3. Clustering of Genomic Sequences
In this section, we propose a method for clustering genomic sequences based on variations in local entropy. The analysis includes several viruses from the family of
Phycodnaviridae as well as eukaryotic organisms. The latter comprise higher and lower plants. The algae, which represent the lower plants, are specific hosts of the viruses [
4,
5]. For the relationship between the lower and the higher plants, it is worth noting that the Chlorella species belong to the green algae and are hence considered ancestors of the higher plants. The brown alga
Ectocarpus, on the other hand, belongs to the
Heterocontophyta, a group of algae, which separated very early from the green algae [
6].
Viruses in the family of the
Phycodnaviridae are huge, icosahedral viruses with large double-stranded DNA genomes. They replicate in a host specific manner in algae [
4]. Virus EsV1 has, as a specific host, the brown alga
Ectocarpus siliculosus. The viruses PBCV-1, NY2A and AR158 replicate exclusively in the chlorella species C. NC64A. The hosts for the remaining viruses are closely related Chlorella species for which we have no sequence information yet [
4]. The viruses considered here are very suitable for this analysis. The annotation of the 330 kbp genome of the prototype virus
Paramecium bursaria chlorella virus 1 (PBCV-1) identified
ca. 366 protein-encoding genes and 11
tRNA genes. More than half of the predicted gene products resemble proteins from pro- and eukaryotes with a known function [
4]. It is interesting to note is that these virus-encoded proteins are either the smallest or among the smallest proteins of their class; some are so much reduced that they represent not more than the minimal catalytic unit. A further interesting feature of these viruses is that they also have, unlike most other viruses, introns. Virus PBCV-1 for example has three types of introns: a self-splicing intron, a spliceosomal processed intron, and a small tRNA intron.
Accumulating evidence indicates that the chlorella viruses have a very long evolutionary history possibly dating back to the time when eukaryotes arose from prokaryotes [
7,
8,
9]. They are predicted to have a common ancestor with the poxviruses (e.g., vaccinia virus), asfarvirues, iridoviruses, ascoviruses and mimiviruses [
