Putative Fusion-Associated Small Transmembrane (FAST) Proteins Encoded by Viruses of Pistolviridae, Order Ghabrivirales, Identified from In Silico Analyses
Abstract
1. Introduction
2. Materials and Methods
2.1. Protein Sequence Analyses
2.2. Phylogenetic Analyses
2.3. Sample Origin and Tissue Preparation for Histological Analyses of Atlantic Salmon Heart Tissue
3. Results
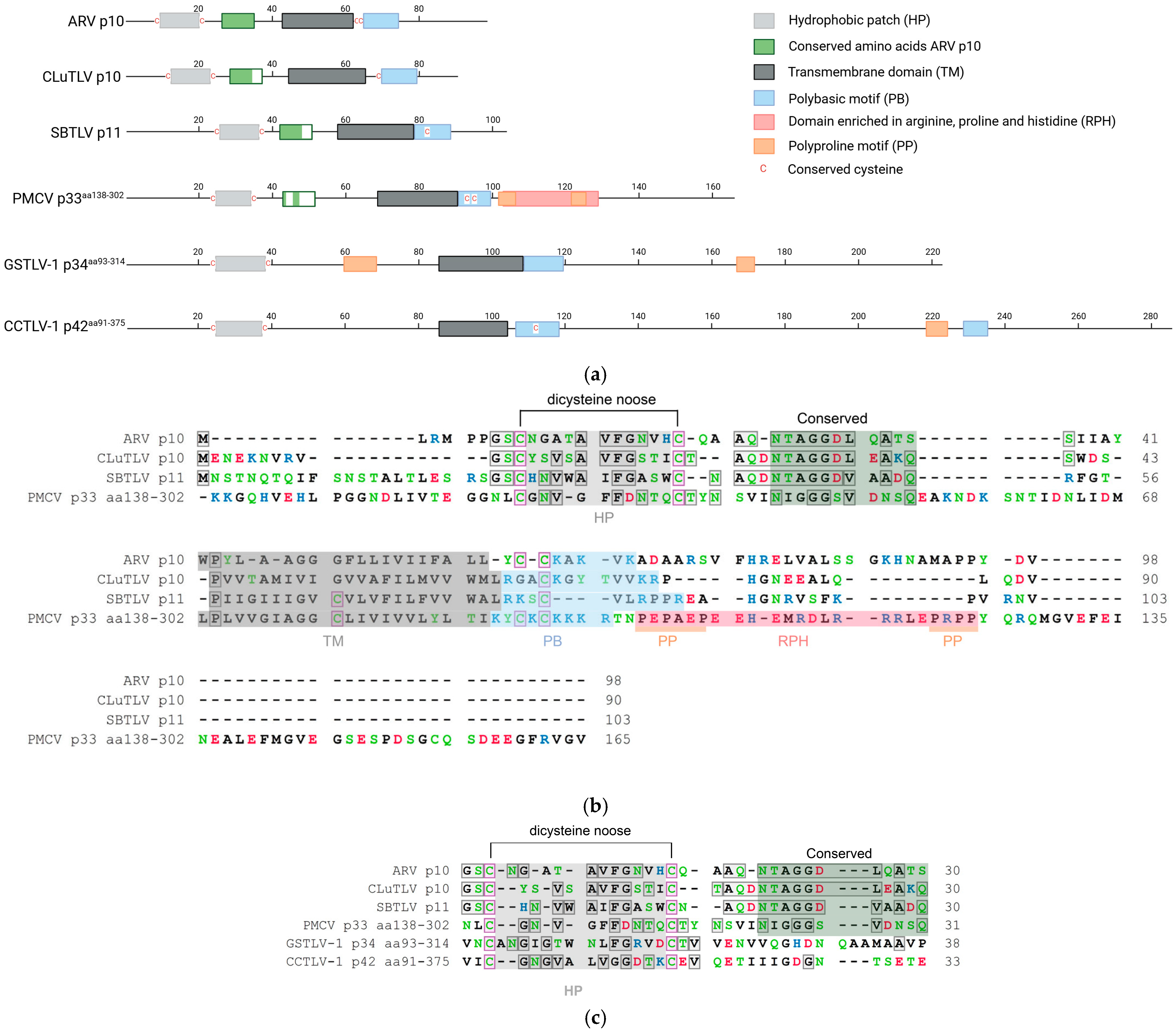
