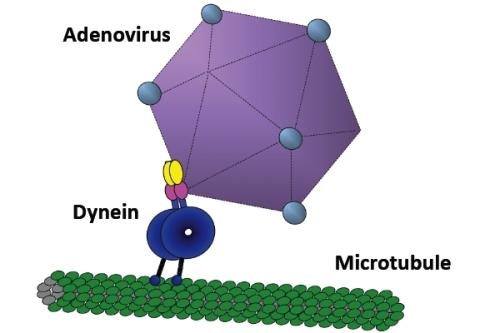
Adenovirus Recruits Dynein by an Evolutionary Novel Mechanism Involving Direct Binding to pH-Primed Hexon
Abstract
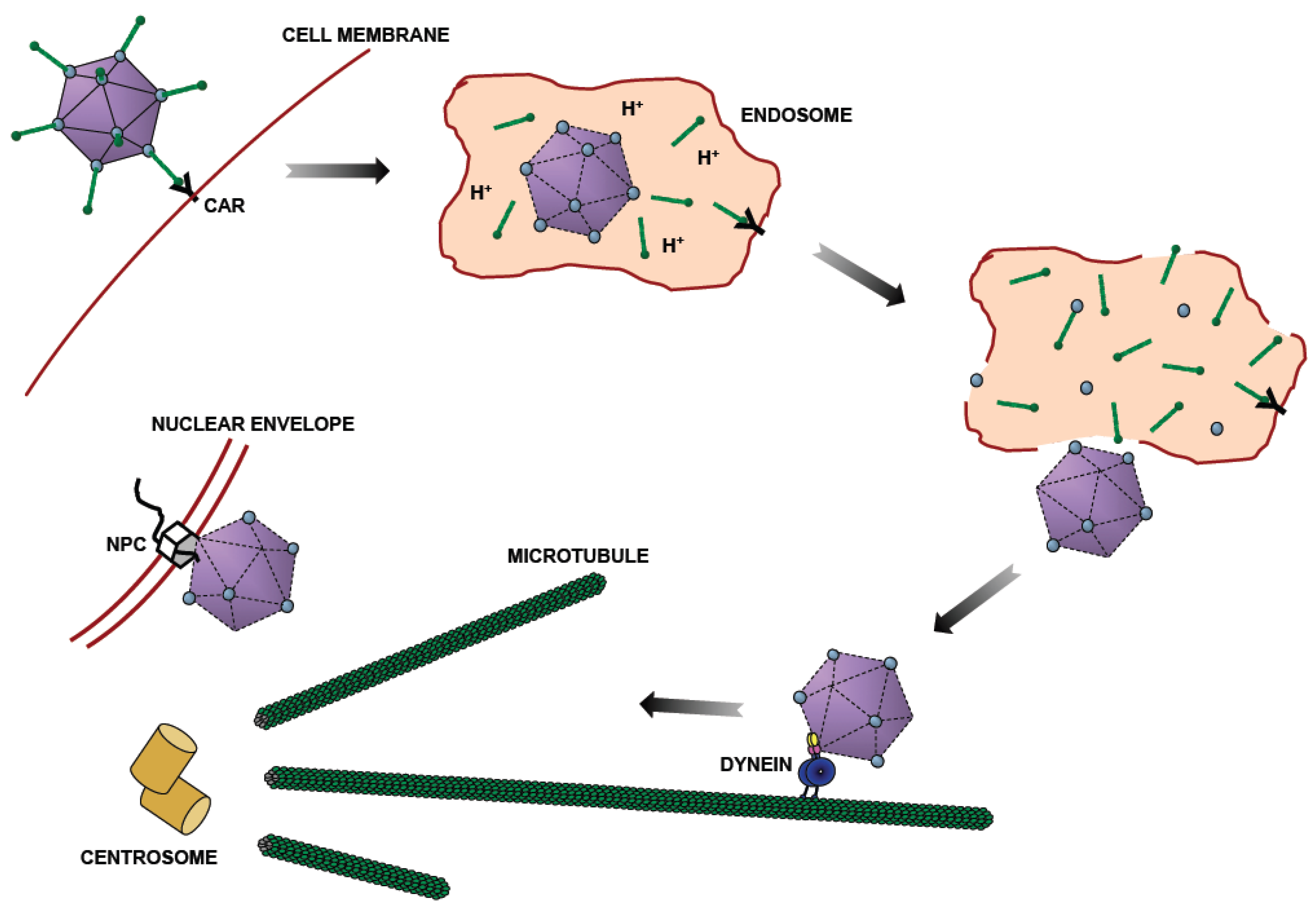
:1. Introduction—Role of Microtubule Motors in Adenovirus Transport
2. Identification of Viral Capsid Proteins Involved in Cytoplasmic Dynein-Mediated Motility
3. pH-Dependent Biochemical Characteristics of the Adenovirus Capsid and Hexon
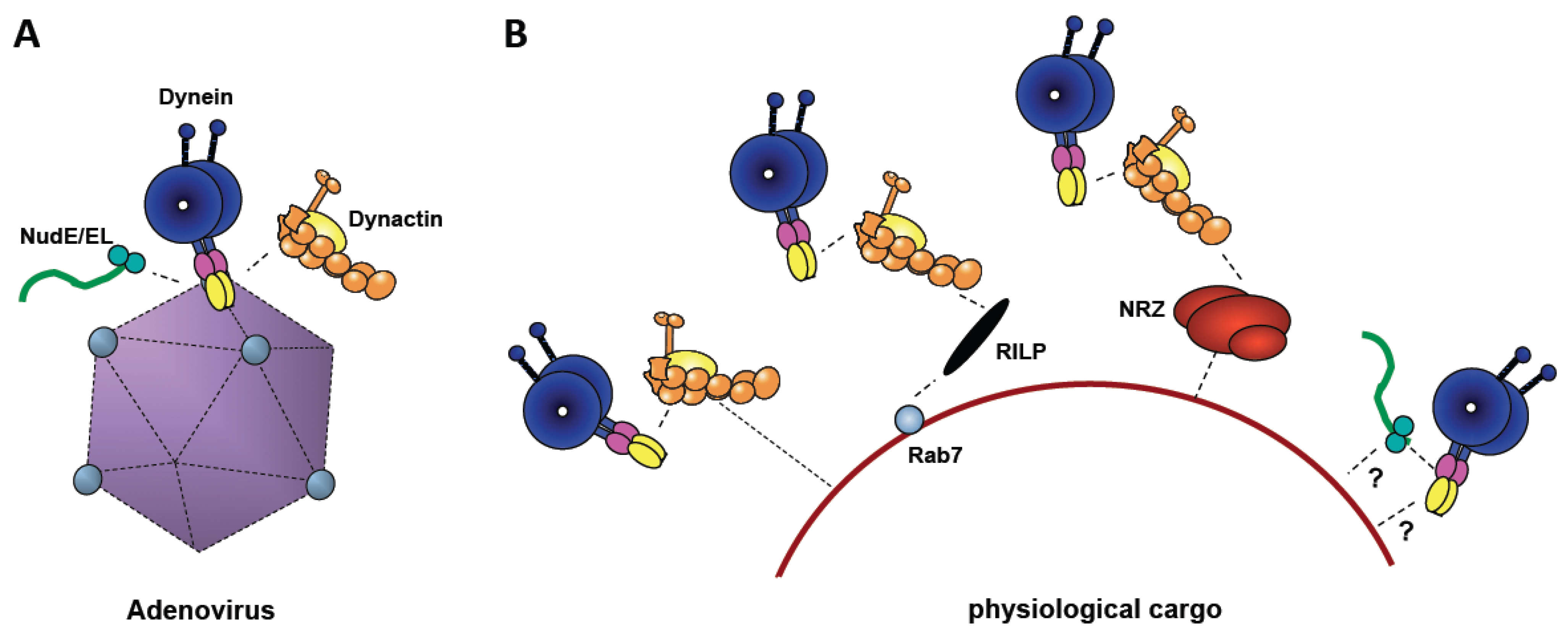
4. Identification of Hexon-Interacting Cytoplasmic Dynein Subunit
5. Involvement of Cytoplasmic Dynein Regulators in Adenovirus Transport
6. Evolutionary Origin of Dynein Recruitment by Viruses
7. Bidirectional Intracellular Adenovirus Motility
8. Conclusions
Conflict of Interest
References and Notes
- Rowe, W.P.; Huebner, R.J.; Gilmore, L.K.; Parrott, R.H.; Ward, T.G. Isolation of a cytopathogenic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 1953, 84, 570–573. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.S.; Natchiar, S.K.; Stewart, P.L.; Nemerow, G.R. Crystal structure of human adenovirus at 3.5 A resolution. Science 2010, 329, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Jin, L.; Koh, S.B.; Atanasov, I.; Schein, S.; Wu, L.; Zhou, Z.H. Atomic structure of human adenovirus by cryo-EM reveals interactions among protein networks. Science 2010, 329, 1038–1043. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.L.; Fuller, S.D.; Burnett, R.M. Difference imaging of adenovirus: Bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 1993, 12, 2589–2599. [Google Scholar] [CrossRef]
- Rexroad, J.; Wiethoff, C.M.; Green, A.P.; Kierstead, T.D.; Scott, M.O.; Middaugh, C.R. Structural stability of adenovirus type 5. J. Pharm. Sci. 2003, 92, 665–678. [Google Scholar] [CrossRef]
- Rux, J.J.; Kuser, P.R.; Burnett, R.M. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J. Virol. 2003, 77, 9553–9566. [Google Scholar] [CrossRef]
- Vellinga, J.; Van der Heijdt, S.; Hoeben, R.C. The adenovirus capsid: Major progress in minor proteins. J. Gen. Virol. 2005, 86, 1581–1588. [Google Scholar] [CrossRef]
- Patterson, S.; Russell, W.C. Ultrastructural and immunofluorescence studies of early events in adenovirus-HeLa cell interactions. J. Gen. Virol. 1983, 64, 1091–1099. [Google Scholar] [CrossRef]