7,
8]. Collectively, these viruses are referred to as nucleocytoplasmic large DNA viruses (NCLDVs). We now show that the local variations in entropy are not only very useful for clustering viruses and plant genomes, but may also suggest host specificity of viruses.
We start with analyzing the superinformation content of viruses and plant genomes. The choice of block size
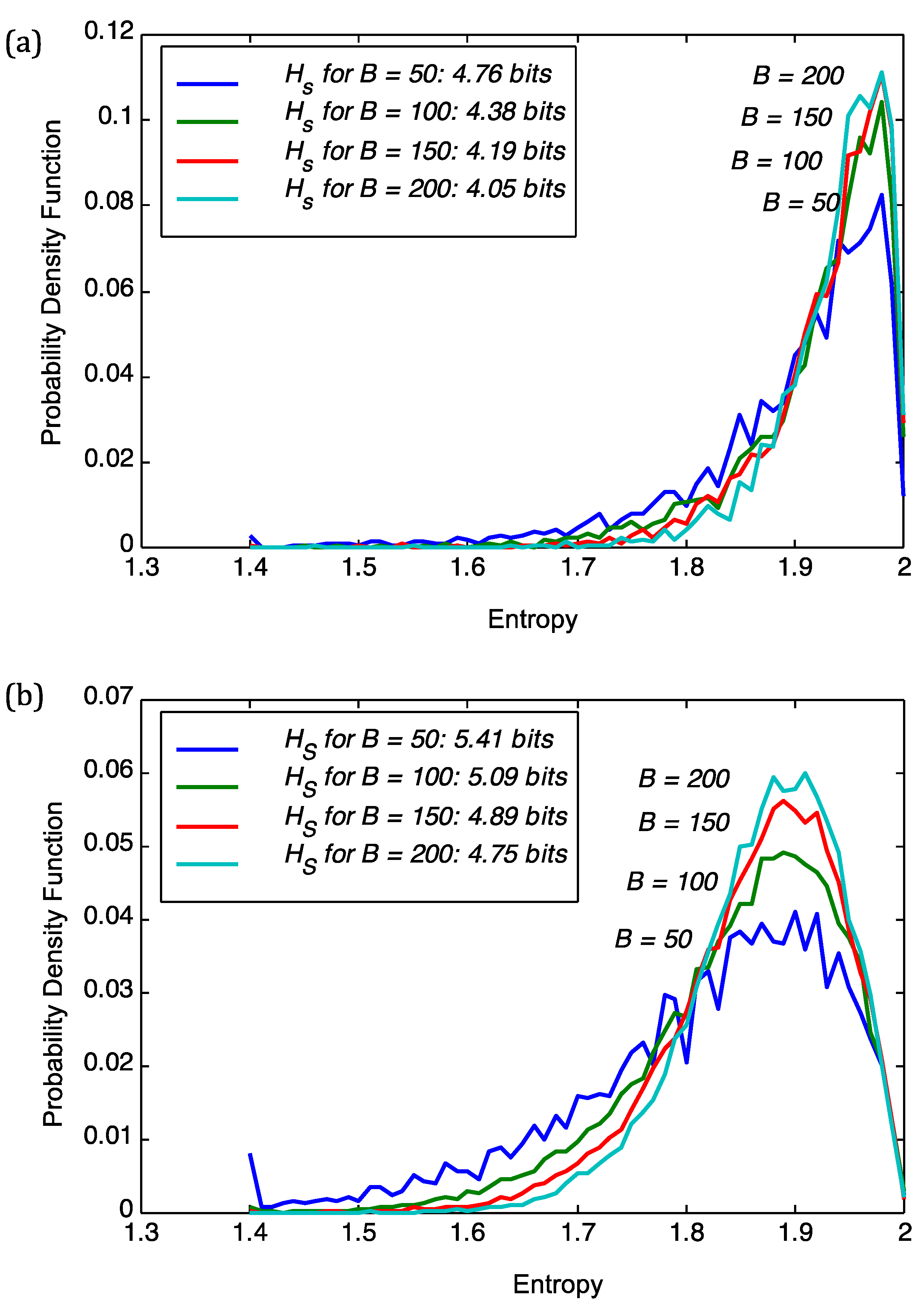
B in Equation (3) was adjusted to stabilize the results for the superinformation, as shown in
Figure 1. Intuitively,
B defines the resolution at which superinformation is calculated. The figure shows the sensitivity of the probability density functions (pdfs) of sequence entropies with respect to block size
B for the viral genome of PBCV1 and
Chlorella NC64A. From
Figure 1 we deduce that a choice of
B = 100 implies stability of the subsequent analysis. For
B = 50 we obtained severe fluctuations in the pdfs as is the case for both PBCV1 and
Chlorella NC64A in
Figure 1. For
B = 100,
B = 150, and
B = 200 we obtain more regular histograms and therefore more stable superinformation values. However, the smaller the
B the better the resolution and this we have opted to use
B = 100 in the subsequent parts as it provides stability and good resolution at the same time.
Similar sensitivities have been observed for the other seven plant viral genomes (AR158, ATCV, CVM1, FR483, MR325, NY2A, TN603),
Arabidopsis thaliana,
Ectocarpus siliculosus and EsV-1. Thus, we use this value of
B in the subsequent parts of this study. It should be noted that the block-size (
B = 100) is much smaller than the typical length of isochores (homogeneous domains) [
10]. For example, in
Arabidopsis genome, the length of GC isochore is of the order of 1 million base pairs [
11].
Figure 1.
Sensitivity of the probability density function (pdf) of sequence entropies with respect to block size B for (a) the viral genome of PBCV1; and (b) Chlorella NC64A. We also show the superinformation Hs.
Figure 1.
Sensitivity of the probability density function (pdf) of sequence entropies with respect to block size B for (a) the viral genome of PBCV1; and (b) Chlorella NC64A. We also show the superinformation Hs.
