Herpesviruses and Intermediate Filaments: Close Encounters with the Third Type
Abstract
:1. Introduction
1.1. IF: An Overview
1.2. Human Herpesviruses and Their Tropisms
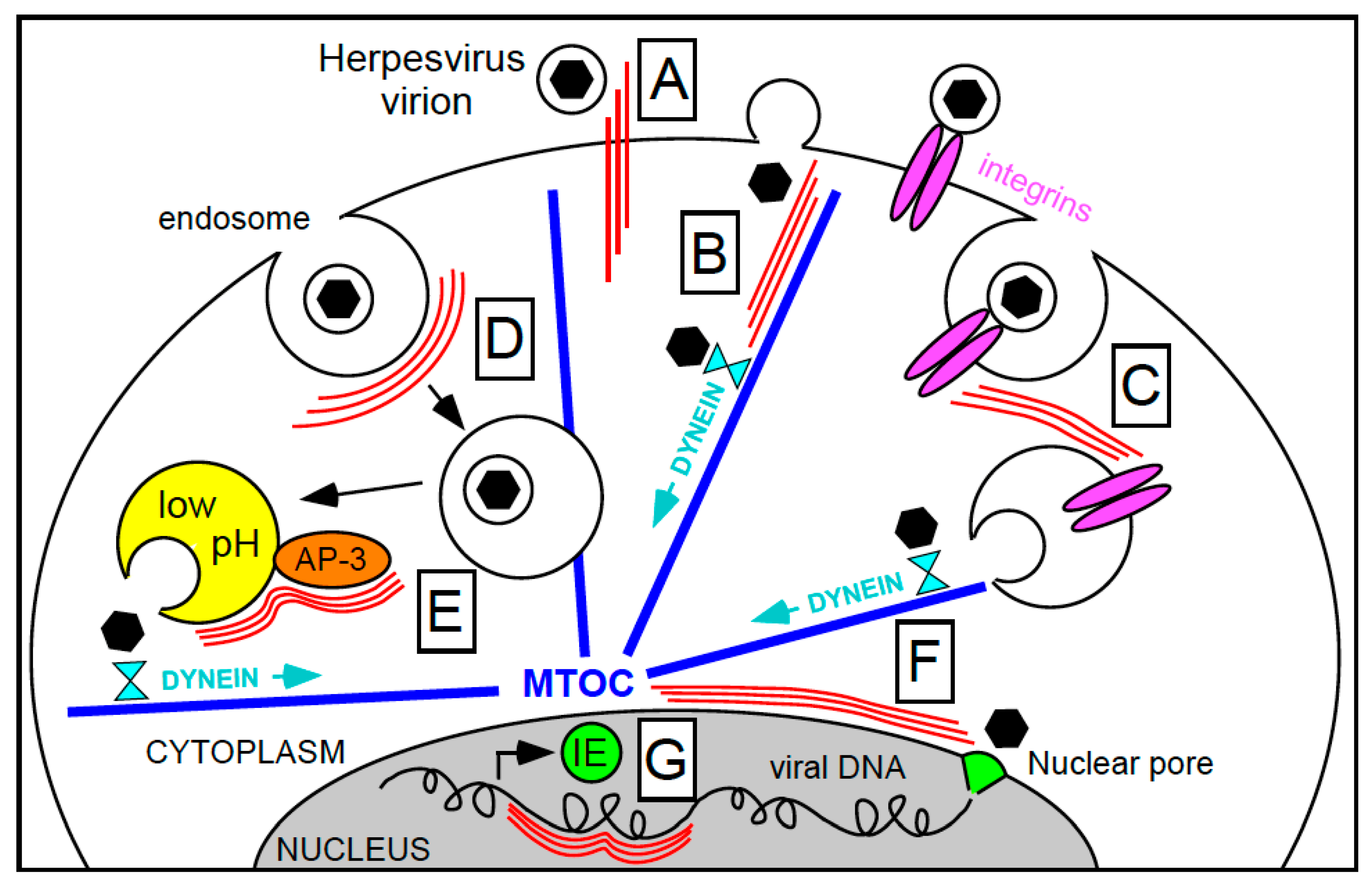
2. Intermediate Filaments in Herpesvirus Entry
2.1. Virion Binding at the Cell Surface
2.2. Virion Penetration
2.3. Viral Genome Deposition into the Nucleus
3. Intermediate Filaments in Herpesvirus Replication
4. Intermediate Filaments in Herpesvirus Egress
4.1. Nucleocapsid Egress from the Nucleus
4.2. Subviral Particle Trafficking from the Nucleus to the Cell Surface
5. Concluding Remarks
References and Notes
- Liem, R.K.; Messing, A. Dysfunctions of neuronal and glial intermediate filaments in disease. J. Clin. Invest. 2009, 119, 1814–1824. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, J.E.; Dechat, T.; Grin, B.; Helfand, B.; Mendez, M.; Pallari, H.M.; Goldman, R.D. Introducing intermediate filaments: From discovery to disease. J. Clin. Invest. 2009, 119, 1763–1771. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Strelkov, S.V.; Burkhard, P.; Aebi, U. Intermediate filaments: Primary determinants of cell architecture and plasticity. J. Clin. Invest. 2009, 119, 1772–1783. [Google Scholar] [CrossRef] [PubMed]
- Omary, M.B. "IF-pathies": A broad spectrum of intermediate filament-associated diseases. J. Clin. Invest. 2009, 119, 1756–1762. [Google Scholar] [CrossRef] [PubMed]
- Szeverenyi, I.; Cassidy, A.J.; Chung, C.W.; Lee, B.T.; Common, J.E.; Ogg, S.C.; Chen, H.; Sim, S.Y.; Goh, W.L.; Ng, K.W.; et al. The Human Intermediate Filament Database: Comprehensive information on a gene family involved in many human diseases. Hum. Mutat. 2008, 29, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Bar, H.; Kreplak, L.; Strelkov, S.V.; Aebi, U. Intermediate filaments: From cell architecture to nanomechanics. Nat. Rev. Mol. Cell Biol. 2007, 8, 562–573. [Google Scholar] [CrossRef]
- Kim, S.; Coulombe, P.A. Intermediate filament scaffolds fulfill mechanical, organizational, and signaling functions in the cytoplasm. Genes Dev. 2007, 21, 1581–1597. [Google Scholar] [CrossRef]
- Godsel, L.M.; Hobbs, R.P.; Green, K.J. Intermediate filament assembly: Dynamics to disease. Trends Cell Biol. 2008, 18, 28–37. [Google Scholar] [CrossRef]
- Amann, K.J.; Pollard, T.D. Cellular regulation of actin network assembly. Curr. Biol. 2000, 10, R728–R730. [Google Scholar] [CrossRef]
- Margolis, R.L.; Wilson, L. Microtubule treadmills—Possible molecular machinery. Nature 1981, 293, 705–711. [Google Scholar] [CrossRef]
- Strelkov, S.V.; Herrmann, H.; Aebi, U. Molecular architecture of intermediate filaments. Bioessays 2003, 25, 243–251. [Google Scholar] [CrossRef]
- Nguyen, M.D.; Shu, T.; Sanada, K.; Lariviere, R.C.; Tseng, H.C.; Park, S.K.; Julien, J.P.; Tsai, L.H. A NUDEL-dependent mechanism of neurofilament assembly regulates the integrity of CNS neurons. Nat. Cell Biol. 2004, 6, 595–608. [Google Scholar] [CrossRef]
- Omary, M.B.; Ku, N.O.; Tao, G.Z.; Toivola, D.M.; Liao, J. "Heads and tails" of intermediate filament phosphorylation: Multiple sites and functional insights. Trends Biochem. Sci. 2006, 31, 383–394. [Google Scholar] [CrossRef]
- Wagner, O.I.; Rammensee, S.; Korde, N.; Wen, Q.; Leterrier, J.F.; Janmey, P.A. Softness, strength and self-repair in intermediate filament networks. Exp. Cell Res. 2007, 313, 2228–2235. [Google Scholar] [CrossRef]
- Herrmann, H.; Aebi, U. Intermediate filaments: Molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu. Rev. Biochem. 2004, 73, 749–789. [Google Scholar] [CrossRef]
- Kolsch, A.; Windoffer, R.; Wurflinger, T.; Aach, T.; Leube, R.E. The keratin-filament cycle of assembly and disassembly. J. Cell Sci. 2010, 123, 2266–2272. [Google Scholar] [CrossRef]
- Prahlad, V.; Yoon, M.; Moir, R.D.; Vale, R.D.; Goldman, R.D. Rapid movements of vimentin on microtubule tracks: Kinesin-dependent assembly of intermediate filament networks. J. Cell Biol. 1998, 143, 159–170. [Google Scholar] [CrossRef]
- Martys, J.L.; Ho, C.L.; Liem, R.K.; Gundersen, G.G. Intermediate filaments in motion: Observations of intermediate filaments in cells using green fluorescent protein-vimentin. Mol. Biol. Cell 1999, 10, 1289–1295. [Google Scholar] [CrossRef]
- Yabe, J.T.; Pimenta, A.; Shea, T.B. Kinesin-mediated transport of neurofilament protein oligomers in growing axons. J. Cell Sci. 1999, 112, 3799–3814. [Google Scholar] [CrossRef]
- Motil, J.; Chan, W.K.; Dubey, M.; Chaudhury, P.; Pimenta, A.; Chylinski, T.M.; Ortiz, D.T.; Shea, T.B. Dynein mediates retrograde neurofilament transport within axons and anterograde delivery of NFs from perikarya into axons: Regulation by multiple phosphorylation events. Cell Motil. Cytoskeleton 2006, 63, 266–286. [Google Scholar] [CrossRef]
- Shah, J.V.; Flanagan, L.A.; Janmey, P.A.; Leterrier, J.F. Bidirectional translocation of neurofilaments along microtubules mediated in part by dynein/dynactin. Mol. Biol. Cell 2000, 11, 3495–3508. [Google Scholar] [CrossRef] [PubMed]
- Helfand, B.T.; Loomis, P.; Yoon, M.; Goldman, R.D. Rapid transport of neural intermediate filament protein. J. Cell Sci. 2003, 116, 2345–2359. [Google Scholar] [CrossRef] [PubMed]
- Goldman, R.D. The role of three cytoplasmic fibers in BHK-21 cell motility. I. Microtubules and the effects of colchicine. J. Cell Biol. 1971, 51, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Franke, W.W.; Schmid, E.; Osborn, M.; Weber, K. Different intermediate-sized filaments distinguished by immunofluorescence microscopy. Proc. Natl. Acad. Sci. U. S. A. 1978, 75, 5034–5038. [Google Scholar] [CrossRef]
- Aggeler, J.; Seely, K. Cytoskeletal dynamics in rabbit synovial fibroblasts: I. Effects of acrylamide on intermediate filaments and microfilaments. Cell Motil. Cytoskeleton 1990, 16, 110–120. [Google Scholar] [CrossRef]