- Chardonnet, Y.; Dales, S. Early events in the interaction of adenoviruses with HeLa cells. I. Penetration of type 5 and intracellular release of the DNA genome. Virology 1970, 40, 462–477. [Google Scholar] [CrossRef]
- Nakano, M.Y.; Boucke, K.; Suomalainen, M.; Stidwill, R.P.; Greber, U.F. The first step of adenovirus type 2 disassembly occurs at the cell surface, independently of endocytosis and escape to the cytosol. J. Virol. 2000, 74, 7085–7095. [Google Scholar] [CrossRef]
- Wiethoff, C.M.; Wodrich, H.; Gerace, L.; Nemerow, G.R. Adenovirus protein VI mediates membrane disruption following capsid disassembly. J. Virol. 2005, 79, 1992–2000. [Google Scholar] [CrossRef] [PubMed]
- Greber, U.F.; Willetts, M.; Webster, P.; Helenius, A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 1993, 75, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Suomalainen, M.; Nakano, M.Y.; Keller, S.; Boucke, K.; Stidwill, R.P.; Greber, U.F. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 1999, 144, 657–672. [Google Scholar] [CrossRef] [PubMed]
- Trotman, L.C.; Mosberger, N.; Fornerod, M.; Stidwill, R.P.; Greber, U.F. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 2001, 3, 1092–1100. [Google Scholar] [CrossRef]
- Suomalainen, M.; Nakano, M.Y.; Boucke, K.; Keller, S.; Greber, U.F. Adenovirus-activated PKA and p38/MAPK pathways boost microtubule-mediated nuclear targeting of virus. EMBO J. 2001, 20, 1310–1319. [Google Scholar] [CrossRef] [PubMed]
- Martin-Fernandez, M.; Longshaw, S.V.; Kirby, I.; Santis, G.; Tobin, M.J.; Clarke, D.T.; Jones, G.R. Adenovirus type-5 entry and disassembly followed in living cells by FRET, fluorescence anisotropy, and FLIM. Biophys. J. 2004, 87, 1316–1327. [Google Scholar] [CrossRef] [PubMed]
- Seth, P.; Willingham, M.C.; Pastan, I. Binding of adenovirus and its external proteins to Triton X-114. Dependence on pH. J. Biol. Chem. 1985, 260, 14431–14434. [Google Scholar] [CrossRef]
- Maier, O.; Galan, D.L.; Wodrich, H.; Wiethoff, C.M. An N-terminal domain of adenovirus protein VI fragments membranes by inducing positive membrane curvature. Virology 2010, 402, 11–19. [Google Scholar] [CrossRef]
- Gastaldelli, M.; Imelli, N.; Boucke, K.; Amstutz, B.; Meier, O.; Greber, U.F. Infectious adenovirus type 2 transport through early but not late endosomes. Traffic 2008, 9, 2265–2278. [Google Scholar] [CrossRef]