In
Table 1 we show the superinformation values for the genomic data of two plant hosts and their respective viruses. These results suggest the existence of a distinct pattern in these genomic sequences, implying differing selective pressures that shape the range of
local variability (block entropies) and of
global variability (superinformation). To derive evolutionary distances between all the sequences, the superinformation
Hs is, however, unsuitable. It should be noted that due to Chargaff's second parity G%~C%, A%~C%, plus the fact that GC% + AT% = 1, four base compositions (three are independent) in a block are reduced to one variable: GC%. So the entropy of a block, more or less has, a one-to-one correspondence with the GC%, and distribution of block-entropies are similar to distribution of a function of GC%. Thus the superinformation for the whole sequence corresponds to some measure of the window-GC% distribution (e.g., variance).
Table 1.
Superinformation values Hs for various genomic sequences. Note the tendency of increased Hs for organisms in comparison to viruses.
Table 1.
Superinformation values Hs for various genomic sequences. Note the tendency of increased Hs for organisms in comparison to viruses.
| Genomic Sequence | Hs [Bit] |
|---|
| AR158 | 4.21 |
| ATCV | 3.62 |
| CVM1 | 3.87 |
| FR483 | 3.83 |
| MT325 | 3.77 |
| NY2A | 4.22 |
| PBCV1 | 4.38 |
| TN603 | 3.65 |
| EsV-1 | 3.54 |
| Arabidopsis thaliana | 5.27 |
| Chlorella NC64A | 5.10 |
| Oryza sativa | 4.74 |
| Ectocarpus siliculosus | 4.10 |
Continuing with the rationale above that the distribution of
local sequence entropies is a signal for evolutionary pressure of the environment, we decided to take one step back and use these distributions of block entropies
pj(
Xi,
M) instead. These distributions describe, in detail, the
local variability of the sequences and thus any distance of such
pj(
Xi,
M) clusters entities based on the variability of entropies. The distributions of the block entropies of the genomic sequences are shown in
Figure 2. Upon visual inspection, there appears to be a distinct pattern for viruses and plant genomes. The distributions for plant genomes show a larger variance as compared to that of the viruses. This motivates us to explore clustering based on the distributions of block entropies. We note that characterization of sequences based on the distribution of their sub-sequences have been explored earlier [
12,
13,
14].
Figure 2.
Plot of the probability density function (pdf) of the entropy for 8 plant viral genomes (AR158, ATCV, CVM1, FR483, MR325, NY2A, PBCV1, TN603), Arabidopsis thaliana, Chlorella NC64A, Ectocarpus siliculosus and EsV-1. The plots were obtained for a block size of B = 100.
Figure 2.
Plot of the probability density function (pdf) of the entropy for 8 plant viral genomes (AR158, ATCV, CVM1, FR483, MR325, NY2A, PBCV1, TN603), Arabidopsis thaliana, Chlorella NC64A, Ectocarpus siliculosus and EsV-1. The plots were obtained for a block size of B = 100.
We can quantify the differences in the respective
pj(
Xi,
M) by generalized Kullback-Leibler divergences [
15], the Jensen-Shannon-distances in particular [
16] as discussed in [
17,
18]. The Jensen-Shannon-divergence,
DJS(
p,
q), between the entropy distributions
pi(
Xi,
M) and
qj(
Xi,
M) for two data sources is given by
Here,
mj(
Xi,
M) is an “intermediate” distribution and the
DKL(
p||
m) and
DKL(
q||
m) are the Kullback-Leibler distances between
p and
m or between
q and
m, respectively. A suitable metric for clustering is then
d (
p,q) [
16]. The application of Jensen-Shannon distance to DNA has also been carried out by previous researchers [
19].