- Klymkowsky, M.W. Intermediate filaments in 3T3 cells collapse after intracellular injection of a monoclonal anti-intermediate filament antibody. Nature 1981, 291, 249–251. [Google Scholar] [CrossRef]
- Lin, J.J.; Feramisco, J.R. Disruption of the in vivo distribution of the intermediate filaments in fibroblasts through the microinjection of a specific monoclonal antibody. Cell 1981, 24, 185–193. [Google Scholar] [CrossRef]
- Gawlitta, W.; Osborn, M.; Weber, K. Coiling of intermediate filaments induced by microinjection of a vimentin-specific antibody does not interfere with locomotion and mitosis. Eur. J. Cell Biol. 1981, 26, 83–90. [Google Scholar]
- Toivola, D.M.; Tao, G.Z.; Habtezion, A.; Liao, J.; Omary, M.B. Cellular integrity plus: Organelle-related and protein-targeting functions of intermediate filaments. Trends Cell Biol. 2005, 15, 608–617. [Google Scholar] [CrossRef]
- Pallari, H.M.; Eriksson, J.E. Intermediate filaments as signaling platforms. Sci. STKE 2006, 2006, pe53. [Google Scholar] [CrossRef]
- Sarria, A.J.; Lieber, J.G.; Nordeen, S.K.; Evans, R.M. The presence or absence of a vimentin-type intermediate filament network affects the shape of the nucleus in human SW-13 cells. J. Cell Sci. 1994, 107, 1593–1607. [Google Scholar] [CrossRef]
- Shah, S.B.; Davis, J.; Weisleder, N.; Kostavassili, I.; McCulloch, A.D.; Ralston, E.; Capetanaki, Y.; Lieber, R.L. Structural and functional roles of desmin in mouse skeletal muscle during passive deformation. Biophys. J. 2004, 86, 2993–3008. [Google Scholar] [CrossRef]
- Gerashchenko, M.V.; Chernoivanenko, I.S.; Moldaver, M.V.; Minin, A.A. Dynein is a motor for nuclear rotation while vimentin IFs is a "brake". Cell Biol. Int. 2009, 33, 1057–1064. [Google Scholar] [CrossRef]
- Liu, F.; Hong Zhou, Z. Comparative virion structures of human herpesviruses. In Human Herpesviruses: Biology, Therapy and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Mori, I.; Nishiyama, Y. Herpes simplex virus and varicella-zoster virus: Why do these human alphaherpesviruses behave so differently from one another? Rev. Med. Virol. 2005, 15, 393–406. [Google Scholar] [CrossRef]
- Campadelli-Fiume, G.; Menotti, L. Entry of alphaherpesviruses into the cell. In Human Herpesviruses: Biology, Therapy and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Arvin, A.M.; Moffat, J.F.; Sommer, M.; Oliver, S.; Che, X.; Vleck, S.; Zerboni, L.; Ku, C.C. Varicella-zoster virus T cell tropism and the pathogenesis of skin infection. Curr. Top. Microbiol. Immunol. 2010, 342, 189–209. [Google Scholar]
- Sinzger, C.; Digel, M.; Jahn, G. Cytomegalovirus cell tropism. Curr. Top. Microbiol. Immunol. 2008, 325, 63–83. [Google Scholar]
- Sinzger, C.; Grefte, A.; Plachter, B.; Gouw, A.S.; The, T.H.; Jahn, G. Fibroblasts, epithelial cells, endothelial cells and smooth muscle cells are major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J. Gen. Virol. 1995, 76, 741–750. [Google Scholar] [CrossRef]
- Bissinger, A.L.; Sinzger, C.; Kaiserling, E.; Jahn, G. Human cytomegalovirus as a direct pathogen: correlation of multiorgan involvement and cell distribution with clinical and pathological findings in a case of congenital inclusion disease. J. Med. Virol. 2002, 67, 200–206. [Google Scholar] [CrossRef]
- Rolle, A.; Olweus, J. Dendritic cells in cytomegalovirus infection: Viral evasion and host countermeasures. Apmis 2009, 117, 413–426. [Google Scholar] [CrossRef]
- Sinclair, J. Manipulation of dendritic cell functions by human cytomegalovirus. Expert Rev. Mol. Med. 2008, 10, e35. [Google Scholar] [CrossRef]
- Hahn, G.; Revello, M.G.; Patrone, M.; Percivalle, E.; Campanini, G.; Sarasini, A.; Wagner, M.; Gallina, A.; Milanesi, G.; Koszinowski, U.; et al. Human cytomegalovirus UL131–128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J. Virol. 2004, 78, 10023–10033. [Google Scholar] [CrossRef] [PubMed]
- Adler, B.; Sinzger, C. Endothelial cells in human cytomegalovirus infection: One host cell out of many or a crucial target for virus spread? Thromb. Haemost. 2009, 102, 1057–1063. [Google Scholar] [PubMed]
- Emery, V.C.; Cope, A.V.; Bowen, E.F.; Gor, D.; Griffiths, P.D. The dynamics of human cytomegalovirus replication in vivo. J. Exp. Med. 1999, 190, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, J. Human cytomegalovirus: Latency and reactivation in the myeloid lineage. J. Clin. Virol. 2008, 41, 180–185. [Google Scholar] [CrossRef] [PubMed]
- De Bolle, L.; Naesens, L.; De Clercq, E. Update on human herpesvirus 6 biology, clinical features, and therapy. Clin. Microbiol. Rev. 2005, 18, 217–245. [Google Scholar] [CrossRef]
- Mori, Y. Recent topics related to human herpesvirus 6 cell tropism. Cell. Microbiol. 2009, 11, 1001–1006. [Google Scholar] [CrossRef]
- Wang, F.-Z.; Pellett, P.E. HHV-6A, 6B, and 7: Immunobiology and host response. In Human Herpesviruses: Biology, Therapy and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Ganem, D. Kaposi’s sarcoma-associated herpesvirus. In Fields Virology, 5th ed.; Knipe, D., Howley, P., Griffin, D., Lamb, R., Martin, M., Roizman, B., Straus, S., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; pp. 2875–2888. [Google Scholar]
- Chandran, B.; Hutt-Fletcher., L. Gammaherpesviruses entry and early events during infection. In Human Herpesviruses: Biology, Therapy and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Mocarski, E.S. Comparative analysis of herpesvirus-common proteins. In Human Herpesviruses: Biology, Therapy and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Ogawa-Goto, K.; Tanaka, K.; Gibson, W.; Moriishi, E.; Miura, Y.; Kurata, T.; Irie, S.; Sata, T. Microtubule network facilitates nuclear targeting of human cytomegalovirus capsid. J. Virol. 2003, 77, 8541–8547. [Google Scholar] [CrossRef]
- Mabit, H.; Nakano, M.Y.; Prank, U.; Saam, B.; Dohner, K.; Sodeik, B.; Greber, U.F. Intact microtubules support adenovirus and herpes simplex virus infections. J. Virol. 2002, 76, 9962–9971. [Google Scholar] [CrossRef]