3.1. Pistolviruses Encode Proteins with Sequence Features Characteristic of FAST Proteins
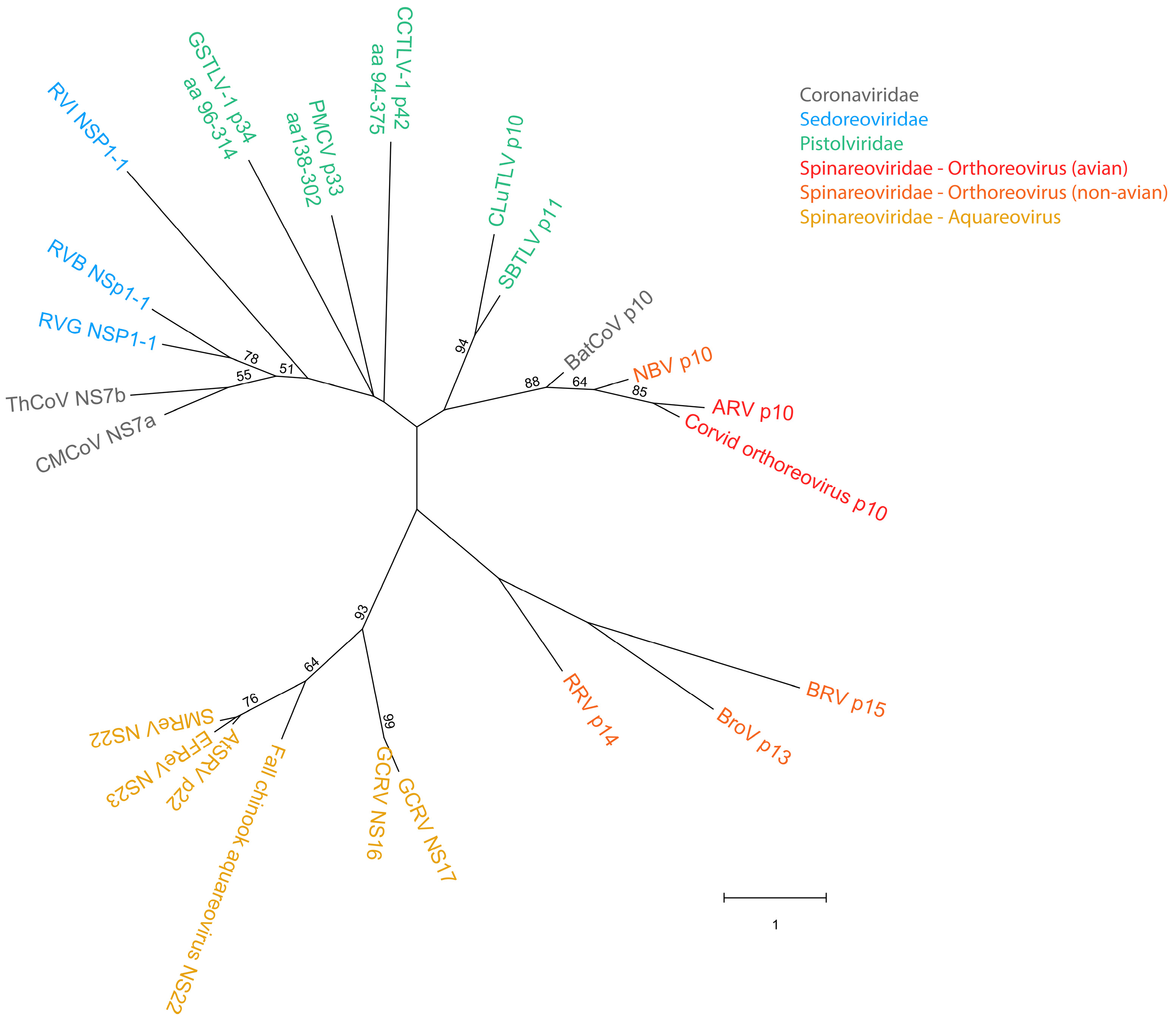
3.2. Phylogenetic Analysis of Putative FAST Protein Sequences from Pistolviruses with FAST Proteins from Selected Orthoreoviruses, Aquareoviruses, Rotaviruses, and Coronaviruses
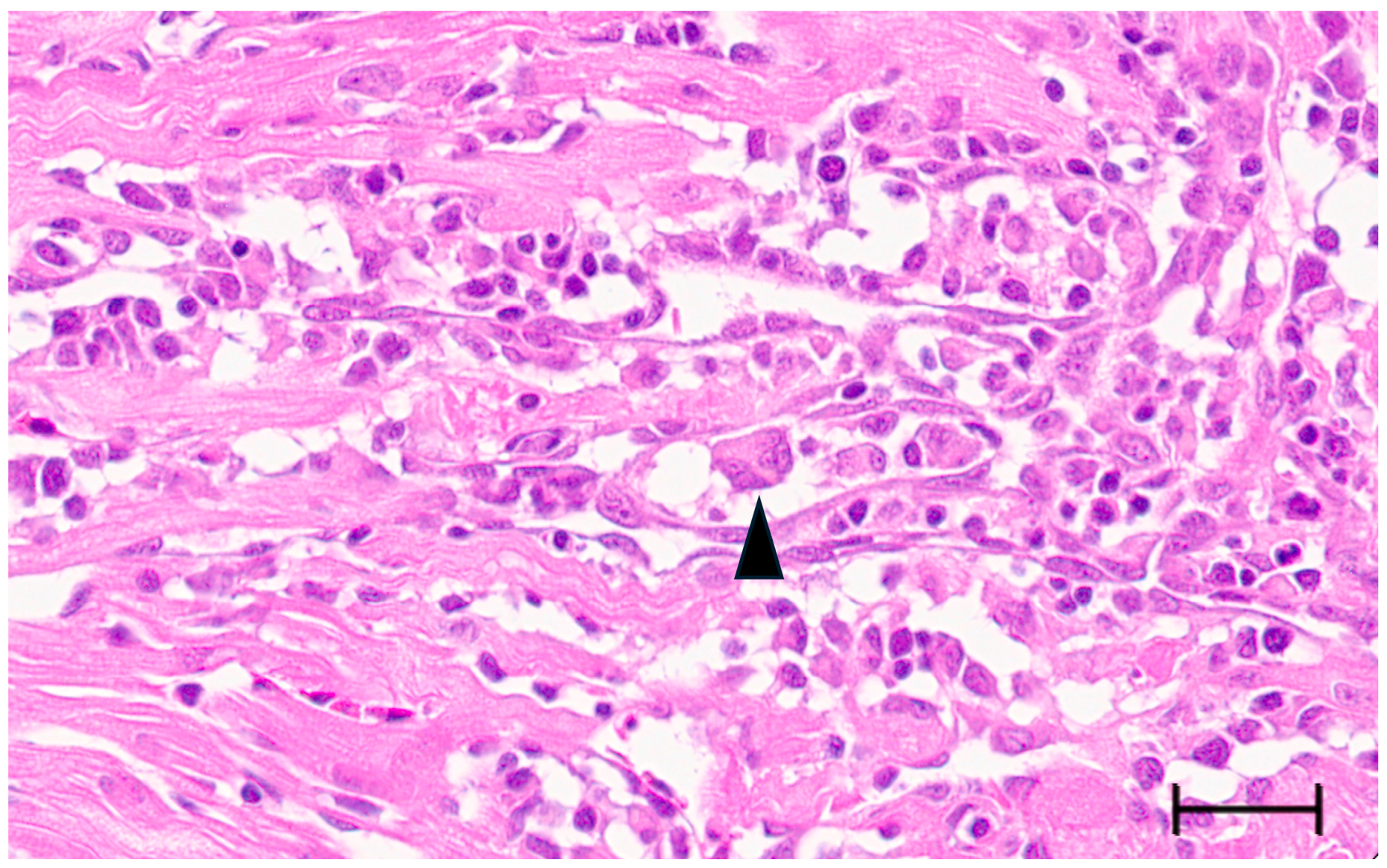
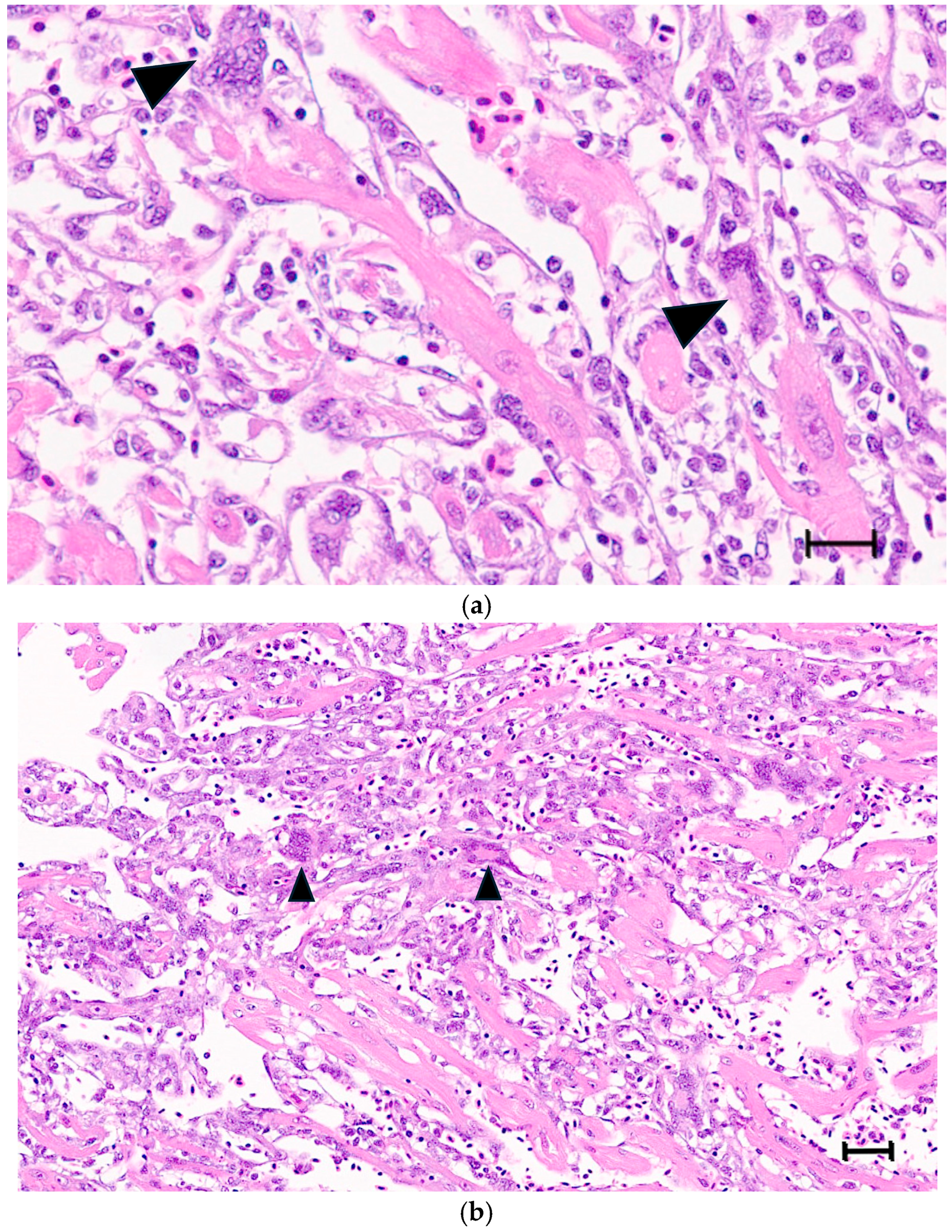
3.3. Multinucleated Cells Indicative of Cell Fusion Are Found Correlated with SBTLV Infection In Vitro and PMCV In Vivo, Supporting Presence of Functional FAST Proteins
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ARV | Avian reovirus |
| CCTLV-1 | Common carp toti-like virus 1 |
| CLuTLV | Cyclopterus lumpus toti-like virus |
| CMS | Cardiomyopathy syndrome |
| FAST | Fusion-associated small transmembrane proteins |
| GSTLV-1 | Golden shiner toti-like virus 1 |
| HP | Hydrophobic patch |
| kDa | Kilodalton |
| ML | Maximum likelihood |
| ORF | Open reading frame |
| PB | Polybasic motif |
| PMCV | Piscine myocarditis virus |
| PP | Polyproline motif |
| RdRp | RNA-dependent RNA polymerase |
| RPH | Motif enriched in arginine, proline and histidine |
| SBTLV | Sea bass toti-like virus |
| TM | Transmembrane domain |
References
- ICTV. ICTV Taxonomy Release 2023. Available online: https://ictv.global/taxonomy (accessed on 24 October 2025).
