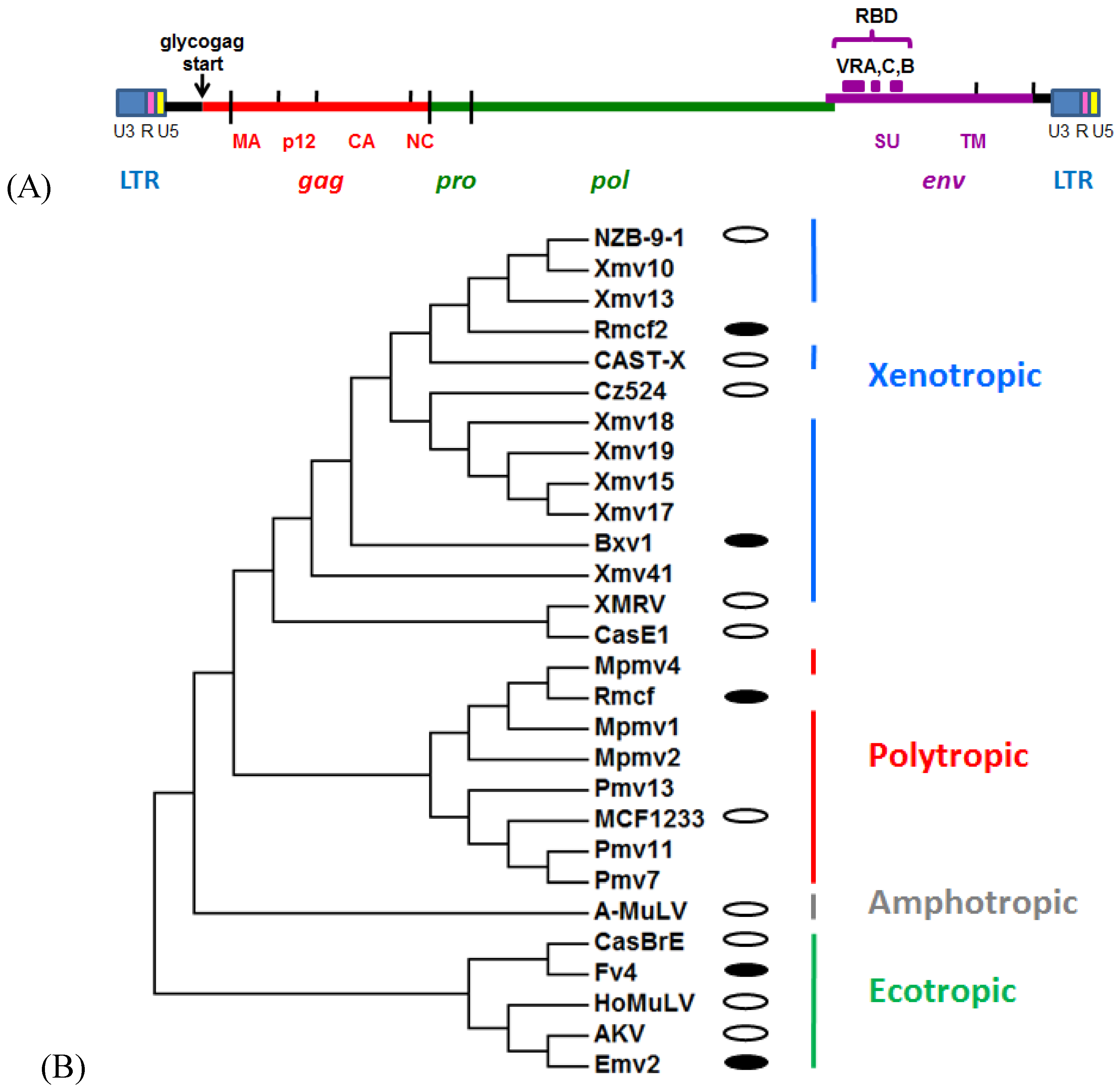
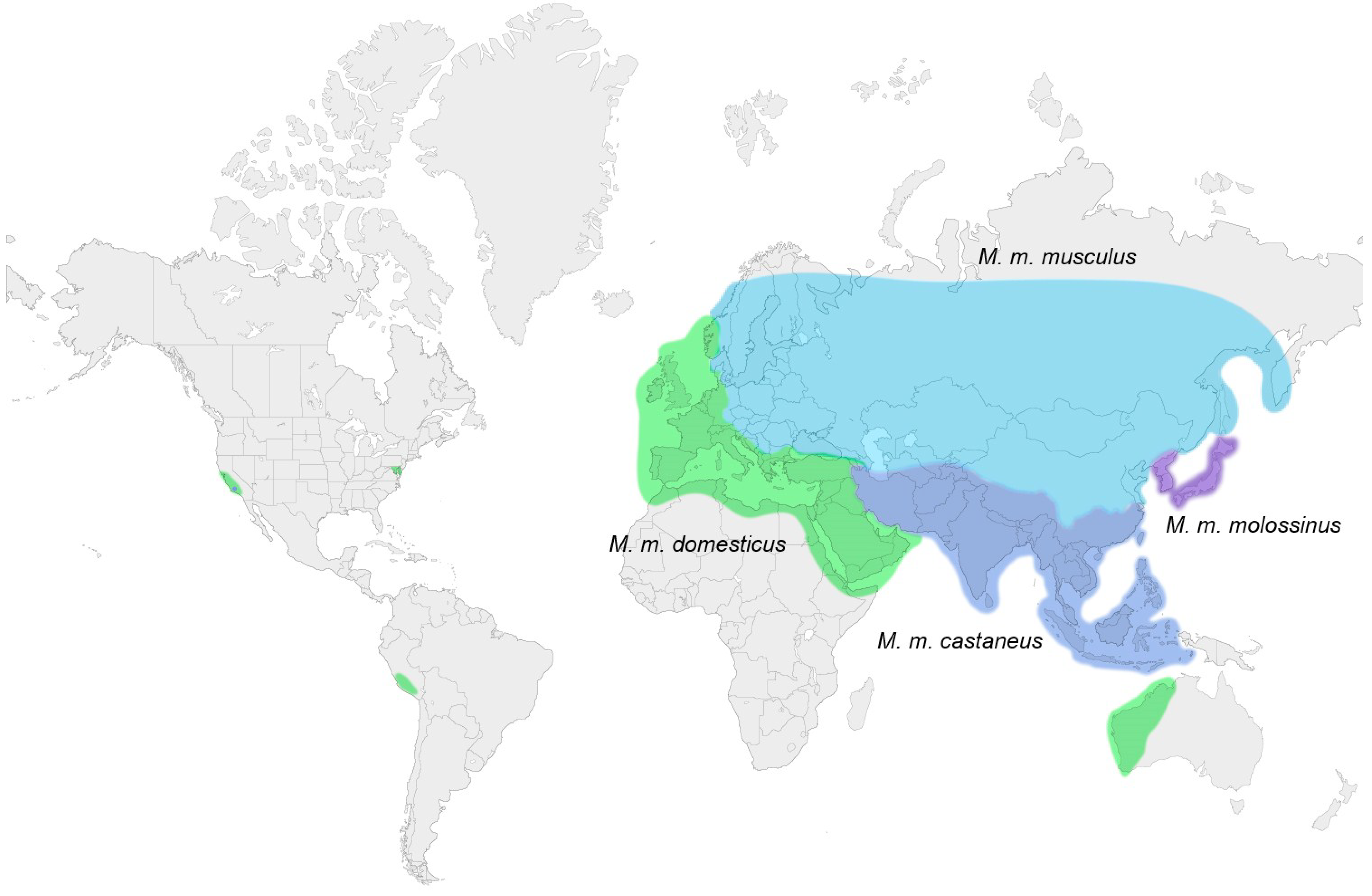
3. Laboratory Strains and Wild Mouse Species that Carry MuLV ERVs
3.1. Ecotropic MuLV ERVs
3.2. Xenotropic/Polytropic MuLV ERVs
| Type | Number of C57BL ERVs | Number Present in House Mouse Subspecies * | |||
|---|---|---|---|---|---|
| M. m. molossinus | M. m. castaneus | M. m. musculus | M. m. domesticus | ||
| Xmvs | 12 | 12 | 2 | 0 | 0 |
| Pmvs | 19 | 0 | 0 | 0 | 0 |
| Mpmvs | 12 | 0 | 0 | 0 | 0 |
4. Origins of Infectious MuLVs
4.1. Active E-MuLV ERVs (Emvs)
4.2. Active X-MuLV ERVs (Xmvs)
| Type | ERV * | Expression Level | Mouse Strains | Chromosome | Defect | Reference |
|---|---|---|---|---|---|---|
| Ecotropic | Emv1 | Low | BALB/c,CBA,C3H | 5 | Env: furin cleavage site | [102,103,104] |
| Emv2 | Low | C57BL | 8 | Pol mutation | [92,105] | |
| Emv3 | Low | DBA | 9 | Gag: myristylation site | [106] | |
| Emv10 | Low | SJL | ? | None | [107] | |
| Emv11 | High | AKR | 7 | None | [108] | |
| Emv12 | High | AKR | 16 | None | [109] | |
| Emv13 | Low | AKR | 2 | Env: C-terminus | [110] | |
| Emv14 | High | AKR | 11 | None | [111] | |
| Emv26 | High | C58 | 8 | None | [105] | |
| Emv30 | Low | NOD | 11 | None | [112] | |
| Xenotropic | Bxv1 | Low High | BALB,C57BL,AKR F/St | 1 | None | [75,113] |
| Mxv1 | Low | MA/My | ? | ? | [74] | |
| Nzv1 | Low | NZB | ? | ? | [114] | |
| Nzv2 | High | NZB | ? | None | [114] |
4.3. Recombinant P-MuLVs Generated during Leukemogenesis
4.4. XRVs in Xenografts and Immunosuppressed Mice
4.5. The Generation of Acute Transforming Viruses
6. Horizontal and Trans-Species Transmission
7. Conclusions
Acknowledgments
Conflicts of Interest
References and Notes
- Gross, L. “Spontaneous” leukemia developing in C3H mice following inoculation in infancy, with AK-leukemic extracts, or AK-embryos. Proc. Soc. Exp. Biol Med. 1951, 76, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.A.; Pincus, T. Demonstration of biological activity of a murine leukemia virus of New Zealand Black mice. Science 1970, 170, 326–327. [Google Scholar] [CrossRef] [PubMed]
- Oie, H.K.; Russell, E.K.; Dotson, J.H.; Rhoads, J.M.; Gazdar, A.F. Host range properties of murine xenotropic and ecotropic type-C viruses. J. Natl. Cancer Inst. 1976, 56, 423–426. [Google Scholar] [PubMed]
- Levy, J.A. Host range of murine xenotropic virus: Replication in avian cells. Nature 1975, 253, 140–142. [Google Scholar] [CrossRef] [PubMed]
- Kozak, C.A. The mouse “xenotropic” gammaretroviruses and their XPR1 receptor. Retrovirology 2010, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Fischinger, P.J.; Nomura, S.; Bolognesi, D.P. A novel murine oncornavirus with dual eco- and xenotropic properties. Proc. Natl. Acad. Sci. USA 1975, 72, 5150–5155. [Google Scholar] [CrossRef] [PubMed]
- Hartley, J.W.; Wolford, N.K.; Old, L.J.; Rowe, W.P. New class of murine leukemia-virus associated with development of spontaneous lymphomas. Proc. Natl. Acad. Sci. USA 1977, 74, 789–792. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Liu, Q.; Wollenberg, K.; Martin, C.; Buckler-White, A.; Kozak, C.A. Evolution of functional and sequence variants of the mammalian XPR1 receptor for mouse xenotropic gammaretroviruses and the human-derived XMRV. J. Virol. 2010, 84, 11970–11980. [Google Scholar] [CrossRef] [PubMed]
- Vogt, V. Retroviral virions and genomes. In Retroviruses; Coffin, J.A., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 1997; pp. 27–70. [Google Scholar]
- Edwards, S.A.; Fan, H. gag-Related polyproteins of Moloney murine leukemia virus: Evidence for independent synthesis of glycosylated and unglycosylated forms. J. Virol. 1979, 30, 551–563. [Google Scholar] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Vogt, M.; Haggblom, C.; Swift, S.; Haas, M. Specific sequences of the env gene determine the host range of two XC-negative viruses of the Rauscher virus complex. Virology 1986, 154, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Heard, J.M.; Danos, O. An amino-terminal fragment of the Friend murine leukemia virus envelope glycoprotein binds the ecotropic receptor. J. Virol. 1991, 65, 4026–4032. [Google Scholar] [PubMed]
- Battini, J.L.; Heard, J.M.; Danos, O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J. Virol. 1992, 66, 1468–1475. [Google Scholar] [PubMed]
- Albritton, L.M.; Tseng, L.; Scadden, D.; Cunningham, J.M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 1989, 57, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Tailor, C.S.; Nouri, A.; Lee, C.G.; Kozak, C.; Kabat, D. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc. Natl. Acad. Sci. USA 1999, 96, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Battini, J.-L.; Rasko, J.E.J.; Miller, A.D. A human cell-surface receptor for xenotropic and polytropic murine leukemia viruses: Possible role in G protein-coupled signal transduction. Proc. Natl. Acad. Sci. USA 1999, 96, 1385–1390. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-L.; Guo, L.; Xu, S.; Holland, C.A.; Kitamura, T.; Hunter, K.; Cunningham, J.M. Receptors for polytropic and xenotropic mouse leukaemia viruses encoded by a single gene at Rmc1. Nat. Genet. 1999, 21, 216–219. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Closs, E.I.; Albritton, L.M.; Cunningham, J.M. Transport of cationic amino acids by the mouse ecotropic retrovirus receptor. Nature 1991, 352, 725–728. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, A.E.; Mendoza, R.; Aranda, R.; Battini, J.L.; Miller, A.D. Xpr1 is an atypical G-protein-coupled receptor that mediates xenotropic and polytropic murine retrovirus neurotoxicity. J. Virol 2012, 86, 1661–1669. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, D.; Touhami, J.; Charnet, P.; Sitbon, M.; Battini, J.L. Inorganic phosphate export by the retrovirus receptor XPR1 in metazoans. Cell. Rep. 2013, 3, 1866–1873. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, S.; Duch, M.; Pedersen, F.S. Change of tropism of SL3–2 murine leukemia virus, using random mutational libraries. J. Virol. 2004, 78, 9343–9351. [Google Scholar] [CrossRef] [PubMed]