- Naranatt, P.P.; Krishnan, H.H.; Smith, M.S.; Chandran, B. Kaposi's sarcoma-associated herpesvirus modulates microtubule dynamics via RhoA-GTP-diaphanous 2 signaling and utilizes the dynein motors to deliver its DNA to the nucleus. J. Virol. 2005, 79, 1191–1206. [Google Scholar] [CrossRef]
- Adams, D. Keratinization of the oral epithelium. Ann. R. Coll. Surg. Engl. 1976, 58, 351–358. [Google Scholar]
- Batchelor, M.; Guignot, J.; Patel, A.; Cummings, N.; Cleary, J.; Knutton, S.; Holden, D.W.; Connerton, I.; Frankel, G. Involvement of the intermediate filament protein cytokeratin-18 in actin pedestal formation during EPEC infection. EMBO Rep. 2004, 5, 104–110. [Google Scholar] [CrossRef]
- O'Brien, L.M.; Walsh, E.J.; Massey, R.C.; Peacock, S.J.; Foster, T.J. Staphylococcus aureus clumping factor B (ClfB) promotes adherence to human type I cytokeratin 10: Implications for nasal colonization. Cell. Microbiol. 2002, 4, 759–770. [Google Scholar] [CrossRef]
- Samen, U.; Eikmanns, B.J.; Reinscheid, D.J.; Borges, F. The surface protein Srr-1 of Streptococcus agalactiae binds human keratin 4 and promotes adherence to epithelial HEp-2 cells. Infect. Immun. 2007, 75, 5405–5414. [Google Scholar] [CrossRef]
- Carlson, S.A.; Omary, M.B.; Jones, B.D. Identification of cytokeratins as accessory mediators of Salmonella entry into eukaryotic cells. Life Sci. 2002, 70, 1415–1426. [Google Scholar] [CrossRef]
- Tamura, G.S.; Nittayajarn, A. Group B streptococci and other gram-positive cocci bind to cytokeratin 8. Infect. Immun. 2000, 68, 2129–2134. [Google Scholar] [CrossRef]
- Kim, J.K.; Fahad, A.M.; Shanmukhappa, K.; Kapil, S. Defining the cellular target(s) of porcine reproductive and respiratory syndrome virus blocking monoclonal antibody 7G10. J. Virol. 2006, 80, 689–696. [Google Scholar] [CrossRef]
- Thomas, E.K.; Connelly, R.J.; Pennathur, S.; Dubrovsky, L.; Haffar, O.K.; Bukrinsky, M.I. Anti-idiotypic antibody to the V3 domain of gp120 binds to vimentin: A possible role of intermediate filaments in the early steps of HIV-1 infection cycle. Viral. Immunol. 1996, 9, 73–87. [Google Scholar] [CrossRef]
- Nedellec, P.; Vicart, P.; Laurent-Winter, C.; Martinat, C.; Prevost, M.C.; Brahic, M. Interaction of Theiler's virus with intermediate filaments of infected cells. J. Virol. 1998, 72, 9553–9560. [Google Scholar] [CrossRef]
- Spear, P.G.; Longnecker, R. Herpesvirus entry: An update. J. Virol. 2003, 77, 10179–10185. [Google Scholar] [CrossRef]
- Spruance, S.L. Pathogenesis of herpes simplex labialis: Excretion of virus in the oral cavity. J. Clin. Microbiol. 1984, 19, 675–679. [Google Scholar] [CrossRef]
- Visalli, R.J.; Courtney, R.J.; Meyers, C. Infection and replication of herpes simplex virus type 1 in an organotypic epithelial culture system. Virology 1997, 230, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Ryckman, B.J.; Jarvis, M.A.; Drummond, D.D.; Nelson, J.A.; Johnson, D.C. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J. Virol. 2006, 80, 710–722. [Google Scholar] [CrossRef] [PubMed]
- Ryckman, B.J.; Rainish, B.L.; Chase, M.C.; Borton, J.A.; Nelson, J.A.; Jarvis, M.A.; Johnson, D.C. Characterization of the human cytomegalovirus gH/gL/UL128–131 complex that mediates entry into epithelial and endothelial cells. J. Virol. 2008, 82, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Sinzger, C. Entry route of HCMV into endothelial cells. J. Clin. Virol. 2008, 41, 174–179. [Google Scholar] [CrossRef] [PubMed]
- Patrone, M.; Secchi, M.; Bonaparte, E.; Milanesi, G.; Gallina, A. Cytomegalovirus UL131–128 products promote gB conformational transition and gB-gH interaction during entry into endothelial cells. J. Virol. 2007, 81, 11479–11488. [Google Scholar] [CrossRef]
- Wang, D.; Shenk, T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 18153–18158. [Google Scholar] [CrossRef]
- Wang, D.; Shenk, T. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J. Virol. 2005, 79, 10330–10338. [Google Scholar] [CrossRef]
- Nicola, A.V.; McEvoy, A.M.; Straus, S.E. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J. Virol. 2003, 77, 5324–5332. [Google Scholar] [CrossRef]
- Nicola, A.V.; Straus, S.E. Cellular and viral requirements for rapid endocytic entry of herpes simplex virus. J. Virol. 2004, 78, 7508–7517. [Google Scholar] [CrossRef]
- Nicola, A.V.; Hou, J.; Major, E.O.; Straus, S.E. Herpes simplex virus type 1 enters human epidermal keratinocytes, but not neurons, via a pH-dependent endocytic pathway. J. Virol. 2005, 79, 7609–7616. [Google Scholar] [CrossRef]
- Miller, N.; Hutt-Fletcher, L.M. Epstein-Barr virus enters B cells and epithelial cells by different routes. J. Virol. 1992, 66, 3409–3414. [Google Scholar] [CrossRef]
- Roberts, K.L.; Baines, J.D. Actin in Herpesvirus Infection. Viruses 2011, 3, 336–346. [Google Scholar] [CrossRef]
- Arcangeletti, M.C.; Pinardi, F.; Medici, M.C.; Pilotti, E.; De Conto, F.; Ferraglia, F.; Landini, M.P.; Chezzi, C.; Dettori, G. Cytoskeleton involvement during human cytomegalovirus replicative cycle in human embryo fibroblasts. New Microbiol. 2000, 23, 241–256. [Google Scholar]
- Jones, N.L.; Lewis, J.C.; Kilpatrick, B.A. Cytoskeletal disruption during human cytomegalovirus infection of human lung fibroblasts. Eur. J. Cell Biol. 1986, 41, 304–312. [Google Scholar]
- Losse, D.; Lauer, R.; Weder, D.; Radsak, K. Actin distribution and synthesis in human fibroblasts infected by cytomegalovirus. Arch. Virol. 1982, 71, 353–359. [Google Scholar] [CrossRef]
- Miller, M.S.; Furlong, W.E.; Pennell, L.; Geadah, M.; Hertel, L. RASCAL is a new human cytomegalovirus-encoded protein that localizes to the nuclear lamina and in cytoplasmic vesicles at late times postinfection. J. Virol. 2010, 84, 6483–6496. [Google Scholar] [CrossRef]