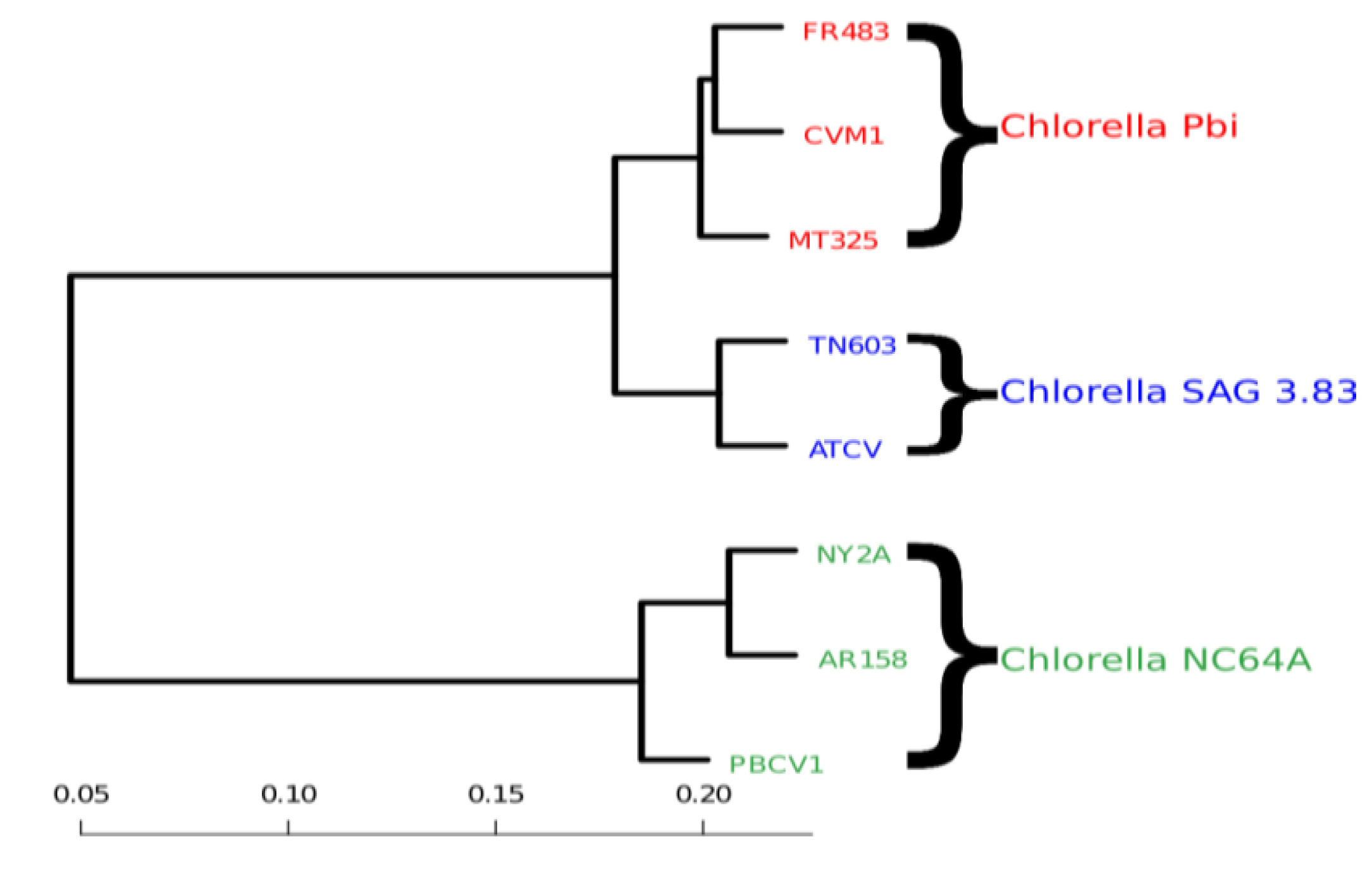
We first computed the distances
d(
p,q) between all genomes that infest chlorella algae. We then used distance-matrix based clustering [
20] as implemented in the statistical software R (R2009). In
Figure 3, we clearly see that the genomic variability is able to reflect the host specificity of the viruses. The viruses that have a common host are closer to one another in terms of their block-entropy distributions. For example, the viruses FR483, CVM1 and MT325 that specifically infect Chlorella Pbi, are clustered together. The viruses PBCV-1, NY2A and AR158 that replicate exclusively in the chlorella species C. NC64A, are clustered together. This is a valuable finding and probably suggests that the respective host environment gives rise to unique selective pressures. It should be noted that (i) MT325 and FR483 are strongly related genomes sharing 94% of their genes with an average 86% amino-acid sequence identity and an almost identical gene order [
21] and that (ii) both of these genomes are related to PBCV-1, but to a lesser extent (82%, 50%, weak gene order conservation). Hence, the identification of MT325 and FR483 (infecting the same host) to be more closely related than they are to PBCV-1 would be captured by sequence alignment tools.