- Simmonds, P.; Adriaenssens, E.M.; Lefkowitz, E.J.; Oksanen, H.M.; Siddell, S.G.; Zerbini, F.M.; Alfenas-Zerbini, P.; Aylward, F.O.; Dempsey, D.M.; Dutilh, B.E.; et al. Changes to virus taxonomy and the ICTV Statutes ratified by the International Committee on Taxonomy of Viruses (2024). Arch. Virol. 2024, 169, 236. [Google Scholar] [CrossRef]
- Sandlund, L.; Mor, S.K.; Singh, V.K.; Padhi, S.K.; Phelps, N.B.D.; Nylund, S.; Mikalsen, A.B. Comparative molecular characterization of novel and known piscine toti-like viruses. Viruses 2021, 13, 1063. [Google Scholar] [CrossRef]
- Louboutin, L.; Cabon, J.; Beven, V.; Hirchaud, E.; Blanchard, Y.; Morin, T. Characterization of a new toti-like virus in sea bass, Dicentrarchus labrax. Viruses 2023, 15, 2423. [Google Scholar] [CrossRef]
- Fritsvold, C.; Kongtorp, R.T.; Taksdal, T.; Orpetveit, I.; Heum, M.; Poppe, T.T. Experimental transmission of cardiomyopathy syndrome (CMS) in Atlantic salmon Salmo salar. Dis. Aquat. Organ. 2009, 87, 225–234. [Google Scholar] [CrossRef]
- Fritsvold, C.; Mikalsen, A.B.; Haugland, Ø.; Tartor, H.; Sindre, H. Characterization of early phases of cardiomyopathy syndrome pathogenesis in Atlantic salmon (Salmo salar L.) through various diagnostic methods. J. Fish Dis. 2022, 45, 1267–1279. [Google Scholar] [CrossRef]
- Amono, R.; Fredlund, S.A.T.N.; Chesnais, M.; Thiede, B.; Markussen, T.; Evensen, Ø.; Mikalsen, A.B. Defined domains and cleavage determine the diverse functions of piscine myocarditis virus p33 protein. Front. Microbiol. 2025, 16, 1633241. [Google Scholar] [CrossRef] [PubMed]
- Haugland, Ø.; Mikalsen, A.B.; Nilsen, P.; Lindmo, K.; Thu, B.J.; Eliassen, T.M.; Roos, N.; Rode, M.; Evensen, Ø. Cardiomyopathy syndrome of Atlantic salmon (Salmo salar L.) is caused by a dsRNA virus of the Totiviridae family. J. Virol. 2011, 85, 5275–5286. [Google Scholar] [CrossRef] [PubMed]
- Diller, J.R.; Parrington, H.M.; Patton, J.T.; Ogden, K.M. Rotavirus Species B Encodes a Functional Fusion-Associated Small Transmembrane Protein. J. Virol. 2019, 93, e00813-19. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R. Fusogenic reoviruses and their fusion-associated small transmembrane (FAST) proteins. Annu. Rev. Virol. 2019, 6, 341–363. [Google Scholar] [CrossRef]
- Veletanlic, V.; Sartalamacchia, K.; Diller, J.R.; Ogden, K.M. Multiple rotavirus species encode fusion-associated small transmembrane (FAST) proteins with cell type-specific activity. bioRxiv 2023. [Google Scholar] [CrossRef]
- Huang, C.; Liu, W.J.; Xu, W.; Jin, T.; Zhao, Y.; Song, J.; Shi, Y.; Ji, W.; Jia, H.; Zhou, Y.; et al. A Bat-Derived Putative Cross-Family Recombinant Coronavirus with a Reovirus Gene. PLoS Pathog. 2016, 12, e1005883. [Google Scholar] [CrossRef] [PubMed]
- Sartalamacchia, K.; Porter, M.S.; Veletanlic, V.; Ogden, K.M. Avian deltacoronaviruses encode fusion-associated small transmembrane proteins that can induce syncytia formation. Virology 2024, 600, 110258. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Kawagishi, T.; Sakai, Y.; Nouda, R.; Shimojima, M.; Saijo, M.; Matsuura, Y.; Kobayashi, T. Cell-cell fusion induced by reovirus FAST proteins enhances replication and pathogenicity of non-enveloped dsRNA viruses. PLoS Pathog. 2019, 15, e1007675. [Google Scholar] [CrossRef] [PubMed]