- Miller, M.S.; Hertel, L. Onset of human cytomegalovirus replication in fibroblasts requires the presence of an intact vimentin cytoskeleton. J. Virol. 2009, 83, 7015–7028. [Google Scholar] [CrossRef]
- Norrild, B.; Lehto, V.P.; Virtanen, I. Organization of cytoskeleton elements during herpes simplex virus type 1 infection of human fibroblasts: An immunofluorescence study. J. Gen. Virol. 1986, 67, 97–105. [Google Scholar] [CrossRef]
- Dienes, H.P.; Hiller, G.; Muller, S.; Falke, D. Microtubules and intermediate filaments of herpes simplex virus infected cells. Arch. Virol. 1987, 94, 15–28. [Google Scholar] [CrossRef]
- Lyman, M.G.; Enquist, L.W. Herpesvirus interactions with the host cytoskeleton. J. Virol. 2008, 83, 2058–2066. [Google Scholar] [CrossRef]
- Chandran, B. Early events in Kaposi's sarcoma-associated herpesvirus infection of target cells. J. Virol. 2010, 84, 2188–2199. [Google Scholar] [CrossRef] [PubMed]
- Walter, I.; Nowotny, N. Equine herpes virus type 1 (EHV-1) infection induces alterations in the cytoskeleton of vero cells but not apoptosis. Arch. Virol. 1999, 144, 1827–1836. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; Desloges, N.; Rahaus, M.; Wolff, M.H. Varicella-zoster virus infection influences expression and organization of actin and alpha-tubulin but does not affect lamin A and vimentin. Intervirology 2005, 48, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Compton, T.; Nepomuceno, R.R.; Nowlin, D.M. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology 1992, 191, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Ivaska, J.; Pallari, H.M.; Nevo, J.; Eriksson, J.E. Novel functions of vimentin in cell adhesion, migration, and signaling. Exp. Cell Res. 2007, 313, 2050–2062. [Google Scholar] [CrossRef]
- Ivaska, J.; Vuoriluoto, K.; Huovinen, T.; Izawa, I.; Inagaki, M.; Parker, P.J. PKCepsilon-mediated phosphorylation of vimentin controls integrin recycling and motility. EMBO J. 2005, 24, 3834–3845. [Google Scholar] [CrossRef]
- Feire, A.L.; Koss, H.; Compton, T. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 15470–15475. [Google Scholar] [CrossRef]
- Wang, X.; Huang, D.Y.; Huong, S.M.; Huang, E.S. Integrin alphavbeta3 is a coreceptor for human cytomegalovirus. Nat. Med. 2005, 11, 515–521. [Google Scholar] [CrossRef]
- Isaacson, M.K.; Juckem, L.K.; Compton, T. Virus entry and innate immune activation. Curr. Top. Microbiol. Immunol. 2008, 325, 85–100. [Google Scholar]
- Akula, S.M.; Pramod, N.P.; Wang, F.Z.; Chandran, B. Integrin alpha3beta1 (CD 49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 2002, 108, 407–419. [Google Scholar] [CrossRef]
- Veettil, M.V.; Sadagopan, S.; Sharma-Walia, N.; Wang, F.Z.; Raghu, H.; Varga, L.; Chandran, B. Kaposi's sarcoma-associated herpesvirus forms a multimolecular complex of integrins (alphaVbeta5, alphaVbeta3, and alpha3beta1) and CD98-xCT during infection of human dermal microvascular endothelial cells, and CD98-xCT is essential for the postentry stage of infection. J. Virol. 2008, 82, 12126–12144. [Google Scholar]
- Chesnokova, L.S.; Nishimura, S.L.; Hutt-Fletcher, L.M. Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins alphavbeta6 or alphavbeta8. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 20464–20469. [Google Scholar] [CrossRef]
- Herman, B.; Albertini, D.F. The intracellular movement of endocytic vesicles in cultured granulosa cells. Cell Motil. 1982, 2, 583–597. [Google Scholar] [CrossRef]
- Potokar, M.; Kreft, M.; Li, L.; Daniel Andersson, J.; Pangrsic, T.; Chowdhury, H.H.; Pekny, M.; Zorec, R. Cytoskeleton and vesicle mobility in astrocytes. Traffic 2007, 8, 12–20. [Google Scholar] [CrossRef]
- Styers, M.L.; Kowalczyk, A.P.; Faundez, V. Intermediate filaments and vesicular membrane traffic: the odd couple's first dance? Traffic 2005, 6, 359–365. [Google Scholar] [CrossRef]
- Akula, S.M.; Naranatt, P.P.; Walia, N.S.; Wang, F.Z.; Fegley, B.; Chandran, B. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) infection of human fibroblast cells occurs through endocytosis. J. Virol. 2003, 77, 7978–7990. [Google Scholar] [CrossRef]
- Styers, M.L.; Salazar, G.; Love, R.; Peden, A.A.; Kowalczyk, A.P.; Faundez, V. The endo-lysosomal sorting machinery interacts with the intermediate filament cytoskeleton. Mol. Biol. Cell 2004, 15, 5369–5382. [Google Scholar] [CrossRef]
- Robinson, M.S. Adaptable adaptors for coated vesicles. Trends Cell Biol. 2004, 14, 167–174. [Google Scholar] [CrossRef]
- Kantheti, P.; Qiao, X.; Diaz, M.E.; Peden, A.A.; Meyer, G.E.; Carskadon, S.L.; Kapfhamer, D.; Sufalko, D.; Robinson, M.S.; Noebels, J.L.; et al. Mutation in AP-3 delta in the mocha mouse links endosomal transport to storage deficiency in platelets, melanosomes, and synaptic vesicles. Neuron 1998, 21, 111–122. [Google Scholar] [CrossRef]
- Salazar, G.; Love, R.; Styers, M.L.; Werner, E.; Peden, A.; Rodriguez, S.; Gearing, M.; Wainer, B.H.; Faundez, V. AP-3-dependent mechanisms control the targeting of a chloride channel (ClC-3) in neuronal and non-neuronal cells. J. Biol. Chem. 2004, 279, 25430–25439. [Google Scholar] [CrossRef]
- Li, X.; Wang, T.; Zhao, Z.; Weinman, S.A. The ClC-3 chloride channel promotes acidification of lysosomes in CHO-K1 and Huh-7 cells. Am. J. Physiol. Cell Physiol. 2002, 282, C1483–C1491. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, J.J.; Filali, M.S.; Collins, M.M.; Volk, K.A.; Lamb, F.S. The ClC-3 Cl-/H+ antiporter becomes uncoupled at low extracellular pH. J. Biol. Chem. 2010, 285, 2569–2579. [Google Scholar] [CrossRef] [PubMed]
- Inoue, N.; Winter, J.; Lal, R.B.; Offermann, M.K.; Koyano, S. Characterization of entry mechanisms of human herpesvirus 8 by using an Rta-dependent reporter cell line. J. Virol. 2003, 77, 8147–8152. [Google Scholar] [CrossRef] [PubMed]