- Miyazawa, N.; Crystal, R.G.; Leopold, P.L. Adenovirus serotype 7 retention in a late endosomal compartment prior to cytosol escape is modulated by fiber protein. J. Virol. 2001, 75, 1387–1400. [Google Scholar] [CrossRef]
- Miyazawa, N.; Leopold, P.L.; Hackett, N.R.; Ferris, B.; Worgall, S.; Falck-Pedersen, E.; Crystal, R.G. Fiber swap between adenovirus subgroups B and C alters intracellular trafficking of adenovirus gene transfer vectors. J. Virol. 1999, 73, 6056–6065. [Google Scholar] [CrossRef]
- Miles, B.D.; Luftig, R.B.; Weatherbee, J.A.; Weihing, R.R.; Weber, J. Quantitation of the interaction between adenovirus types 2 and 5 and microtubules inside infected cells. Virology 1980, 105, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Dales, S.; Chardonnet, Y. Early events in the interaction of adenoviruses with HeLa cells. IV. Association with microtubules and the nuclear pore complex during vectorial movement of the inoculum. Virology 1973, 56, 465–483. [Google Scholar] [CrossRef] [PubMed]
- Leopold, P.L.; Kreitzer, G.; Miyazawa, N.; Rempel, S.; Pfister, K.K.; Rodriguez-Boulan, E.; Crystal, R.G. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum. Gene Ther. 2000, 11, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Bremner, K.H.; Scherer, J.; Yi, J.L.; Vershinin, M.; Gross, S.P.; Vallee, R.B. Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe 2009, 6, 523–535. [Google Scholar] [CrossRef]
- Mabit, H.; Nakano, M.Y.; Prank, U.; Saam, B.; Dohner, K.; Sodeik, B.; Greber, U.F. Intact microtubules support adenovirus and herpes simplex virus infections. J. Virol. 2002, 76, 9962–9971. [Google Scholar] [CrossRef]
- Bailey, C.J.; Crystal, R.G.; Leopold, P.L. Association of adenovirus with the microtubule organizing center. J. Virol. 2003, 77, 13275–13287. [Google Scholar] [CrossRef]
- Wodrich, H.; Henaff, D.; Jammart, B.; Segura-Morales, C.; Seelmeir, S.; Coux, O.; Ruzsics, Z.; Wiethoff, C.M.; Kremer, E.J. A capsid-encoded PPxY-motif facilitates adenovirus entry. PLoS Pathog. 2010, 6, e1000808. [Google Scholar] [CrossRef]
- Meulenbroek, R.A.; Sargent, K.L.; Lunde, J.; Jasmin, B.J.; Parks, R.J. Use of adenovirus protein IX (pIX) to display large polypeptides on the virion—Generation of fluorescent virus through the incorporation of pIX-GFP. Mol. Ther. 2004, 9, 617–624. [Google Scholar] [CrossRef]