Figure 3.
Clustering of viral genomes, annotated by their respective hosts—in this case variants of the Chlorella algae. The branch lengths are proportional to the phylogenetic Jensen-Shannon distance of Equation (4), the scale is indicated at the bottom.
Figure 3.
Clustering of viral genomes, annotated by their respective hosts—in this case variants of the Chlorella algae. The branch lengths are proportional to the phylogenetic Jensen-Shannon distance of Equation (4), the scale is indicated at the bottom.
These findings motivated us to augment the data set by including: (a) the host genome itself (
Chlorella NC64A, [
22]); (b) an independent host-virus genome pair (
Ectocarpus siliculosus, [
5]); and (c) other plant genomes, of which only a few are available up to this day. The plant genomes that we have included in our study belong to Populus trichocarpa (black cottonwood),
Oryza sativa (Asian rice) and
Arabidopsis (a small flowering plant). This set-up allows us to investigate whether the evolution of the host genomes is subject to the same evolutionary dynamics, leading to similar variabilities in the
local entropies, as is the case for their respective viruses. By this, we can judge whether there exists differential evolutionary pressure on host and pathogenic genomes.
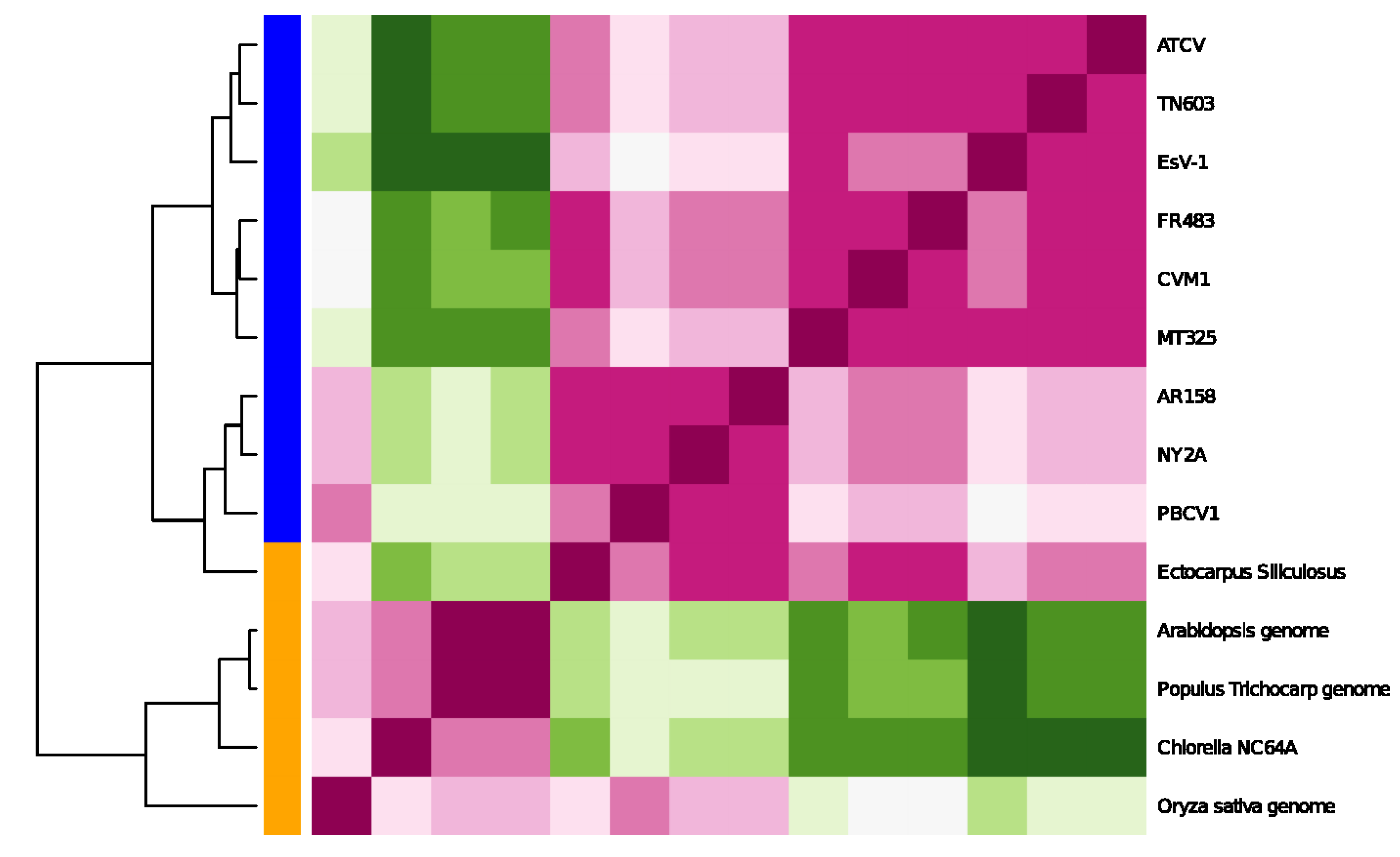
Figure 4 shows the results of this experiment.
Figure 4.
Clustering by the entropy-variability distance of Equation (1) for the extended genomic sequence set. We show the distance matrix d(p,q) for any genomic sequence pairing (p,q) for each genomic sequence with respect to each sequence (center plane, red = small values, green = high values). The bar on the left indicates whether the sequence is of viral origin (blue) or a living organism (orange). Note that the tree is not rooted.
Figure 4.
Clustering by the entropy-variability distance of Equation (1) for the extended genomic sequence set. We show the distance matrix d(p,q) for any genomic sequence pairing (p,q) for each genomic sequence with respect to each sequence (center plane, red = small values, green = high values). The bar on the left indicates whether the sequence is of viral origin (blue) or a living organism (orange). Note that the tree is not rooted.