- Nemerow, G.R.; Cooper, N.R. Early events in the infection of human B lymphocytes by Epstein-Barr virus: The internalization process. Virology 1984, 132, 186–198. [Google Scholar] [CrossRef]
- Seglen, P.O.; Berg, T.O.; Blankson, H.; Fengsrud, M.; Holen, I.; Stromhaug, P.E. Structural aspects of autophagy. Adv. Exp. Med. Biol. 1996, 389, 103–111. [Google Scholar]
- Lee, D.Y.; Lee, J.; Sugden, B. The unfolded protein response and autophagy: Herpesviruses rule! J. Virol. 2009, 83, 1168–1172. [Google Scholar] [CrossRef]
- Lyman, M.G.; Enquist, L.W. Herpesvirus interactions with the host cytoskeleton. J. Virol. 2009, 83, 2058–2066. [Google Scholar] [CrossRef]
- Welte, M.A. Bidirectional transport along microtubules. Curr. Biol. 2004, 14, R525–R537. [Google Scholar] [CrossRef]
- Salpingidou, G.; Smertenko, A.; Hausmanowa-Petrucewicz, I.; Hussey, P.J.; Hutchison, C.J. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J. Cell Biol. 2007, 178, 897–904. [Google Scholar] [CrossRef]
- Renaud, J.; Kerjan, G.; Sumita, I.; Zagar, Y.; Georget, V.; Kim, D.; Fouquet, C.; Suda, K.; Sanbo, M.; Suto, F.; et al. Plexin-A2 and its ligand, Sema6A, control nucleus-centrosome coupling in migrating granule cells. Nat. Neurosci. 2008, 11, 440–449. [Google Scholar] [CrossRef]
- Sodeik, B.; Ebersold, M.W.; Helenius, A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1997, 136, 1007–1021. [Google Scholar] [CrossRef]
- Peng, L.; Ryazantsev, S.; Sun, R.; Zhou, Z.H. Three-dimensional visualization of gammaherpesvirus life cycle in host cells by electron tomography. Structure 2010, 18, 47–58. [Google Scholar] [CrossRef]
- Ojala, P.M.; Sodeik, B.; Ebersold, M.W.; Kutay, U.; Helenius, A. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell Biol. 2000, 20, 4922–4931. [Google Scholar] [CrossRef]
- Shahin, V.; Hafezi, W.; Oberleithner, H.; Ludwig, Y.; Windoffer, B.; Schillers, H.; Kuhn, J.E. The genome of HSV-1 translocates through the nuclear pore as a condensed rod-like structure. J. Cell Sci. 2006, 119, 23–30. [Google Scholar] [CrossRef]
- Copeland, A.M.; Newcomb, W.W.; Brown, J.C. Herpes simplex virus replication: Roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 2009, 83, 1660–1668. [Google Scholar] [CrossRef]
- Djabali, K.; Portier, M.M.; Gros, F.; Blobel, G.; Georgatos, S.D. Network antibodies identify nuclear lamin B as a physiological attachment site for peripherin intermediate filaments. Cell 1991, 64, 109–121. [Google Scholar] [CrossRef]
- Georgatos, S.D.; Blobel, G. Lamin B constitutes an intermediate filament attachment site at the nuclear envelope. J. Cell Biol. 1987, 105, 117–125. [Google Scholar] [CrossRef]
- Georgatos, S.D.; Weber, K.; Geisler, N.; Blobel, G. Binding of two desmin derivatives to the plasma membrane and the nuclear envelope of avian erythrocytes: evidence for a conserved site-specificity in intermediate filament-membrane interactions. Proc. Natl. Acad. Sci. U. S. A. 1987, 84, 6780–6784. [Google Scholar] [CrossRef]
- Capco, D.G.; Wan, K.M.; Penman, S. The nuclear matrix: Three-dimensional architecture and protein composition. Cell 1982, 29, 847–858. [Google Scholar] [CrossRef]
- Katsuma, Y.; Swierenga, S.H.; Marceau, N.; French, S.W. Connections of intermediate filaments with the nuclear lamina and the cell periphery. Biol. Cell 1987, 59, 193–203. [Google Scholar] [CrossRef]
- Carmo-Fonseca, M.; Cidadao, A.J.; David-Ferreira, J.F. Filamentous cross-bridges link intermediate filaments to the nuclear pore complexes. Eur. J. Cell Biol. 1988, 45, 282–290. [Google Scholar] [PubMed]
- French, S.W.; Kawahara, H.; Katsuma, Y.; Ohta, M.; Swierenga, S.H. Interaction of intermediate filaments with nuclear lamina and cell periphery. Electron. Microsc. Rev. 1989, 2, 17–51. [Google Scholar] [CrossRef] [PubMed]
- Starr, D.A. Communication between the cytoskeleton and the nuclear envelope to position the nucleus. Mol. Biosyst. 2007, 3, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Starr, D.A.; Fridolfsson, H.N. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 2010, 26, 421–444. [Google Scholar] [CrossRef] [PubMed]
- Maniotis, A.J.; Chen, C.S.; Ingber, D.E. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. U. S. A. 1997, 94, 849–854. [Google Scholar] [CrossRef]
- Traub, P. Intermediate filaments and gene regulation. Physiol. Chem. Phys. Med. NMR 1995, 27, 377–400. [Google Scholar]
- Shaklai, S.; Amariglio, N.; Rechavi, G.; Simon, A.J. Gene silencing at the nuclear periphery. FEBS J. 2007, 274, 1383–1392. [Google Scholar] [CrossRef]
- Lee, D.C.; Welton, K.L.; Smith, E.D.; Kennedy, B.K. A-type nuclear lamins act as transcriptional repressors when targeted to promoters. Exp. Cell Res. 2009, 315, 996–1007. [Google Scholar] [CrossRef]
- Somech, R.; Shaklai, S.; Geller, O.; Amariglio, N.; Simon, A.J.; Rechavi, G.; Gal-Yam, E.N. The nuclear-envelope protein and transcriptional repressor LAP2beta interacts with HDAC3 at the nuclear periphery, and induces histone H4 deacetylation. J. Cell Sci. 2005, 118, 4017–4025. [Google Scholar] [CrossRef]
- Polioudaki, H.; Kourmouli, N.; Drosou, V.; Bakou, A.; Theodoropoulos, P.A.; Singh, P.B.; Giannakouros, T.; Georgatos, S.D. Histones H3/H4 form a tight complex with the inner nuclear membrane protein LBR and heterochromatin protein 1. EMBO Rep. 2001, 2, 920–925. [Google Scholar] [CrossRef]
- Paulus, C.; Nitzsche, A.; Nevels, M. Chromatinisation of herpesvirus genomes. Rev. Med. Virol. 2010, 20, 34–50. [Google Scholar] [CrossRef]
- Silva, L.; Cliffe, A.; Chang, L.; Knipe, D.M. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog. 2008, 4, e1000071. [Google Scholar] [CrossRef]
- Zhou, Q.; Ji, X.; Chen, L.; Greenberg, H.B.; Lu, S.C.; Omary, M.B. Keratin mutation primes mouse liver to oxidative injury. Hepatology 2005, 41, 517–525. [Google Scholar] [CrossRef]