- Gazzola, M.; Burckhardt, C.J.; Bayati, B.; Engelke, M.; Greber, B.S.; Koumoutsakos, P. A stochastic model for microtubule motors describes the in vivo cytoplasmic transport of human adenovirus. PLoS Comput. Biol. 2009, 5, e1000623. [Google Scholar] [CrossRef]
- Saphire, A.C.; Guan, T.; Schirmer, E.C.; Nemerow, G.R.; Gerace, L. Nuclear import of adenovirus DNA in vitro involves the nuclear protein import pathway and hsc70. J. Biol. Chem. 2000, 275, 4298–4304. [Google Scholar] [CrossRef] [PubMed]
- Greber, U.F.; Suomalainen, M.; Stidwill, R.P.; Boucke, K.; Ebersold, M.W.; Helenius, A. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 1997, 16, 5998–6007. [Google Scholar] [CrossRef] [PubMed]
- Kelkar, S.A.; Pfister, K.K.; Crystal, R.G.; Leopold, P.L. Cytoplasmic dynein mediates adenovirus binding to microtubules. J. Virol. 2004, 78, 10122–10132. [Google Scholar] [CrossRef] [PubMed]
- Tycko, B.; Maxfield, F.R. Rapid acidification of endocytic vesicles containing alpha 2-macroglobulin. Cell 1982, 28, 643–651. [Google Scholar] [CrossRef]
- Cain, C.C.; Sipe, D.M.; Murphy, R.F. Regulation of endocytic pH by the Na+,K+-ATPase in living cells. Proc. Natl. Acad. Sci. USA 1989, 86, 544–548. [Google Scholar] [CrossRef]
- Saftig, P.; Klumperman, J. Lysosome biogenesis and lysosomal membrane proteins: Trafficking meets function. Nat. Rev. Mol. Cell Biol. 2009, 10, 623–635. [Google Scholar] [CrossRef]
- Waris, M.; Halonen, P. Purification of adenovirus hexon protein by high-performance liquid chromatography. J. Chromatogr. 1987, 397, 321–325. [Google Scholar] [CrossRef]
- Seth, P.; Pastan, I.; Willingham, M.C. Adenovirus-dependent increase in cell membrane permeability. J. Biol. Chem. 1985, 260, 9598–9602. [Google Scholar] [CrossRef]
- Smith, J.G.; Wiethoff, C.M.; Stewart, P.L.; Nemerow, G.R. Adenovirus. Curr. Top. Microbiol. Immunol. 2010, 343, 195–224. [Google Scholar]
- Wodrich, H.; Guan, T.; Cingolani, G.; Von Seggern, D.; Nemerow, G.; Gerace, L. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 2003, 22, 6245–6255. [Google Scholar] [CrossRef]
- Tynan, S.H.; Gee, M.A.; Vallee, R.B. Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J. Biol. Chem. 2000, 275, 32769–32774. [Google Scholar] [CrossRef] [PubMed]