- Cloyd, M.W.; Chattopadhyay, S.K. A new class of retrovirus present in many murine leukemia systems. Virology 1986, 151, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Eiden, M.V.; Farrell, K.; Warsowe, J.; Mahan, L.C.; Wilson, C.A. Characterization of a naturally occurring ecotropic receptor that does not facilitate entry of all ecotropic murine retroviruses. J. Virol. 1993, 67, 4056–4061. [Google Scholar] [PubMed]
- Marin, M.; Tailor, C.S.; Nouri, A.; Kozak, S.L.; Kabat, D. Polymorphisms of the cell surface receptor control mouse susceptibilities to xenotropic and polytropic leukemia viruses. J. Virol. 1999, 73, 9362–9368. [Google Scholar] [PubMed]
- Perryman, S.M.; McAtee, F.J.; Portis, J.L. Complete nucleotide sequence of the neurotropic murine retrovirus CAS-BR-E. Nucleic Acids Res. 1991, 19, 1707. [Google Scholar] [CrossRef] [PubMed]
- Voytek, P.; Kozak, C.A. Nucleotide sequence and mode of transmission of the wild mouse ecotropic virus, HoMuLV. Virology 1989, 173, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Cloyd, M.W.; Thompson, M.M.; Hartley, J.W. Host range of mink cell focus-inducing viruses. Virology 1985, 140, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Liu, Q.; Kozak, C.A. Six host range variants of the xenotropic/polytropic gammaretroviruses define determinants for entry in the XPR1 cell surface receptor. Retrovirology 2009, 6, 87. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.G.; Edwards, R.H.; Miller, A.D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc. Natl. Acad. Sci. USA 1994, 91, 78–82. [Google Scholar] [CrossRef] [PubMed]
- van Zeijl, M.; Johann, S.V.; Closs, E.; Cunningham, J.; Eddy, R.; Shows, T.B.; O'Hara, B. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc. Natl. Acad. Sci. USA 1994, 91, 1168–1172. [Google Scholar]
- Miller, D.G.; Miller, A.D. A family of retroviruses that utilize related phosphate transporters for cell entry. J. Virol. 1994, 68, 8270–8276. [Google Scholar] [PubMed]
- Gardner, M.B. Type C viruses of wild mice: Characterization and natural history of amphotropic, ecotropic, and xenotropic MuLV. Curr. Top. Microbiol. Immunol. 1978, 79, 215–259. [Google Scholar] [PubMed]
- Stocking, C.; Kozak, C. Murine endogenous retroviruses. Cell. Mol. Life Sci. 2008, 65, 3383–3398. [Google Scholar] [CrossRef] [PubMed]
- Tomonaga, K.; Coffin, J.M. Structures of endogenous nonecotropic murine leukemia virus (MLV) long terminal repeats in wild mice: Implication for evolution of MLVs. J. Virol. 1999, 73, 4327–4340. [Google Scholar] [PubMed]
- Benveniste, R.E.; Callahan, R.; Sherr, C.J.; Chapman, V.; Todaro, G.J. Two distinct endogenous type C viruses isolated from the asian rodent Mus cervicolor: Conservation of virogene sequences in related rodent species. J. Virol. 1977, 21, 849–862. [Google Scholar] [PubMed]
- Prassolov, V.; Hein, S.; Ziegler, M.; Ivanov, D.; Munk, C.; Lohler, J.; Stocking, C. Mus cervicolor murine leukemia virus isolate M813 belongs to a unique receptor interference group. J. Virol. 2001, 75, 4490–4498. [Google Scholar] [CrossRef] [PubMed]
- Hein, S.; Prassolov, V.; Zhang, Y.; Ivanov, D.; Lohler, J.; Ross, S.R.; Stocking, C. Sodium-dependent myo-inositol transporter 1 is a cellular receptor for Mus cervicolor M813 murine leukemia virus. J. Virol. 2003, 77, 5926–5932. [Google Scholar] [CrossRef] [PubMed]
- Tipper, C.H.; Cingoz, O.; Coffin, J.M. Mus spicilegus endogenous retrovirus HEMV uses murine sodium-dependent myo-inositol transporter 1 as a receptor. J. Virol. 2012, 86, 6341–6344. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.D.; Bergholz, U.; Ziegler, M.; Stocking, C. Identification of the myelin protein plasmolipin as the cell entry receptor for Mus caroli endogenous retrovirus. J. Virol. 2008, 82, 6862–6868. [Google Scholar] [CrossRef] [PubMed]
- Wolgamot, G.; Bonham, L.; Miller, A.D. Sequence analysis of Mus dunni endogenous virus reveals a hybrid VL30/gibbon ape leukemia virus-like structure and a distinct envelope. J. Virol. 1998, 72, 7459–7466. [Google Scholar] [PubMed]
- Ribet, D.; Harper, F.; Esnault, C.; Pierron, G.; Heidmann, T. The GLN family of murine endogenous retroviruses contains an element competent for infectious viral particle formation. J. Virol. 2008, 82, 4413–4419. [Google Scholar] [CrossRef] [PubMed]
- Guenet, J.L.; Bonhomme, F. Wild mice: An ever-increasing contribution to a popular mammalian model. Trends Genet. 2003, 19, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Sage, R.; Atchley, W.R.; Capanna, E. House mice as models in systematic biology. Syst. Biol. 1993, 42, 523–561. [Google Scholar] [CrossRef]
- Boursot, P.; Din, W.; Anand, R.; Darviche, D.; Dod, B.; VonDeimling, F.; Talwar, G.P.; Bonhomme, F. Origin and radiation of the house mouse: Mitochondrial DNA phylogeny. J. Evolut. Biol. 1996, 9, 391–415. [Google Scholar] [CrossRef]
- Marshall, J. Taxonomy. In The mouse in Biomedical Research, vol. 1; Foster HL, S.J., Fox, J.G., Eds.; Academic Press: New York, NY, USA, 1981; pp. 17–26. [Google Scholar]
- Yang, H.; Bell, T.A.; Churchill, G.A.; Pardo-Manuel de Villena, F. On the subspecific origin of the laboratory mouse. Nat. Genet. 2007, 39, 1100–1107. [Google Scholar] [CrossRef] [PubMed]
- Morse, H.C., III. Introduction. In Origins of Inbred Mice; Morse, H.C., III, Ed.; Academic Press: New York, NY, USA, 1978; pp. 1–31. [Google Scholar]