We see a clear separation of the genomic sequences when we cluster based on local entropy of the genomic data. In particular, there is a clear separation between viral and host genomes. This is a very interesting result and shows that clustering based on local entropy is able to assign the plants and viruses to different groups. The green alga and the higher plants form a sub-clade, while the plant viruses form a separate one. This comes out very clearly in
Figure 4. The other interesting observation is that the brown alga (
Ectocarpus siliculosus) is separate from the green alga (
Chlorella) and other higher plants (Populus trichocarpa,
Oryza sativa and
Arabidopsis). This is consistent with the fact that these plants separated very early in evolution [
6]. The fact that the genome of
Ectocarpus silicolosus includes the entire genome of the respective virus EsV1 [
5] may contribute to this separate position. Interestingly, the viruses infecting
Chlorella subspecies include the
Ectocarpus silicolosus virus; this occurs even though the viruses have very different lifestyles [
4].
 , where, X is a random variable with realizations {x1, x2, …, xN} drawn from a discrete sampling space S. The p(xi) is the probability that xi occurs. In the case of genomes, S is the nucleotide alphabet of the DNA. When applied at the nucleotide level to genomic sequences, the sequence entropy for the full genome can be reduced to the question of CG-content [1,2]. This is a global property and related to the mutation rate, while the selective advantage reveals itself locally in, e.g., gene products such as regions coding for proteins.
, where, X is a random variable with realizations {x1, x2, …, xN} drawn from a discrete sampling space S. The p(xi) is the probability that xi occurs. In the case of genomes, S is the nucleotide alphabet of the DNA. When applied at the nucleotide level to genomic sequences, the sequence entropy for the full genome can be reduced to the question of CG-content [1,2]. This is a global property and related to the mutation rate, while the selective advantage reveals itself locally in, e.g., gene products such as regions coding for proteins.