- Everitt, E.; Persson, M.J.; Wohlfart, C. Ph-Dependent Exposure of endoproteolytic cleavage sites of the Adenovirus-2 hexon protein. Fems. Microbiol. Lett. 1988, 49, 229–233. [Google Scholar] [CrossRef]
- Vallee, R.B.; Wall, J.S.; Paschal, B.M.; Shpetner, H.S. Microtubule-associated protein 1C from brain is a two-headed cytosolic dynein. Nature 1988, 332, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Pfister, K.K.; Fisher, E.M.; Gibbons, I.R.; Hays, T.S.; Holzbaur, E.L.; McIntosh, J.R.; Porter, M.E.; Schroer, T.A.; Vaughan, K.T.; Witman, G.B.; et al. Cytoplasmic dynein nomenclature. J. Cell Biol. 2005, 171, 411–413. [Google Scholar] [CrossRef]
- Williams, J.C.; Roulhac, P.L.; Roy, A.G.; Vallee, R.B.; Fitzgerald, M.C.; Hendrickson, W.A. Structural and thermodynamic characterization of a cytoplasmic dynein light chain-intermediate chain complex. Proc. Natl. Acad. Sci. USA 2007, 104, 10028–10033. [Google Scholar] [CrossRef] [PubMed]
- Barbar, E. Dynein light chain LC8 is a dimerization hub essential in diverse protein networks. Biochemistry 2008, 47, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Ligon, L.A.; Karki, S.; Tokito, M.; Holzbaur, E.L. Dynein binds to beta-catenin and may tether microtubules at adherens junctions. Nat. Cell Biol. 2001, 3, 913–917. [Google Scholar] [CrossRef]
- Vaughan, K.T.; Vallee, R.B. Cytoplasmic dynein binds dynactin through a direct interaction between the intermediate chains and p150Glued. J. Cell Biol. 1995, 131, 1507–1516. [Google Scholar] [CrossRef]
- Karki, S.; Holzbaur, E.L. Affinity chromatography demonstrates a direct binding between cytoplasmic dynein and the dynactin complex. J. Biol. Chem. 1995, 270, 28806–28811. [Google Scholar] [CrossRef]
- Stehman, S.A.; Chen, Y.; McKenney, R.J.; Vallee, R.B. NudE and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J. Cell Biol. 2007, 178, 583–594. [Google Scholar] [CrossRef]
- Schmoranzer, J.; Fawcett, J.P.; Segura, M.; Tan, S.; Vallee, R.B.; Pawson, T.; Gundersen, G.G. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr. Biol. 2009, 19, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- Tynan, S.H.; Purohit, A.; Doxsey, S.J.; Vallee, R.B. Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J. Biol. Chem. 2000, 275, 32763–32768. [Google Scholar] [CrossRef] [PubMed]