- Hagemann, T.L.; Gaeta, S.A.; Smith, M.A.; Johnson, D.A.; Johnson, J.A.; Messing, A. Gene expression analysis in mice with elevated glial fibrillary acidic protein and Rosenthal fibers reveals a stress response followed by glial activation and neuronal dysfunction. Hum. Mol. Genet. 2005, 14, 2443–2458. [Google Scholar] [CrossRef]
- Li, H.; Choudhary, S.K.; Milner, D.J.; Munir, M.I.; Kuisk, I.R.; Capetanaki, Y. Inhibition of desmin expression blocks myoblast fusion and interferes with the myogenic regulators MyoD and myogenin. J. Cell Biol. 1994, 124, 827–841. [Google Scholar] [CrossRef]
- Jones, J.O.; Arvin, A.M. Microarray analysis of host cell gene transcription in response to varicella-zoster virus infection of human T cells and fibroblasts in vitro and SCIDhu skin xenografts in vivo. J. Virol. 2003, 77, 1268–1280. [Google Scholar] [CrossRef]
- Hertel, L.; Lacaille, V.G.; Strobl, H.; Mellins, E.D.; Mocarski, E.S. Susceptibility of immature and mature Langerhans cell-type dendritic cells to infection and immunomodulation by human cytomegalovirus. J. Virol. 2003, 77, 7563–7574. [Google Scholar] [CrossRef]
- Luo, M.H.; Hannemann, H.; Kulkarni, A.S.; Schwartz, P.H.; O'Dowd, J.M.; Fortunato, E.A. Human cytomegalovirus infection causes premature and abnormal differentiation of human neural progenitor cells. J. Virol. 2010, 84, 3528–3541. [Google Scholar] [CrossRef]
- Cornelissen, M.; van der Kuyl, A.C.; van den Burg, R.; Zorgdrager, F.; van Noesel, C.J.; Goudsmit, J. Gene expression profile of AIDS-related Kaposi's sarcoma. BMC Cancer 2003, 3, 7. [Google Scholar] [CrossRef]
- Uozaki, H.; Chong, J.M.; Fujimoto, E.; Itoh, M.; Saito, M.; Sakuma, K.; Sudo, M.; Ushiku, T.; Niki, T.; Nagai, H.; et al. Soft and hard keratin expression in Epstein-Barr-virus-associated gastric carcinoma. Anticancer Res. 2005, 25, 3183–3190. [Google Scholar]
- Nishikawa, J.; Kiss, C.; Imai, S.; Takada, K.; Okita, K.; Klein, G.; Szekely, L. Upregulation of the truncated basic hair keratin 1(hHb1-DeltaN) in carcinoma cells by Epstein-Barr virus (EBV). Int. J. Cancer 2003, 107, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Birkenbach, M.; Liebowitz, D.; Wang, F.; Sample, J.; Kieff, E. Epstein-Barr virus latent infection membrane protein increases vimentin expression in human B-cell lines. J. Virol. 1989, 63, 4079–4084. [Google Scholar] [CrossRef] [PubMed]
- Allday, M.J.; Crawford, D.H.; Thomas, J.A. Epstein-Barr virus (EBV) nuclear antigen 6 induces expression of the EBV latent membrane protein and an activated phenotype in Raji cells. J. Gen. Virol. 1993, 74, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Silins, S.L.; Sculley, T.B. Modulation of vimentin, the CD40 activation antigen and Burkitt's lymphoma antigen (CD77) by the Epstein-Barr virus nuclear antigen EBNA-4. Virology 1994, 202, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Ray, N.; Enquist, L.W. Transcriptional response of a common permissive cell type to infection by two diverse alphaherpesviruses. J. Virol. 2004, 78, 3489–3501. [Google Scholar] [CrossRef]
- Taddeo, B.; Esclatine, A.; Roizman, B. The patterns of accumulation of cellular RNAs in cells infected with a wild-type and a mutant herpes simplex virus 1 lacking the virion host shutoff gene. Proc. Natl. Acad. Sci. U. S. A. 2002, 99, 17031–17036. [Google Scholar] [CrossRef]
- Mettenleiter, T.C.; Klupp, B.G.; Granzow, H. Herpesvirus assembly: An update. Virus Res. 2009, 143, 222–234. [Google Scholar] [CrossRef]
- Lee, C.P.; Chen, M.R. Escape of herpesviruses from the nucleus. Rev. Med. Virol. 2010, 20, 214–230. [Google Scholar] [CrossRef]
- Britt, B. Maturation and egress. In Human Herpesviruses: Biology, Therapy and Immunoprophylaxis; Arvin, A., Campadelli-Fiume, G., Mocarski, E., Moore, P.S., Roizman, B., Whitley, R., Yamanishi, K., Eds.; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- Prokocimer, M.; Davidovich, M.; Nissim-Rafinia, M.; Wiesel-Motiuk, N.; Bar, D.; Barkan, R.; Meshorer, E.; Gruenbaum, Y. Nuclear lamins: Key regulators of nuclear structure and activities. J. Cell. Mol. Med. 2009, 13, 1059–1085. [Google Scholar] [CrossRef]
- Scott, E.S.; O'Hare, P. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 2001, 75, 8818–8830. [Google Scholar] [CrossRef]
- Park, R.; Baines, J.D. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 2006, 80, 494–504. [Google Scholar] [CrossRef]
- Reynolds, A.E.; Liang, L.; Baines, J.D. Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J. Virol. 2004, 78, 5564–5575. [Google Scholar] [CrossRef]
- Simpson-Holley, M.; Baines, J.; Roller, R.; Knipe, D.M. Herpes simplex virus 1 U(L)31 and U(L)34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 2004, 78, 5591–5600. [Google Scholar] [CrossRef]
- Morris, J.B.; Hofemeister, H.; O'Hare, P. Herpes simplex virus infection induces phosphorylation and delocalization of emerin, a key inner nuclear membrane protein. J. Virol. 2007, 81, 4429–4437. [Google Scholar] [CrossRef]
- Leach, N.; Bjerke, S.L.; Christensen, D.K.; Bouchard, J.M.; Mou, F.; Park, R.; Baines, J.; Haraguchi, T.; Roller, R.J. Emerin is hyperphosphorylated and redistributed in herpes simplex virus type 1-infected cells in a manner dependent on both UL34 and US3. J. Virol. 2007, 81, 10792–10803. [Google Scholar] [CrossRef]
- Mou, F.; Forest, T.; Baines, J.D. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 2007, 81, 6459–6470. [Google Scholar] [CrossRef]
- Cano-Monreal, G.L.; Wylie, K.M.; Cao, F.; Tavis, J.E.; Morrison, L.A. Herpes simplex virus 2 UL13 protein kinase disrupts nuclear lamins. Virology 2009, 392, 137–147. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Shiba, C.; Goshima, F.; Nawa, A.; Murata, T.; Nishiyama, Y. Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J. Gen. Virol. 2001, 82, 1423–1428. [Google Scholar] [CrossRef]
- Simpson-Holley, M.; Colgrove, R.C.; Nalepa, G.; Harper, J.W.; Knipe, D.M. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 2005, 79, 12840–12851. [Google Scholar] [CrossRef]