- Waterston, R.H.; Lindblad-Toh, K.; Birney, E.; Rogers, J.; Abril, J.F.; Agarwal, P.; Agarwala, R.; Ainscough, R.; Alexandersson, M.; An, P.; et al. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, N.A.; Copeland, N.G.; Taylor, B.A.; Lee, B.K. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J. Virol. 1982, 43, 26–36. [Google Scholar] [PubMed]
- O'Neill, R.R.; Khan, A.S.; Hoggan, M.D.; Hartley, J.W.; Martin, M.A.; Repaske, R. Specific hybridization probes demonstrate fewer xenotropic than mink cell focus-forming murine leukemia virus env-related sequences in DNAs from inbred laboratory mice. J. Virol. 1986, 58, 359–366. [Google Scholar] [PubMed]
- Stoye, J.P.; Coffin, J.M. The four classes of endogenous murine leukemia virus: Structural relationships and potential for recombination. J. Virol. 1987, 61, 2659–2669. [Google Scholar] [PubMed]
- Hoggan, M.D.; Buckler, C.E.; Sears, J.F.; Rowe, W.P.; Martin, M.A. Organization and stability of endogenous xenotropic murine leukemia virus proviral DNA in mouse genomes. J. Virol. 1983, 45, 473–477. [Google Scholar] [PubMed]
- Kozak, C.A.; O'Neill, R.R. Diverse wild mouse origins of xenotropic, mink cell focus-forming, and two types of ecotropic proviral genes. J. Virol. 1987, 61, 3082–3088. [Google Scholar] [PubMed]
- Yonekawa, H.; Moriwaki, K.; Gotoh, O.; Miyashita, N.; Matsushima, Y.; Shi, L.; Cho, W.; Zhen, X.; Tagashira, Y. Hybrid origin of Japanese mice “Mus musculus molossinus”: Evidence from restriction analysis of mitochondrial DNA. Mol. Biol. Evol. 1988, 5, 63–78. [Google Scholar] [PubMed]
- Inaguma, Y.; Miyashita, N.; Moriwaki, K.; Huai, W.C.; Jin, M.L.; He, X.Q.; Ikeda, H. Acquisition of two endogenous ecotropic murine leukemia viruses in distinct Asian wild mouse populations. J. Virol. 1991, 65, 1796–1802. [Google Scholar] [PubMed]
- Ikeda, H.; Laigret, F.; Martin, M.A.; Repaske, R. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J. Virol. 1985, 55, 768–777. [Google Scholar] [PubMed]
- Rassart, E.; Nelbach, L.; Jolicoeur, P. Cas-Br-E murine leukemia virus: Sequencing of the paralytogenic region of its genome and derivation of specific probes to study its origin and the structure of its recombinant genomes in leukemic tissues. J. Virol. 1986, 60, 910–919. [Google Scholar] [PubMed]
- Ikeda, H.; Kato, K.; Kitani, H.; Suzuki, T.; Yoshida, T.; Inaguma, Y.; Yamamoto, N.; Suh, J.G.; Hyun, B.H.; Yamagata, T.; et al. Virological properties and nucleotide sequences of cas-E-type endogenous ecotropic murine leukemia viruses in south Asian wild mice, Mus musculus castaneus. J. Virol. 2001, 75, 5049–5058. [Google Scholar] [CrossRef] [PubMed]
- Dandekar, S.; Rossitto, P.; Pickett, S.; Mockli, G.; Bradshaw, H.; Cardiff, R.; Gardner, M. Molecular characterization of the Akvr-1 restriction gene: A defective endogenous retrovirus-borne gene identical to Fv-4r. J. Virol. 1987, 61, 308–314. [Google Scholar] [PubMed]
- Voytek, P.; Kozak, C. HoMuLV: A novel pathogenic ecotropic virus isolated from the European mouse, Mus hortulanus. Virology 1988, 165, 469–475. [Google Scholar] [CrossRef] [PubMed]
- Stoye, J.P.; Coffin, J.M. Polymorphism of murine endogenous proviruses revealed by using virus class-specific oligonucleotide probes. J. Virol. 1988, 62, 168–175. [Google Scholar] [PubMed]
- Frankel, W.N.; Stoye, J.P.; Taylor, B.A.; Coffin, J.M. A linkage map of endogenous murine leukemia proviruses. Genetics 1990, 124, 221–236. [Google Scholar] [PubMed]
- Jung, Y.T.; Lyu, M.S.; Buckler-White, A.; Kozak, C.A. Characterization of a polytropic murine leukemia virus proviral sequence associated with the virus resistance gene Rmcf of DBA/2 mice. J. Virol. 2002, 76, 8218–8224. [Google Scholar] [CrossRef] [PubMed]
- Bamunusinghe, D.; Liu, Q.; Lu, X.; Oler, A.; Kozak, C.A. Endogenous gammaretrovirus acquisition in Mus musculus subspecies carrying functional variants of the XPR1 virus receptor. J. Virol. 2013, 87, 9845–9855. [Google Scholar] [CrossRef] [PubMed]
- Nellaker, C.; Keane, T.M.; Yalcin, B.; Wong, K.; Agam, A.; Belgard, T.G.; Flint, J.; Adams, D.J.; Frankel, W.N.; Ponting, C.P. The genomic landscape shaped by selection on transposable elements across 18 mouse strains. Genome Biol. 2012, 13, R45. [Google Scholar] [CrossRef] [PubMed]
- Jern, P.; Stoye, J.P.; Coffin, J.M. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 2007, 3, e183. [Google Scholar] [CrossRef]
- Lamont, C.; Culp, P.; Talbott, R.L.; Phillips, T.R.; Trauger, R.J.; Frankel, W.N.; Wilson, M.C.; Coffin, J.M.; Elder, J.H. Characterization of endogenous and recombinant proviral elements of a highly tumorigenic AKR cell line. J. Virol. 1991, 65, 4619–4628. [Google Scholar] [PubMed]
- Baudino, L.; Yoshinobu, K.; Morito, N.; Kikuchi, S.; Fossati-Jimack, L.; Morley, B.J.; Vyse, T.J.; Hirose, S.; Jorgensen, T.N.; Tucker, R.M.; et al. Dissection of genetic mechanisms governing the expression of serum retroviral gp70 implicated in murine lupus nephritis. J. Immunol. 2008, 181, 2846–2854. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.T.; Wu, T.; Kozak, C.A. Characterization of recombinant nonecotropic murine leukemia viruses from the wild mouse species Mus spretus. J. Virol. 2003, 77, 12773–12781. [Google Scholar] [CrossRef] [PubMed]
- Orth, A.; Belkhir, K.; Britton-Davidian, J.; Boursot, P.; Benazzou, T.; Bonhomme, F. Natural hybridization between 2 sympatric species of mice, Mus musculus domesticus L. and Mus spretus Lataste. C. R. Biol. 2002, 325, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Tomonaga, K.; Coffin, J.M. Structure and distribution of endogenous nonecotropic murine leukemia viruses in wild mice. J. Virol. 1998, 72, 8289–8300. [Google Scholar] [PubMed]
- Kozak, C.A.; Hartley, J.W.; Morse, H.C., III. Laboratory and wild-derived mice with multiple loci for production of xenotropic murine leukemia virus. J. Virol. 1984, 51, 77–80. [Google Scholar] [PubMed]