- Horgan, C.P.; Hanscom, S.R.; Jolly, R.S.; Futter, C.E.; McCaffrey, M.W. Rab11-FIP3 links the Rab11 GTPase and cytoplasmic dynein to mediate transport to the endosomal-recycling compartment. J. Cell Sci. 2010, 123, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.C.; Scherer, J.; Vallee, R.B. Recruitment of dynein to late endosomes and lysosomes through light intermediate chains. Mol. Biol. Cell 2011, 22, 467–477. [Google Scholar] [CrossRef]
- Palmer, K.J.; Hughes, H.; Stephens, D.J. Specificity of cytoplasmic dynein subunits in discrete membrane-trafficking steps. Mol. Biol. Cell 2009, 20, 2885–2899. [Google Scholar] [CrossRef]
- Burkhardt, J.K.; Echeverri, C.J.; Nilsson, T.; Vallee, R.B. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 1997, 139, 469–484. [Google Scholar] [CrossRef]
- Roghi, C.; Allan, V.J. Dynamic association of cytoplasmic dynein heavy chain 1a with the Golgi apparatus and intermediate compartment. J. Cell Sci. 1999, 112, 4673–4685. [Google Scholar] [CrossRef]
- Vergnolle, M.A.; Taylor, S.S. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr. Biol. 2007, 17, 1173–1179. [Google Scholar] [CrossRef]
- Liang, Y.; Yu, W.; Li, Y.; Yu, L.; Zhang, Q.; Wang, F.; Yang, Z.; Du, J.; Huang, Q.; Yao, X.; et al. Nudel modulates kinetochore association and function of cytoplasmic dynein in M phase. Mol. Biol. Cell 2007, 18, 2656–2666. [Google Scholar] [CrossRef]
- King, S.J.; Schroer, T.A. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat. Cell Biol. 2000, 2, 20–24. [Google Scholar] [CrossRef]
- Kardon, J.R.; Reck-Peterson, S.L.; Vale, R.D. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 5669–5674. [Google Scholar] [CrossRef]
- McKenney, R.J.; Vershinin, M.; Kunwar, A.; Vallee, R.B.; Gross, S.P. LIS1 and NudE induce a persistent dynein force-producing state. Cell 2010, 141, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Bingham, J.B.; King, S.J.; Schroer, T.A. Purification of dynactin and dynein from brain tissue. Methods Enzymol. 1998, 298, 171–184. [Google Scholar] [PubMed]
- Scherer, J. Neither purified dynactin nor NudE interact with acidified hexon. Department of Pathology and Cell Biology, Columbia University, New York, NY, USA. Unpublished work, 2011.