- Bjerke, S.L.; Roller, R.J. Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 2006, 347, 261–276. [Google Scholar] [CrossRef]
- Kato, A.; Yamamoto, M.; Ohno, T.; Tanaka, M.; Sata, T.; Nishiyama, Y.; Kawaguchi, Y. Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 2006, 80, 1476–1486. [Google Scholar] [CrossRef] [PubMed]
- Leach, N.R.; Roller, R.J. Significance of host cell kinases in herpes simplex virus type 1 egress and lamin-associated protein disassembly from the nuclear lamina. Virology 2010, 406, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Mou, F.; Wills, E.G.; Park, R.; Baines, J.D. Effects of lamin A/C, lamin B1, and viral US3 kinase activity on viral infectivity, virion egress, and the targeting of herpes simplex virus U(L)34-encoded protein to the inner nuclear membrane. J. Virol. 2008, 82, 8094–8104. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, E.C.; Foisner, R. Proteins that associate with lamins: Many faces, many functions. Exp. Cell Res. 2007, 313, 2167–2179. [Google Scholar] [CrossRef]
- Dechat, T.; Pfleghaar, K.; Sengupta, K.; Shimi, T.; Shumaker, D.K.; Solimando, L.; Goldman, R.D. Nuclear lamins: major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008, 22, 832–853. [Google Scholar] [CrossRef]
- Wild, P.; Engels, M.; Senn, C.; Tobler, K.; Ziegler, U.; Schraner, E.M.; Loepfe, E.; Ackermann, M.; Mueller, M.; Walther, P. Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J. Virol. 2005, 79, 1071–1083. [Google Scholar] [CrossRef]
- Ruebner, B.H.; Miyai, K.; Slusser, R.J.; Wedemeyer, P.; Medearis, D.N., Jr. Mouse cytomegalovirus infection. An electron microscopic study of hepatic parenchymal cells. Am. J. Pathol. 1964, 44, 799–821. [Google Scholar]
- Papadimitriou, J.M.; Shellam, G.R.; Robertson, T.A. An ultrastructural investigation of cytomegalovirus replication in murine hepatocytes. J. Gen. Virol. 1984, 65, 1979–1990. [Google Scholar] [CrossRef]
- Gilloteaux, J.; Nassiri, M.R. Human bone marrow fibroblasts infected by cytomegalovirus: ultrastructural observations. J. Submicrosc. Cytol. Pathol. 2000, 32, 17–45. [Google Scholar]
- Severi, B.; Landini, M.P.; Govoni, E. Human cytomegalovirus morphogenesis: an ultrastructural study of the late cytoplasmic phases. Arch. Virol. 1988, 98, 51–64. [Google Scholar] [CrossRef]
- Buser, C.; Walther, P.; Mertens, T.; Michel, D. Cytomegalovirus primary envelopment occurs at large infoldings of the inner nuclear membrane. J. Virol. 2007, 81, 3042–3048. [Google Scholar] [CrossRef]
- Muranyi, W.; Haas, J.; Wagner, M.; Krohne, G.; Koszinowski, U.H. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 2002, 297, 854–857. [Google Scholar] [CrossRef]
- Milbradt, J.; Auerochs, S.; Sticht, H.; Marschall, M. Cytomegaloviral proteins that associate with the nuclear lamina: Components of a postulated nuclear egress complex. J. Gen. Virol. 2009, 90, 579–590. [Google Scholar] [CrossRef]
- Milbradt, J.; Webel, R.; Auerochs, S.; Sticht, H.; Marschall, M. Novel mode of phosphorylation-triggered reorganization of the nuclear lamina during nuclear egress of human cytomegalovirus. J. Biol. Chem. 2010, 285, 13979–13989. [Google Scholar] [CrossRef]
- Marschall, M.; Marzi, A.; aus dem Siepen, P.; Jochmann, R.; Kalmer, M.; Auerochs, S.; Lischka, P.; Leis, M.; Stamminger, T. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 2005, 280, 33357–33367. [Google Scholar] [CrossRef]
- Radsak, K.; Schneider, D.; Jost, E.; Brucher, K.H. Alteration of nuclear lamina protein in human fibroblasts infected with cytomegalovirus (HCMV). Arch. Virol. 1989, 105, 103–112. [Google Scholar] [CrossRef]
- Camozzi, D.; Pignatelli, S.; Valvo, C.; Lattanzi, G.; Capanni, C.; Dal Monte, P.; Landini, M.P. Remodelling of the nuclear lamina during human cytomegalovirus infection: Role of the viral proteins pUL50 and pUL53. J. Gen. Virol. 2008, 89, 731–740. [Google Scholar] [CrossRef]
- Hamirally, S.; Kamil, J.P.; Ndassa-Colday, Y.M.; Lin, A.J.; Jahng, W.J.; Baek, M.C.; Noton, S.; Silva, L.A.; Simpson-Holley, M.; Knipe, D.M.; et al. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 2009, 5, e1000275. [Google Scholar] [CrossRef]
- Milbradt, J.; Auerochs, S.; Marschall, M. Cytomegaloviral proteins pUL50 and pUL53 are associated with the nuclear lamina and interact with cellular protein kinase C. J. Gen. Virol. 2007, 88, 2642–2650. [Google Scholar] [CrossRef]
- Prichard, M.N. Function of human cytomegalovirus UL97 kinase in viral infection and its inhibition by maribavir. Rev. Med. Virol. 2009, 19, 215–229. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Kato, K. Protein kinases conserved in herpesviruses potentially share a function mimicking the cellular protein kinase cdc2. Rev. Med. Virol. 2003, 13, 331–340. [Google Scholar] [CrossRef] [PubMed]
- Peter, M.; Nakagawa, J.; Doree, M.; Labbe, J.C.; Nigg, E.A. In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell 1990, 61, 591–602. [Google Scholar] [CrossRef] [PubMed]
- Hertel, L.; Mocarski, E.S. Global analysis of host cell gene expression late during cytomegalovirus infection reveals extensive dysregulation of cell cycle gene expression and induction of Pseudomitosis independent of US28 function. J. Virol. 2004, 78, 11988–12011. [Google Scholar] [CrossRef] [PubMed]
- Hertel, L.; Chou, S.; Mocarski, E.S. Viral and cell cycle-regulated kinases in cytomegalovirus-induced pseudomitosis and replication. PLoS Pathog. 2007, 3, e6. [Google Scholar] [CrossRef]
- Torrisi, M.R.; Cirone, M.; Pavan, A.; Zompetta, C.; Barile, G.; Frati, L.; Faggioni, A. Localization of Epstein-Barr virus envelope glycoproteins on the inner nuclear membrane of virus-producing cells. J. Virol. 1989, 63, 828–832. [Google Scholar] [CrossRef]
- Farina, A.; Santarelli, R.; Gonnella, R.; Bei, R.; Muraro, R.; Cardinali, G.; Uccini, S.; Ragona, G.; Frati, L.; Faggioni, A.; et al. The BFRF1 gene of Epstein-Barr virus encodes a novel protein. J. Virol. 2000, 74, 3235–3244. [Google Scholar] [CrossRef]