- Baliji, S.; Liu, Q.; Kozak, C.A. Common inbred strains of the laboratory mouse that are susceptible to infection by mouse xenotropic gammaretroviruses and the human-derived retrovirus XMRV. J. Virol. 2010, 84, 12841–12849. [Google Scholar] [CrossRef] [PubMed]
- Cingoz, O.; Paprotka, T.; Delviks-Frankenberry, K.A.; Wildt, S.; Hu, W.S.; Pathak, V.K.; Coffin, J.M. Characterization, mapping, and distribution of the two XMRV parental proviruses. J. Virol. 2012, 86, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Lathrop, A.E.; Loeb, L. Further investigations on the origin of tumors in mice : V. the tumor rate in hybrid strains. J. Exp. Med. 1918, 28, 475–500. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, E. Origin of the Japanese waltzing mouse. Science 1942, 95, 46. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, J.R.; Didion, J.P.; Buus, R.J.; Bell, T.A.; Welsh, C.E.; Bonhomme, F.; Yu, A.H.; Nachman, M.W.; Pialek, J.; et al. Subspecific origin and haplotype diversity in the laboratory mouse. Nat. Genet. 2011, 43, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Rowe, W.P.; Pincus, T. Quantitative studies of naturally occurring murine leukemia virus infection of AKR mice. J. Exp. Med. 1972, 135, 429–436. [Google Scholar] [CrossRef] [PubMed]
- O'Brien, S.J.; Moore, J.L.; Martin, M.A.; Womack, J.E. Evidence for the horizontal acquisition of murine AKR virogenes by recent horizontal infection of the germ line. J. Exp. Med. 1982, 155, 1120–1123. [Google Scholar] [CrossRef] [PubMed]
- Buckler, C.E.; Staal, S.P.; Rowe, W.P.; Martin, M.A. Variation in the number of copies and in the genomic organization of ecotropic murine leukemia virus proviral sequences in sublines of AKR mice. J. Virol. 1982, 43, 629–640. [Google Scholar] [PubMed]
- Steffen, D.L.; Taylor, B.A.; Weinberg, R.A. Continuing germ line integration of AKV proviruses during the breeding of AKR mice and derivative recombinant inbred strains. J. Virol. 1982, 42, 165–175. [Google Scholar] [PubMed]
- Herr, W.; Gilbert, W. Germ-line MuLV reintegrations in AKR/J mice. Nature 1982, 296, 865–868. [Google Scholar] [CrossRef] [PubMed]
- Lock, L.F.; Keshet, E.; Gilbert, D.J.; Jenkins, N.A.; Copeland, N.G. Studies of the mechanism of spontaneous germline ecotropic provirus acquisition in mice. EMBO J. 1988, 7, 4169–4177. [Google Scholar] [PubMed]
- Lowy, D.R.; Rowe, W.P.; Teich, N.; Hartley, J.W. Murine leukemia virus: High-frequency activation in vitro by 5-iododeoxyuridine and 5-bromodeoxyuridine. Science 1971, 174, 155–156. [Google Scholar] [CrossRef] [PubMed]
- Aaronson, S.A.; Todaro, G.J.; Scolnick, E.M. Induction of murine C-type viruses from clonal lines of virus-free BALB/3T3 Cells. Science 1971, 174, 157–159. [Google Scholar] [CrossRef] [PubMed]
- Odaka, T. Genetic transmission of endogenous N- and B-tropic murine leukemia viruses in low-leukemic strain C57BL/6. J. Virol. 1975, 15, 332–337. [Google Scholar] [PubMed]
- Turturro, A.; Blank, K.; Murasko, D.; Hart, R. Mechanisms of caloric restriction affecting aging and disease. Ann. N. Y. Acad. Sci. 1994, 719, 159–170. [Google Scholar] [CrossRef] [PubMed]
- McCubrey, J.; Risser, R. Genetic interactions in the spontaneous production of endogenous murine leukemia virus in low leukemic mouse strains. J. Exp. Med. 1982, 156, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Beemon, K.; Duesberg, P.; Vogt, P. Evidence for crossing-over between avian tumor viruses based on analysis of viral RNAs. Proc. Natl. Acad. Sci. USA 1974, 71, 4254–4258. [Google Scholar] [CrossRef] [PubMed]
- King, S.R.; Berson, B.J.; Risser, R. Mechanism of interaction between endogenous ecotropic murine leukemia viruses in (BALB/c X C57BL/6) hybrid cells. Virology 1988, 162, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bartman, T.; Murasko, D.M.; Sieck, T.G.; Turturro, A.; Hart, R.; Blank, K.J. A murine leukemia virus expressed in aged DBA/2 mice is derived by recombination of the Emv-3 locus and another endogenous gag sequence. Virology 1994, 203, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Robbins, K.C.; Cabradilla, C.D.; Stephenson, J.R.; Aaronson, S.A. Segregation of genetic information for a B-tropic leukemia virus with the structural locus for BALB:virus-1. Proc. Natl. Acad. Sci. USA 1977, 74, 2953–2957. [Google Scholar] [CrossRef] [PubMed]
- McCubrey, J.; Risser, R. Genetic interactions in induction of endogenous murine leukemia virus from low leukemic mice. Cell 1982, 28, 881–888. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.Y.; Khiroya, R.; Schwartz, R.S.; Coffin, J.M. Role of recombinant ecotropic and polytropic viruses in the development of spontaneous thymic lymphomas in HRS/J mice. J. Virol. 1984, 50, 397–407. [Google Scholar] [PubMed]
- Morse, H.C., III; Kozak, C.A.; Yetter, R.A.; Hartley, J.W. Unique features of retrovirus expression in F/St mice. J. Virol. 1982, 43, 1–7. [Google Scholar] [PubMed]
- East, J.; Tilly, R.J.; Tuffrey, M.; Harvey, J.J. The early appearance and subsequent distribution of murine leukaemia virus in NZB embryos. Int. J. Cancer 1978, 22, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.M.; Sherr, C.J.; Todaro, G.J. S-tropic murine type-C viruses: Frequency of isolation from continuous cell lines, leukemia virus preparations and normal spleens. Int. J. Cancer 1974, 13, 587–598. [Google Scholar] [CrossRef] [PubMed]
- Greenberger, J.S.; Phillips, S.M.; Stephenson, J.R.; Aaronson, S.A. Induction of mouse type-C RNA virus by lipopolysaccharide. J. Immunol. 1975, 115, 317–320. [Google Scholar] [PubMed]
- Sherr, C.J.; Lieber, M.M.; Todaro, G.J. Mixed splenocyte cultures and graft versus host reactions selectively induce an S-tropic murine type C virus. Cell 1974, 1, 55–58. [Google Scholar] [CrossRef]