- Engelke, M.F.; Burckhardt, C.J.; Morf, M.K.; Greber, U.F. The dynactin complex enhances the speed of microtubule-dependent motions of adenovirus both towards and away from the nucleus. Viruses 2011, 3, 233–253. [Google Scholar] [CrossRef] [PubMed]
- Whyte, J.; Bader, J.R.; Tauhata, S.B.; Raycroft, M.; Hornick, J.; Pfister, K.K.; Lane, W.S.; Chan, G.K.; Hinchcliffe, E.H.; Vaughan, P.S.; et al. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J. Cell Biol. 2008, 183, 819–834. [Google Scholar] [CrossRef]
- Varma, D.; Dujardin, D.L.; Stehman, S.A.; Vallee, R.B. Role of the kinetochore/cell cycle checkpoint protein ZW10 in interphase cytoplasmic dynein function. J. Cell Biol. 2006, 172, 655–662. [Google Scholar] [CrossRef]
- Starr, D.A.; Williams, B.C.; Hays, T.S.; Goldberg, M.L. ZW10 helps recruit dynactin and dynein to the kinetochore. J. Cell Biol. 1998, 142, 763–774. [Google Scholar] [CrossRef]
- Alonso, C.; Miskin, J.; Hernaez, B.; Fernandez-Zapatero, P.; Soto, L.; Canto, C.; Rodriguez-Crespo, I.; Dixon, L.; Escribano, J.M. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J. Virol. 2001, 75, 9819–9827. [Google Scholar] [CrossRef]
- Radtke, K.; Kieneke, D.; Wolfstein, A.; Michael, K.; Steffen, W.; Scholz, T.; Karger, A.; Sodeik, B. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 2010, 6, e1000991. [Google Scholar] [CrossRef]
- Hernaez, B.; Tarrago, T.; Giralt, E.; Escribano, J.M.; Alonso, C. Small peptide inhibitors disrupt a high-affinity interaction between cytoplasmic dynein and a viral cargo protein. J. Virol. 2010, 84, 10792–10801. [Google Scholar] [CrossRef]
- Douglas, M.W.; Diefenbach, R.J.; Homa, F.L.; Miranda-Saksena, M.; Rixon, F.J.; Vittone, V.; Byth, K.; Cunningham, A.L. Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J. Biol. Chem. 2004, 279, 28522–28530. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.J.; Vaughan, K.T.; Vallee, R.B.; Roizman, B. The herpes simplex virus 1 U(L)34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 2000, 74, 1355–1363. [Google Scholar] [CrossRef] [PubMed]
- Döhner, K.; Radtke, K.; Schmidt, S.; Sodeik, B. Eclipse phase of herpes simplex virus type 1 infection: Efficient dynein-mediated capsid transport without the small capsid protein VP26. J. Virol. 2006, 80, 8211–8224. [Google Scholar] [CrossRef]
- Scherer, J.; Vallee, R. Mechanism of cytoplasmic dynein recruitment by adenovirus. Mol. Biol. Cell 2010, 21, Abstract No. 1683. [Google Scholar]
- Döhner, K.; Wolfstein, A.; Prank, U.; Echeverri, C.; Dujardin, D.; Vallee, R.; Sodeik, B. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 2002, 13, 2795–2809. [Google Scholar] [CrossRef]