- Farina, A.; Feederle, R.; Raffa, S.; Gonnella, R.; Santarelli, R.; Frati, L.; Angeloni, A.; Torrisi, M.R.; Faggioni, A.; Delecluse, H.J. BFRF1 of Epstein-Barr virus is essential for efficient primary viral envelopment and egress. J. Virol. 2005, 79, 3703–3712. [Google Scholar] [CrossRef]
- Gonnella, R.; Farina, A.; Santarelli, R.; Raffa, S.; Feederle, R.; Bei, R.; Granato, M.; Modesti, A.; Frati, L.; Delecluse, H.J.; et al. Characterization and intracellular localization of the Epstein-Barr virus protein BFLF2: Interactions with BFRF1 and with the nuclear lamina. J. Virol. 2005, 79, 3713–3727. [Google Scholar] [CrossRef]
- Lee, C.P.; Huang, Y.H.; Lin, S.F.; Chang, Y.; Chang, Y.H.; Takada, K.; Chen, M.R. Epstein-Barr virus BGLF4 kinase induces disassembly of the nuclear lamina to facilitate virion production. J. Virol. 2008, 82, 11913–11926. [Google Scholar] [CrossRef]
- Norregard Nielsen, L.; Forchhammer, J.; Dabelsteen, E.; Jepsen, A.; Stubbe Teglbjaerg, C.; Norrild, B. Herpes simplex virus-induced changes of the keratin type intermediate filament in rat epithelial cells. J. Gen. Virol. 1987, 68, 737–748. [Google Scholar] [CrossRef]
- Murata, T.; Goshima, F.; Nishizawa, Y.; Daikoku, T.; Takakuwa, H.; Ohtsuka, K.; Yoshikawa, T.; Nishiyama, Y. Phosphorylation of cytokeratin 17 by herpes simplex virus type 2 US3 protein kinase. Microbiol. Immunol. 2002, 46, 707–719. [Google Scholar] [CrossRef]
- Linden, M.; Li, Z.; Paulin, D.; Gotow, T.; Leterrier, J.F. Effects of desmin gene knockout on mice heart mitochondria. J. Bioenerg. Biomembr. 2001, 33, 333–341. [Google Scholar] [CrossRef]
- Milner, D.J.; Mavroidis, M.; Weisleder, N.; Capetanaki, Y. Desmin cytoskeleton linked to muscle mitochondrial distribution and respiratory function. J. Cell Biol. 2000, 150, 1283–1298. [Google Scholar] [CrossRef]
- Brownlees, J.; Ackerley, S.; Grierson, A.J.; Jacobsen, N.J.; Shea, K.; Anderton, B.H.; Leigh, P.N.; Shaw, C.E.; Miller, C.C. Charcot-Marie-Tooth disease neurofilament mutations disrupt neurofilament assembly and axonal transport. Hum. Mol. Genet. 2002, 11, 2837–2844. [Google Scholar] [CrossRef]
- Magin, T.M.; Vijayaraj, P.; Leube, R.E. Structural and regulatory functions of keratins. Exp. Cell Res. 2007, 313, 2021–2032. [Google Scholar] [CrossRef]
- Labudova, M.; Tomaskova, J.; Skultety, L.; Pastorek, J.; Pastorekova, S. The nucleoprotein of lymphocytic choriomeningitis virus facilitates spread of persistent infection through stabilization of the keratin network. J. Virol. 2009, 83, 7842–7849. [Google Scholar] [CrossRef]

| Type | Protein Name | Cell Type | Tissue Type |
|---|---|---|---|
| I | Acidic keratins: K9-K28 K31-K40 | Epithelial cells, keratinocytes | Mucosae, epidermis Hair, epidermal appendages |
| II | Basic keratins: K1-8, K71-80 K81-86 | Epithelial cells, keratinocytes | Mucosae, epidermis Hair, epidermal appendages |
| III | Vimentin Peripherin Glial fibrillary acidic protein Syncoilin Desmin | Mesenchymal cells: fibroblasts, endothelial, hematopoietic cells Neuronal cells Astrocytes and glia Muscle cells | Connective tissue, blood, blood vessels Nervous system Nervous system Muscles |
| IV | α-internexin Neurofilament H, L, and M Nestin Synemin α, synemin β | Neuronal cells Neuronal cells Neuroepithelial cells Muscle cells | Nervous system Nervous system Nervous system Muscles |
| V | Lamin A, B1, B2, C1, C2 | Ubiquitous | Ubiquitous |
| VI | CP49/phakinin, filensin/CP115 | Eye lens cells | Eye lens |
| Subfamily | Virus Name | Productive Infection | Latency |
|---|---|---|---|
| Alpha | HSV-1 | Epithelial cells, neurons | Neurons: trigeminal ganglia |
| HSV-2 | |||
| VZV | Epithelial cells, neurons, monocytes, dendritic cells, T and B lymphocytes | Neurons: dorsal root ganglia | |
| Beta | CMV | Most cell types except lymphocytes, eosinophils, basophils, and neutrophils | Myeloid progenitors |
| HHV-6 | CD4+ T cells, neurons, astrocytes, microglia, fibroblasts, epithelial, endothelial and dendritic cells | Lymphocytes, Monocyte/macrophages, other? | |
| HHV-7 | CD4+ T cells | ||
| Gamma | EBV | B and T cells, dendritic, NK and smooth muscle cells | Memory B cells |
| KSHV | B cells, endothelial cells, epithelial cells, keratinocytes and fibroblasts |
| Gene Name | Gene Symbol | VZV in T Cells * | VZV in Skin * | VZV in HF * | HSV in HF & | CMV in HF @ |
|---|---|---|---|---|---|---|
| Keratin 1 Keratin 5 Keratin 6A Keratin 8 Keratin 13 Keratin 17 Keratin 18 Keratin 19 Keratin 33A Keratin 71 Keratin 85 Keratin 86 | KRT1 KRT5 KRT6A KRT8 KRT13 KRT17 KRT18 KRT19 KRT33A KRT71 KRT85 KRT86 | - - - - - - - - UP - - - | DOWN DOWN DOWN - - DOWN - UP - DOWN - - | - - UP - - - - - - - - - | - - - UP - - UP - - - - - |
- UP - - UP - UP DOWN - - UP UP |
| Desmin Peripherin Syncoilin Vimentin | DES PRPH SYNC VIM | - - UP - | UP - - UP | - - - - | - UP - DOWN | - - DOWN DOWN |
| Neurofilament 3 Neurofilament heavy | NEF3 NEFH | - DOWN | DOWN - | - - | - - | UP - |
| Lamin B1 | LMNB1 | - | - | - | - | UP |
© 2011 by the author. licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hertel, L. Herpesviruses and Intermediate Filaments: Close Encounters with the Third Type. Viruses 2011, 3, 1015-1040. https://doi.org/10.3390/v3071015
Hertel L. Herpesviruses and Intermediate Filaments: Close Encounters with the Third Type. Viruses. 2011; 3(7):1015-1040. https://doi.org/10.3390/v3071015
Chicago/Turabian StyleHertel, Laura. 2011. "Herpesviruses and Intermediate Filaments: Close Encounters with the Third Type" Viruses 3, no. 7: 1015-1040. https://doi.org/10.3390/v3071015
APA StyleHertel, L. (2011). Herpesviruses and Intermediate Filaments: Close Encounters with the Third Type. Viruses, 3(7), 1015-1040. https://doi.org/10.3390/v3071015