- Kozak, C.A.; Rowe, W.P. Genetic mapping of the ecotropic murine leukemia virus-inducing locus of BALB/c mouse to chromosome 5. Science 1979, 204, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Ihle, J.N.; Joseph, D.R.; Domotor, J.J., Jr. Genetic linkage of C3H/HeJ and BALB/c endogenous ecotropic C-type viruses to phosphoglucomutase-1 on chromosome 5. Science 1979, 204, 71–73. [Google Scholar] [CrossRef] [PubMed]
- Freed, E.O.; Risser, R. The role of envelope glycoprotein processing in murine leukemia virus infection. J. Virol. 1987, 61, 2852–2856. [Google Scholar] [PubMed]
- Kozak, C.A.; Rowe, W.P. Genetic mapping of ecotropic murine leukemia virus-inducing loci in six inbred strains. J. Exp. Med. 1982, 155, 524–534. [Google Scholar] [CrossRef] [PubMed]
- Copeland, N.G.; Jenkins, N.A.; Nexo, B.; Schultz, A.M.; Rein, A.; Mikkelsen, T.; Jorgensen, P. Poorly expressed endogenous ecotropic provirus of DBA/2 mice encodes a mutant Pr65gag protein that is not myristylated. J. Virol. 1988, 62, 479–487. [Google Scholar] [PubMed]
- Yetter, R.A.; Langdon, W.Y.; Morse, H.C., 3rd. Characterization of ecotropic murine leukemia viruses in SJL/J mice. Virology 1985, 141, 319–321. [Google Scholar] [CrossRef] [PubMed]
- Rowe, W.P.; Hartley, J.W.; Bremner, T. Genetic mapping of a murine leukemia virus-inducing locus of AKR mice. Science 1972, 178, 860–862. [Google Scholar] [CrossRef] [PubMed]
- Kozak, C.A.; Rowe, W.P. Genetic mapping of the ecotropic virus-inducing locus Akv-2 of the AKR mouse. J. Exp. Med. 1980, 152, 1419–1423. [Google Scholar] [CrossRef] [PubMed]
- Copeland, N.G.; Bedigian, H.G.; Thomas, C.Y.; Jenkins, N.A. DNAs of two molecularly cloned endogenous ecotropic proviruses are poorly infectious in DNA transfection assays. J. Virol. 1984, 49, 437–444. [Google Scholar] [PubMed]
- Bedigian, H.G.; Copeland, N.G.; Jenkins, N.A.; Salvatore, K.; Rodick, S. Emv-13 (Akv-3): A noninducible endogenous ecotropic provirus of AKR/J mice. J. Virol. 1983, 46, 490–497. [Google Scholar] [PubMed]
- Serreze, D.V.; Leiter, E.H.; Hanson, M.S.; Christianson, S.W.; Shultz, L.D.; Hesselton, R.M.; Greiner, D.L. Emv30null NOD-scid mice. An improved host for adoptive transfer of autoimmune diabetes and growth of human lymphohematopoietic cells. Diabetes 1995, 44, 1392–1398. [Google Scholar] [CrossRef] [PubMed]
- Kozak, C.; Rowe, W.P. Genetic mapping of xenotropic leukemia virus-inducing loci in two mouse strains. Science 1978, 199, 1448–1449. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.K.; Schwartz, R.S. Mendelian segregation of loci controlling xenotropic virus production in NZB crosses. Virology 1977, 83, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Yetter, R.A.; Hartley, J.W.; Morse, H.C., III. H-2-linked regulation of xenotropic murine leukemia virus expression. Proc. Natl. Acad. Sci. USA 1983, 80, 505–509. [Google Scholar] [CrossRef] [PubMed]
- Elder, J.H.; Gautsch, J.W.; Jensen, F.C.; Lerner, R.A.; Chused, T.M.; Morse, H.C.; Hartley, J.W.; Rowe, W.P. Differential expression of two distinct xenotropic viruses in NZB mice. Clin. Immunol. Immunopathol. 1980, 15, 493–501. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.; Sherr, C.; Potter, M.; Todaro, G. Isolation of type-C viruses from the Asian feral mouse Mus musculus molossinus. Int. J. Cancer 1975, 15, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.K.; Lander, M.R.; Gupta, S.; Rands, E.; Lowy, D.R. Origin of mink cytopathic focus-forming (MCF) viruses: Comparison with ecotropic and xenotropic murine leukemia virus genomes. Virology 1981, 113, 465–483. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Knoper, R.C.; Kozak, C.A. Wild mouse variants of envelope genes of xenotropic/polytropic mouse gammaretroviruses and their XPR1 receptors elucidate receptor determinants of virus entry. J. Virol. 2007, 81, 10550–10557. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.K.; Oliff, A.I.; Linemeyer, D.L.; Lander, M.R.; Lowy, D.R. Genomes of murine leukemia viruses isolated from wild mice. J. Virol 1981, 39, 777–791. [Google Scholar] [PubMed]
- Flanagan, J.R.; Krieg, A.M.; Max, E.E.; Khan, A.S. Negative control region at the 5' end of murine leukemia virus long terminal repeats. Mol. Cell. Biol 1989, 9, 739–746. [Google Scholar] [PubMed]
- Khan, A.S.; Martin, M.A. Endogenous murine leukemia proviral long terminal repeats contain a unique 190-base-pair insert. Proc. Natl. Acad. Sci. USA 1983, 80, 2699–2703. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.E.; McKinnon, R.D.; Brolaski, M.N.; Gautsch, J.W.; Wilson, M.C. The 3' long terminal repeat of a transcribed yet defective endogenous retroviral sequence is a competent promoter of transcription. J. Virol. 1987, 61, 1261–1265. [Google Scholar] [PubMed]
- Nitta, T.; Lee, S.; Ha, D.; Arias, M.; Kozak, C.A.; Fan, H. Moloney murine leukemia virus glyco-gag facilitates xenotropic murine leukemia virus-related virus replication through human APOBEC3-independent mechanisms. Retrovirology 2012, 9, 58. [Google Scholar] [CrossRef] [PubMed]
- Fischinger, P.J.; Blevins, C.S.; Dunlop, N.M. Genomic masking of nondefective recombinant murine leukemia virus in Moloney virus stocks. Science 1978, 201, 457–459. [Google Scholar] [CrossRef] [PubMed]
- Lavignon, M.; Evans, L. A multistep process of leukemogenesis in Moloney murine leukemia virus-infected mice that is modulated by retroviral pseudotyping and interference. J. Virol. 1996, 70, 3852–3862. [Google Scholar] [PubMed]
- Evans, L.H.; Alamgir, A.S.; Owens, N.; Weber, N.; Virtaneva, K.; Barbian, K.; Babar, A.; Malik, F.; Rosenke, K. Mobilization of endogenous retroviruses in mice after infection with an exogenous retrovirus. J. Virol. 2009, 83, 2429–2435. [Google Scholar] [CrossRef] [PubMed]
- Wensel, D.L.; Li, W.; Cunningham, J.M. A virus-virus interaction circumvents the virus receptor requirement for infection by pathogenic retroviruses. J. Virol. 2003, 77, 3460–3469. [Google Scholar] [CrossRef] [PubMed]