- Salsman, J.; Top, D.; Boutilier, J.; Duncan, R. Extensive syncytium formation mediated by the reovirus FAST proteins triggers apoptosis-induced membrane instability. J.Virol. 2005, 79, 8090–8100. [Google Scholar] [CrossRef]
- Boutilier, J.; Duncan, R. The reovirus fusion-associated small transmembrane (FAST) proteins: Virus-encoded cellular fusogens. Curr. Top. Membr. 2011, 68, 107–140. [Google Scholar] [CrossRef]
- Ciechonska, M.; Duncan, R. Reovirus FAST proteins: Virus-encoded cellular fusogens. Trends Microbiol. 2014, 22, 715–724. [Google Scholar] [CrossRef]
- Hofman, K.; Stoffel, W. TMbase—A database of membrane spanning proteins segments. Biol. Chem. Hoppe Seyler 1993, 347:166. [Google Scholar]
- Hallgren, J.; Tsirigos, K.D.; Pedersen, M.D.; Armenteros, J.J.A.; Marcatili, P.; Nielsen, H.; Krogh, A.; Winther, O. DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv 2022. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hillis, D.M.; Bull, J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 1993, 42, 182–192. [Google Scholar] [CrossRef]
- Hungnes, O.; Jonassen, T.O.; Jonassen, C.M.; Grinde, B. Molecular epidemiology of viral infections. How sequence information helps us understand the evolution and dissemination of viruses. APMIS 2000, 108, 81–97. [Google Scholar] [CrossRef]
- Amono, R.; Markussen, T.; Singh, V.K.; Lund, M.; Manji, F.; Mor, S.K.; Evensen, Ø.; Mikalsen, A.B. Unraveling the genomic landscape of piscine myocarditis virus: Mutation frequencies, viral diversity and evolutionary dynamics in Atlantic salmon. Virus Evol. 2024, 10, veae097. [Google Scholar] [CrossRef] [PubMed]
- Barry, C.; Key, T.; Haddad, R.; Duncan, R. Features of a spatially constrained cystine loop in the p10 FAST protein ectodomain define a new class of viral fusion peptides. J. Biol. Chem. 2010, 285, 16424–16433. [Google Scholar] [CrossRef] [PubMed]
- Shmulevitz, M.; Salsman, J.; Duncan, R. Palmitoylation, membrane-proximal basic residues, and transmembrane glycine residues in the reovirus p10 protein are essential for syncytium formation. J. Virol. 2003, 77, 9769–9779. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.L.; Wang, A.L.; Wang, C.C. Purification and characterization of the Giardia lamblia double-stranded RNA virus. Mol. Biochem. Parasitol. 1988, 28, 189–195. [Google Scholar] [CrossRef]
- Dantas, M.D.; Cavalcante, G.H.; Oliveira, R.A.; Lanza, D.C. New insights about ORF1 coding regions support the proposition of a new genus comprising arthropod viruses in the family Totiviridae. Virus Res. 2016, 211, 159–164. [Google Scholar] [CrossRef]
- Poulos, B.T.; Tang, K.F.; Pantoja, C.R.; Bonami, J.R.; Lightner, D.V. Purification and characterization of infectious myonecrosis virus of penaeid shrimp. J. Gen. Virol. 2006, 87, 987–996. [Google Scholar] [CrossRef]
- Nibert, M.L.; Takagi, Y. Fibers come and go: Differences in cell-entry components among related dsRNA viruses. Curr. Opin. Virol. 2013, 3, 20–26. [Google Scholar] [CrossRef]
- Bruno, D.W.; Noguera, P.A.; Poppe, T. A Colour Atlas of Salmonid Diseases, 2nd ed.; Springer: Dordrecht, The Netherlands, 2013. [Google Scholar]