- Sathish, N.; Zhu, F.X.; Yuan, Y. Kaposi's sarcoma-associated herpesvirus ORF45 interacts with kinesin-2 transporting viral capsid-tegument complexes along microtubules. PLoS Pathog. 2009, 5, e1000332. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, R.J.; Miranda-Saksena, M.; Diefenbach, E.; Holland, D.J.; Boadle, R.A.; Armati, P.J.; Cunningham, A.L. Herpes simplex virus tegument protein US11 interacts with conventional kinesin heavy chain. J. Virol. 2002, 76, 3282–3291. [Google Scholar] [CrossRef]
- Morgan, G.W.; Hollinshead, M.; Ferguson, B.J.; Murphy, B.J.; Carpentier, D.C.J.; Smith, G.L. Vaccinia protein F12 has structural similarity to kinesin light chain and contains a motor binding motif required for virion export. PLoS Pathog. 2010, 6, e1000785. [Google Scholar] [CrossRef]
- Suikkanen, S.; Aaltonen, T.; Nevalainen, M.; Valilehto, O.; Lindholm, L.; Vuento, M.; Vihinen-Ranta, M. Exploitation of microtubule cytoskeleton and dynein during parvoviral traffic toward the nucleus. J. Virol. 2003, 77, 10270–10279. [Google Scholar] [CrossRef]
- Lakadamyali, M.; Rust, M.J.; Babcock, H.P.; Zhuang, X. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 9280–9285. [Google Scholar] [CrossRef]
- Petit, C.; Giron, M.L.; Tobaly-Tapiero, J.; Bittoun, P.; Real, E.; Jacob, Y.; Tordo, N.; De The, H.; Saib, A. Targeting of incoming retroviral Gag to the centrosome involves a direct interaction with the dynein light chain 8. J. Cell Sci. 2003, 116, 3433–3442. [Google Scholar] [CrossRef]
- McDonald, D.; Vodicka, M.A.; Lucero, G.; Svitkina, T.M.; Borisy, G.G.; Emerman, M.; Hope, T.J. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 2002, 159, 441–452. [Google Scholar] [CrossRef] [PubMed]
- Döhner, K.; Nagel, C.H.; Sodeik, B. Viral stop-and-go along microtubules: Taking a ride with dynein and kinesins. Trends Microbiol. 2005, 13, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Greber, U.F.; Way, M. A superhighway to virus infection. Cell 2006, 124, 741–754. [Google Scholar] [CrossRef] [PubMed]
- Radtke, K.; Dohner, K.; Sodeik, B. Viral interactions with the cytoskeleton: A hitchhiker's guide to the cell. Cell Microbiol. 2006, 8, 387–400. [Google Scholar] [CrossRef]

© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Scherer, J.; Vallee, R.B. Adenovirus Recruits Dynein by an Evolutionary Novel Mechanism Involving Direct Binding to pH-Primed Hexon. Viruses 2011, 3, 1417-1431. https://doi.org/10.3390/v3081417
Scherer J, Vallee RB. Adenovirus Recruits Dynein by an Evolutionary Novel Mechanism Involving Direct Binding to pH-Primed Hexon. Viruses. 2011; 3(8):1417-1431. https://doi.org/10.3390/v3081417
Chicago/Turabian StyleScherer, Julian, and Richard B Vallee. 2011. "Adenovirus Recruits Dynein by an Evolutionary Novel Mechanism Involving Direct Binding to pH-Primed Hexon" Viruses 3, no. 8: 1417-1431. https://doi.org/10.3390/v3081417
APA StyleScherer, J., & Vallee, R. B. (2011). Adenovirus Recruits Dynein by an Evolutionary Novel Mechanism Involving Direct Binding to pH-Primed Hexon. Viruses, 3(8), 1417-1431. https://doi.org/10.3390/v3081417