- Fan, H. Leukemogenesis by Moloney murine leukemia virus: A multistep process. Trends Microbiol. 1997, 5, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Stoye, J.P.; Moroni, C.; Coffin, J.M. Virological events leading to spontaneous AKR thymomas. J. Virol. 1991, 65, 1273–1285. [Google Scholar] [PubMed]
- Evans, L.H. Characterization of polytropic MuLVs from three-week-old AKR/J mice. Virology 1986, 153, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, N.; Jolicoeur, P. Retroviral Pathogenesis. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: Woodbury, NY, USA, 1997; pp. 475–586. [Google Scholar]
- Suzuki, T.; Shen, H.; Akagi, K.; Morse, H.C.; Malley, J.D.; Naiman, D.Q.; Jenkins, N.A.; Copeland, N.G. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 2002, 32, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Li, J.P.; Baltimore, D. Mechanism of leukemogenesis induced by mink cell focus-forming murine leukemia viruses. J. Virol. 1991, 65, 2408–2414. [Google Scholar] [PubMed]
- Tumas, K.M.; Poszgay, J.M.; Avidan, N.; Ksiazek, S.J.; Overmoyer, B.; Blank, K.J.; Prystowsky, M.B. Loss of antigenic epitopes as the result of env gene recombination in retrovirus-induced leukemia in immunocompetent mice. Virology 1993, 192, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Mangeney, M.; Pothlichet, J.; Renard, M.; Ducos, B.; Heidmann, T. Endogenous retrovirus expression is required for murine melanoma tumor growth in vivo. Cancer Res. 2005, 65, 2588–2591. [Google Scholar] [CrossRef] [PubMed]
- Pothlichet, J.; Heidmann, T.; Mangeney, M. A recombinant endogenous retrovirus amplified in a mouse neuroblastoma is involved in tumor growth in vivo. Int. J. Cancer 2006, 119, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Herr, W.; Gilbert, W. Somatically acquired recombinant murine leukemia proviruses in thymic leukemias of AKR/J mice. J. Virol. 1983, 46, 70–82. [Google Scholar] [PubMed]
- Herr, W.; Gilbert, W. Free and integrated recombinant murine leukemia virus DNAs appear in preleukemic thymuses of AKR/J mice. J. Virol. 1984, 50, 155–162. [Google Scholar] [PubMed]
- Yoshimura, F.K.; Wang, T.; Yu, F.; Kim, H.R.; Turner, J.R. Mink cell focus-forming murine leukemia virus infection induces apoptosis of thymic lymphocytes. J. Virol. 2000, 74, 8119–8126. [Google Scholar] [CrossRef] [PubMed]
- Nanua, S.; Yoshimura, F.K. Mink epithelial cell killing by pathogenic murine leukemia viruses involves endoplasmic reticulum stress. J. Virol. 2004, 78, 12071–12074. [Google Scholar] [CrossRef] [PubMed]
- Cloyd, M.W.; Hartley, J.W.; Rowe, W.P. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J. Exp. Med. 1980, 151, 542–552. [Google Scholar] [CrossRef] [PubMed]
- Rudali, G.; Duplan, J.F.; Latarjet, R. Latency of leukosis in Ak mice injected with leukemic alpha-cellular Ak extract. Comptes Rendus Hebd. Seances l'Academie Sci. 1956, 242, 837–839. [Google Scholar]
- Buller, R.S.; Sitbon, M.; Portis, J.L. The endogenous mink cell focus-forming (MCF) gp70 linked to the Rmcf gene restricts MCF virus replication in vivo and provides partial resistance to erythroleukemia induced by Friend murine leukemia virus. J. Exp. Med. 1988, 167, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Brightman, B.K.; Rein, A.; Trepp, D.J.; Fan, H. An enhancer variant of Moloney murine leukemia virus defective in leukemogenesis does not generate detectable mink cell focus-inducing virus in vivo. Proc. Natl. Acad. Sci. USA 1991, 88, 2264–2268. [Google Scholar] [CrossRef] [PubMed]
- Elder, J.H.; Gautsch, J.W.; Jensen, F.C.; Lerner, R.A.; Hartley, J.W.; Rowe, W.P. Biochemical evidence that MCF murine leukemia viruses are envelope (env) gene recombinants. Proc. Natl. Acad. Sci. USA 1977, 74, 4676–4680. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.S.; Rowe, W.P.; Martin, M.A. Cloning of endogenous murine leukemia virus-related sequences from chromosomal DNA of BALB/c and AKR/J mice: Identification of an env progenitor of AKR-247 mink cell focus-forming proviral DNA. J. Virol. 1982, 44, 625–636. [Google Scholar] [PubMed]
- Evans, L.H.; Lavignon, M.; Taylor, M.; Alamgir, A.S. Antigenic subclasses of polytropic murine leukemia virus (MLV) isolates reflect three distinct groups of endogenous polytropic MLV-related sequences in NFS/N mice. J. Virol. 2003, 77, 10327–10338. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, S.K.; Cloyd, M.W.; Linemeyer, D.L.; Lander, M.R.; Rands, E.; Lowy, D.R. Cellular origin and role of mink cell focus-forming viruses in murine thymic lymphomas. Nature 1982, 295, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Rommelaere, J.; Faller, D.V.; Hopkins, N. Characterization and mapping of RNase T1-resistant oligonucleotides derived from the genomes of Akv and MCF murine leukemia viruses. Proc. Natl. Acad. Sci. USA 1978, 75, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Quint, W.; Boelens, W.; van Wezenbeek, P.; Robanus Maandag, E.; Berns, A. Generation of AKR mink cell focus-forming virus: Nucleotide sequence of the 3' end of a somatically acquired AKR-MCF. Virology 1984, 136, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Hoggan, M.D.; O'Neill, R.R.; Kozak, C.A. Nonecotropic murine leukemia viruses in BALB/c and NFS/N mice: Characterization of the BALB/c Bxv-1 provirus and the single NFS endogenous xenotrope. J. Virol. 1986, 60, 980–986. [Google Scholar] [PubMed]
- Alamgir, A.S.; Owens, N.; Lavignon, M.; Malik, F.; Evans, L.H. Precise identification of endogenous proviruses of NFS/N mice participating in recombination with Moloney ecotropic murine leukemia virus (MuLV) to generate polytropic MuLVs. J. Virol. 2005, 79, 4664–4671. [Google Scholar] [CrossRef] [PubMed]