- Ferguson, H.W.; Bjerkas, E.; Evensen, Ø. Systemic Pathology of Fish, 2nd ed.; Scotian Press: Leicester, UK, 2006. [Google Scholar]
- Fritsvold, C. Pathogenesis Studies of Cardiomyopathy Syndrome (CMS) in Atlantic salmon, Salmo salar L. Ph.D. Thesis, Norwegian University of Life Sciences, Ås, Norway, 2021. [Google Scholar]
- Corcoran, J.A.; Duncan, R. Reptilian reovirus utilizes a small type III protein with an external myristylated amino terminus to mediate cell-cell fusion. J. Virol. 2004, 78, 4342–4351. [Google Scholar] [CrossRef]
- Racine, T.; Hurst, T.; Barry, C.; Shou, J.; Kibenge, F.; Duncan, R. Aquareovirus effects syncytiogenesis by using a novel member of the FAST protein family translated from a noncanonical translation start site. J. Virol. 2009, 83, 5951–5955. [Google Scholar] [CrossRef]
- White, J.M.; Ward, A.E.; Odongo, L.; Tamm, L.K. Viral Membrane Fusion: A Dance Between Proteins and Lipids. Annu. Rev. Virol. 2023, 10, 139–161. [Google Scholar] [CrossRef]
- Clancy, E.K.; Duncan, R. Reovirus FAST protein transmembrane domains function in a modular, primary sequence-independent manner to mediate cell-cell membrane fusion. J. Virol. 2009, 83, 2941–2950. [Google Scholar] [CrossRef]
- Voichek, M.; Bernhard, A.; Novatchkova, M.; Handler, D.; Möseneder, P.; Rafanel, B.; Duchek, P.; Senti, K.A.; Brennecke, J. Direct cell-to-cell transmission of retrotransposons. bioRxiv 2025. [Google Scholar] [CrossRef]
- Nibert, M.L.; Duncan, R. Bioinformatics of recent aqua- and orthoreovirus isolates from fish: Evolutionary gain or loss of FAST and fiber proteins and taxonomic implications. PLoS ONE 2013, 8, e68607. [Google Scholar] [CrossRef]
- Lauber, C.; Seitz, S.; Mattei, S.; Suh, A.; Beck, J.; Herstein, J.; Börold, J.; Salzburger, W.; Kaderali, L.; Briggs, J.A.G.; et al. Deciphering the Origin and Evolution of Hepatitis B Viruses by Means of a Family of Non-enveloped Fish Viruses. Cell Host Microbe 2017, 22, 387–399.e6. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Amono, R.; Markussen, T.; Evensen, Ø.; Mikalsen, A.B. Putative Fusion-Associated Small Transmembrane (FAST) Proteins Encoded by Viruses of Pistolviridae, Order Ghabrivirales, Identified from In Silico Analyses. Viruses 2026, 18, 193. https://doi.org/10.3390/v18020193
Amono R, Markussen T, Evensen Ø, Mikalsen AB. Putative Fusion-Associated Small Transmembrane (FAST) Proteins Encoded by Viruses of Pistolviridae, Order Ghabrivirales, Identified from In Silico Analyses. Viruses. 2026; 18(2):193. https://doi.org/10.3390/v18020193
Chicago/Turabian StyleAmono, Racheal, Turhan Markussen, Øystein Evensen, and Aase B. Mikalsen. 2026. "Putative Fusion-Associated Small Transmembrane (FAST) Proteins Encoded by Viruses of Pistolviridae, Order Ghabrivirales, Identified from In Silico Analyses" Viruses 18, no. 2: 193. https://doi.org/10.3390/v18020193
APA StyleAmono, R., Markussen, T., Evensen, Ø., & Mikalsen, A. B. (2026). Putative Fusion-Associated Small Transmembrane (FAST) Proteins Encoded by Viruses of Pistolviridae, Order Ghabrivirales, Identified from In Silico Analyses. Viruses, 18(2), 193. https://doi.org/10.3390/v18020193