- Jahid, S.; Bundy, L.M.; Granger, S.W.; Fan, H. Chimeras between SRS and Moloney murine leukemia viruses reveal novel determinants in disease specificity and MCF recombinant formation. Virology 2006, 351, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Lung, M.L.; Hartley, J.W.; Rowe, W.P.; Hopkins, N.H. Large RNase T1-resistant oligonucleotides encoding p15E and the U3 region of the long terminal repeat distinguish two biological classes of mink cell focus-forming type C viruses of inbred mice. J. Virol. 1983, 45, 275–290. [Google Scholar] [PubMed]
- Thomas, C.Y.; Coffin, J.M. Genetic alterations of RNA leukemia viruses associated with the development of spontaneous thymic leukemia in AKR/J mice. J. Virol. 1982, 43, 416–426. [Google Scholar] [PubMed]
- Yu, P.; Lubben, W.; Slomka, H.; Gebler, J.; Konert, M.; Cai, C.; Neubrandt, L.; Prazeres da Costa, O.; Paul, S.; Dehnert, S.; et al. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity 2012, 37, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Young, G.R.; Eksmond, U.; Salcedo, R.; Alexopoulou, L.; Stoye, J.P.; Kassiotis, G. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature 2012, 491, 774–778. [Google Scholar] [PubMed]
- Dong, B.; Kim, S.; Hong, S.; Das Gupta, J.; Malathi, K.; Klein, E.A.; Ganem, D.; DeRisi, J.L.; Chow, S.A.; Silverman, R.H. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc. Natl. Acad. Sci. USA 2007, 104, 1655–1660. [Google Scholar] [CrossRef] [PubMed]
- Paprotka, T.; Delviks-Frankenberry, K.A.; Cingoz, O.; Martinez, A.; Kung, H.J.; Tepper, C.G.; Hu, W.S.; Fivash, M.J., Jr.; Coffin, J.M.; Pathak, V.K. Recombinant origin of the retrovirus XMRV. Science 2011, 333, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Sfanos, K.S.; Aloia, A.L.; Hicks, J.L.; Esopi, D.M.; Steranka, J.P.; Shao, W.; Sanchez-Martinez, S.; Yegnasubramanian, S.; Burns, K.H.; Rein, A.; et al. Identification of replication competent murine gammaretroviruses in commonly used prostate cancer cell lines. PLoS One 2011, 6, e20874. [Google Scholar] [CrossRef] [PubMed]
- Triviai, I.; Ziegler, M.; Bergholz, U.; Oler, A.J.; Stubig, T.; Prassolov, V.; Fehse, B.; Kozak, C.A.; Kroger, N.; Stocking, C. Endogenous retrovirus induces leukemia in a xenograft mouse model for primary myelofibrosis. Proc. Natl. Acad. Sci. USA 2014, 111, 8595–8600. [Google Scholar] [CrossRef] [PubMed]
- Mucenski, M.L.; Taylor, B.A.; Ihle, J.N.; Hartley, J.W.; Morse, H.C., 3rd; Jenkins, N.A.; Copeland, N.G. Identification of a common ecotropic viral integration site, Evi-1, in the DNA of AKXD murine myeloid tumors. Mol. Cell. Biol. 1988, 8, 301–308. [Google Scholar] [PubMed]
- Linemeyer, D.L.; Ruscetti, S.K.; Scolnick, E.M.; Evans, L.H.; Duesberg, P.H. Biological activity of the spleen focus-forming virus is encoded by a molecularly cloned subgenomic fragment of spleen focus-forming virus DNA. Proc. Natl. Acad. Sci. USA 1981, 78, 1401–1405. [Google Scholar] [CrossRef] [PubMed]
- Linemeyer, D.L.; Menke, J.G.; Ruscetti, S.K.; Evans, L.H.; Scolnick, E.M. Envelope gene sequences which encode the gp52 protein of spleen focus-forming virus are required for the induction of erythroid cell proliferation. J. Virol. 1982, 43, 223–233. [Google Scholar] [PubMed]
- Ruscetti, S.K.; Cmarik, J.L. Deregulation of signal transduction pathways by oncogenic retroviruses. In Retroviruses and Insights into Cancer; Dudley, J., Ed.; Springer: New York, NY, USA, 2011; pp. 53–94. [Google Scholar]
- Best, S.; LeTissier, P.; Towers, G.; Stoye, J.P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 1996, 382, 826–829. [Google Scholar] [CrossRef] [PubMed]
- Benit, L.; DeParseval, N.; Casella, J.F.; Callebaut, I.; Cordonnier, A.; Heidmann, T. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J. Virol. 1997, 71, 5652–5657. [Google Scholar] [PubMed]
- Lilly, F. Susceptibility to two strains of Friend leukemia virus in mice. Science 1967, 155, 461–462. [Google Scholar] [CrossRef] [PubMed]
- Kozak, C.A.; Chakraborti, A. Single amino acid changes in the murine leukemia virus capsid protein gene define the target of Fv1 resistance. Virology 1996, 225, 300–305. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.T.; Kozak, C.A. A single amino acid change in the murine leukemia virus capsid gene responsible for the Fv1nr phenotype. J. Virol. 2000, 74, 5385–5387. [Google Scholar] [CrossRef] [PubMed]
- Stevens, A.; Bock, M.; Ellis, S.; LeTissier, P.; Bishop, K.N.; Yap, M.W.; Taylor, W.; Stoye, J.P. Retroviral capsid determinants of Fv1 NB and NR tropism. J. Virol. 2004, 78, 9592–9598. [Google Scholar] [CrossRef] [PubMed]
- Yap, M.W.; Colbeck, E.; Ellis, S.A.; Stoye, J.P. Evolution of the retroviral restriction gene Fv1: Inhibition of non-MLV retroviruses. PLoS Pathog. 2014, 10, e1003968. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Yan, Y.; Kozak, C.A. Rmcf2, a xenotropic provirus in the Asian mouse species Mus castaneus, blocks infection by polytropic mouse gammaretroviruses. J. Virol. 2005, 79, 9677–9684. [Google Scholar] [CrossRef] [PubMed]
- Ruscetti, S.; Davis, L.; Feild, J.; Oliff, A. Friend murine leukemia virus-induced leukemia is associated with the formation of mink cell focus-inducing viruses and is blocked in mice expressing endogenous mink cell focus-inducing xenotropic viral envelope genes. J. Exp. Med. 1981, 154, 907–920. [Google Scholar] [CrossRef] [PubMed]
- Hartley, J.W.; Yetter, R.A.; Morse, H.C. A mouse gene on chromosome 5 that restricts infectivity of mink cell focus-forming recombinant murine leukemia viruses. J. Exp. Med. 1983, 158, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, S. FV-4: A new gene affecting the splenomegaly induction by Friend leukemia virus. Jpn. J. Exp. Med. 1975, 45, 473–478. [Google Scholar] [PubMed]
- Taylor, G.M.; Gao, Y.; Sanders, D.A. Fv-4: identification of the defect in Env and the mechanism of resistance to ecotropic murine leukemia virus. J. Virol 2001, 75, 11244–11248. [Google Scholar] [CrossRef] [PubMed]
- Robinson, H.L.; Astrin, S.M.; Senior, A.M.; Salazar, F.H. Host susceptibility to endogenous viruses: defective, glycoprotein-expressing proviruses interfere with infections. J. Virol. 1981, 40, 745–751. [Google Scholar] [PubMed]
- Spencer, T.E.; Mura, M.; Gray, C.A.; Griebel, P.J.; Palmarini, M. Receptor usage and fetal expression of ovine endogenous betaretroviruses: implications for coevolution of endogenous and exogenous retroviruses. J. Virol. 2003, 77, 749–753. [Google Scholar] [CrossRef] [PubMed]
- McDougall, A.S.; Terry, A.; Tzavaras, T.; Cheney, C.; Rojko, J.; Neil, J.C. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for a role in natural resistance to subgroup B feline leukemia viruses. J. Virol. 1994, 68, 2151–2160. [Google Scholar] [PubMed]
- Maksakova, I.A.; Romanish, M.T.; Gagnier, L.; Dunn, C.A.; de Lagemaat, L.N.V.; Mager, D.L. Retroviral elements and their hosts: Insertional mutagenesis in the mouse germ line. PLoS Genet. 2006, 2, 1–10. [Google Scholar] [CrossRef]
- Refsland, E.W.; Harris, R.S. The APOBEC3 family of retroelement restriction factors. Curr. Top. Microbiol. Immunol. 2013, 371, 1–27. [Google Scholar] [PubMed]
- Stavrou, S.; Nitta, T.; Kotla, S.; Ha, D.; Nagashima, K.; Rein, A.R.; Fan, H.; Ross, S.R. Murine leukemia virus glycosylated Gag blocks apolipoprotein B editing complex 3 and cytosolic sensor access to the reverse transcription complex. Proc. Natl. Acad. Sci. USA 2013, 110, 9078–9083. [Google Scholar] [CrossRef] [PubMed]
- Takeda, E.; Tsuji-Kawahara, S.; Sakamoto, M.; Langlois, M.A.; Neuberger, M.S.; Rada, C.; Miyazawa, M. Mouse APOBEC3 restricts Friend leukemia virus infection and pathogenesis in vivo. J. Virol. 2008, 82, 10998–11008. [Google Scholar] [CrossRef] [PubMed]
- Sanville, B.; Dolan, M.A.; Wollenberg, K.; Yan, Y.; Martin, C.; Yeung, M.L.; Strebel, K.; Buckler-White, A.; Kozak, C.A. Adaptive evolution of Mus Apobec3 includes retroviral insertion and positive selection at two clusters of residues flanking the substrate groove. PLoS Pathog. 2010, 6, e1000974. [Google Scholar] [CrossRef] [PubMed]
- Gardner, M.B.; Chiri, A.; Dougherty, M.F.; Casagrande, J.; Estes, J.D. Congenital transmission of murine leukemia virus from wild mice prone to the development of lymphoma and paralysis. J. Natl. Cancer Inst. 1979, 62, 63–70. [Google Scholar] [PubMed]
- Portis, J.L.; McAtee, F.J.; Hayes, S.F. Horizontal transmission of murine retroviruses. J. Virol. 1987, 61, 1037–1044. [Google Scholar] [PubMed]
- Hesse, I.; Luz, A.; Kohleisen, B.; Erfle, V.; Schmidt, J. Prenatal transmission and pathogenicity of endogenous ecotropic murine leukemia virus Akv. Lab. Anim. Sci. 1999, 49, 488–495. [Google Scholar] [PubMed]
- O'Neill, R.R.; Hartley, J.W.; Repaske, R.; Kozak, C.A. Amphotropic proviral envelope sequences are absent from the Mus germ line. J. Virol. 1987, 61, 2225–2231. [Google Scholar] [PubMed]
- Weber, W.J. Diseases Transmitted by Rats and Mice; Thomas Publishers: Fresno, CA, USA, 1982. [Google Scholar]
- Martin, J.; Herniou, E.; Cook, J.; O'Neill, R.W.; Tristem, M. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J. Virol. 1999, 73, 2442–2449. [Google Scholar] [PubMed]
- Hayward, A.; Grabherr, M.; Jern, P. Broad-scale phylogenomics provides insights into retrovirus-host evolution. Proc. Natl. Acad. Sci. USA 2013, 110, 20146–20151. [Google Scholar] [CrossRef] [PubMed]
- Tarlinton, R.E.; Meers, J.; Young, P.R. Retroviral invasion of the koala genome. Nature 2006, 442, 79–81. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M.M.; Sherr, C.J.; Todaro, G.J.; Benveniste, R.E.; Callahan, R.; Coon, H.G. Isolation from the Asian mouse Mus caroli of an endogenous type C virus related to infectious primate type C viruses. Proc. Natl. Acad. Sci. USA 1975, 72, 2315–2319. [Google Scholar] [CrossRef] [PubMed]
- Chaipan, C.; Dilley, K.A.; Paprotka, T.; Delviks-Frankenberry, K.A.; Venkatachari, N.J.; Hu, W.S.; Pathak, V.K. Severe restriction of xenotropic murine leukemia virus-related virus replication and spread in cultured human peripheral blood mononuclear cells. J. Virol. 2011, 85, 4888–4897. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, G.Q.; Kearney, M.F.; Spindler, J.; Wiegand, A.; Chertova, E.; Roser, J.D.; Estes, J.D.; Hao, X.P.; Trubey, C.M.; Lara, A.; et al. Restricted replication of xenotropic murine leukemia virus-related virus in pigtailed macaques. J. Virol. 2012, 86, 3152–3166. [Google Scholar] [CrossRef] [PubMed]
- Kozak, C.A. Evolution of different antiviral strategies in wild mouse populations exposed to different gammaretroviruses. Curr. Opin. Virol. 2013, 3, 657–663. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Buckler-White, A.; Wollenberg, K.; Kozak, C.A. The avian XPR1 gammaretrovirus receptor is under positive selection and is disabled in bird species in contact with virus-infected wild mice. J. Virol. 2013, 87, 10094–10104. [Google Scholar] [CrossRef] [PubMed]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kozak, C.A. Origins of the Endogenous and Infectious Laboratory Mouse Gammaretroviruses. Viruses 2015, 7, 1-26. https://doi.org/10.3390/v7010001
Kozak CA. Origins of the Endogenous and Infectious Laboratory Mouse Gammaretroviruses. Viruses. 2015; 7(1):1-26. https://doi.org/10.3390/v7010001
Chicago/Turabian StyleKozak, Christine A. 2015. "Origins of the Endogenous and Infectious Laboratory Mouse Gammaretroviruses" Viruses 7, no. 1: 1-26. https://doi.org/10.3390/v7010001
APA StyleKozak, C. A. (2015). Origins of the Endogenous and Infectious Laboratory Mouse Gammaretroviruses. Viruses, 7(1), 1-26. https://doi.org/10.3390/v7010001





