Rapid Visual Detection of Senecavirus A Based on RPA-CRISPR/Cas12a System with Canonical or Suboptimal PAM
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses, Antibodies, Plasmids, and Clinical Samples
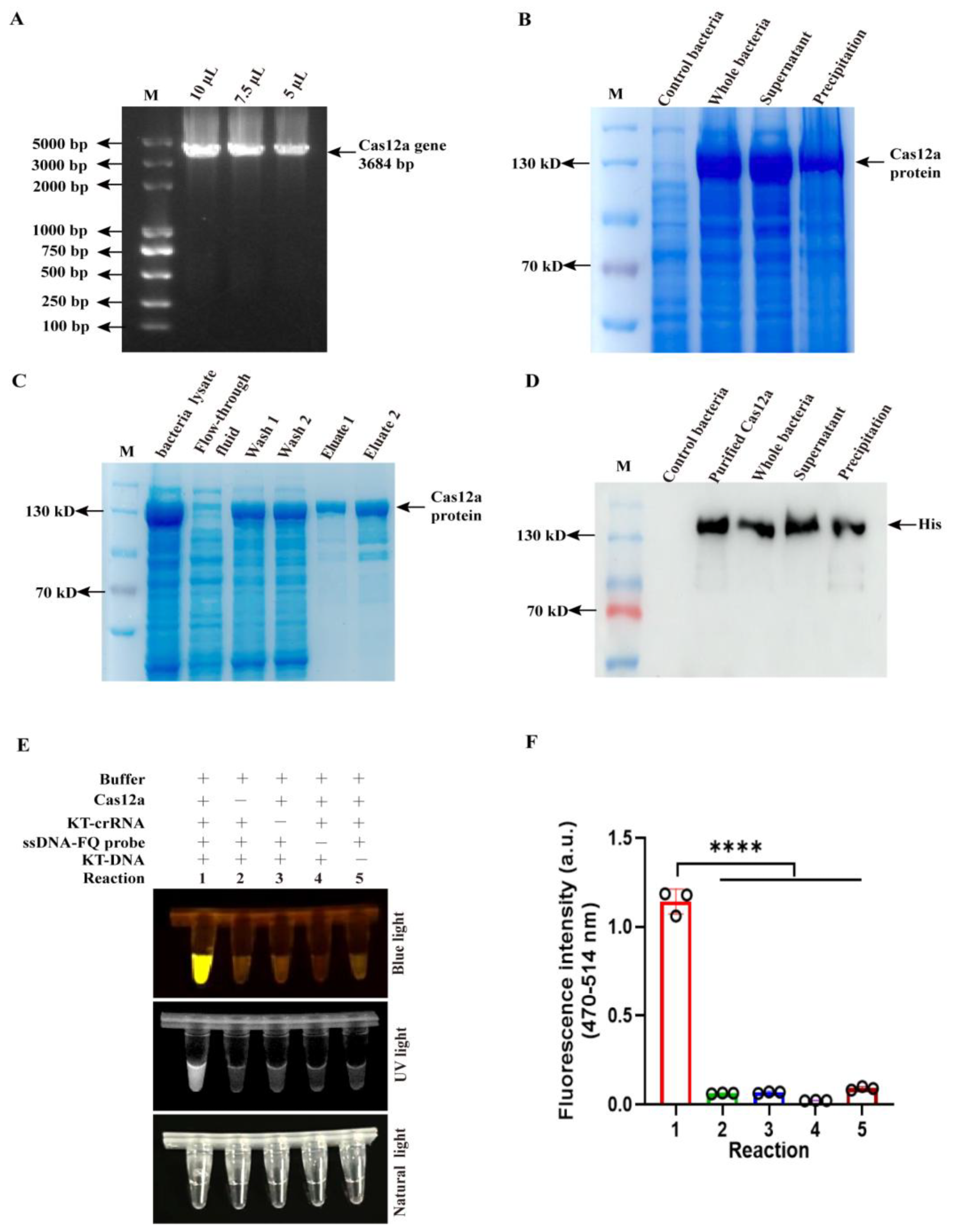
2.2. The Expression and Purification of Cas12a Protein
2.3. Viral cDNA Preparation of Various Viruses and Clinical Samples
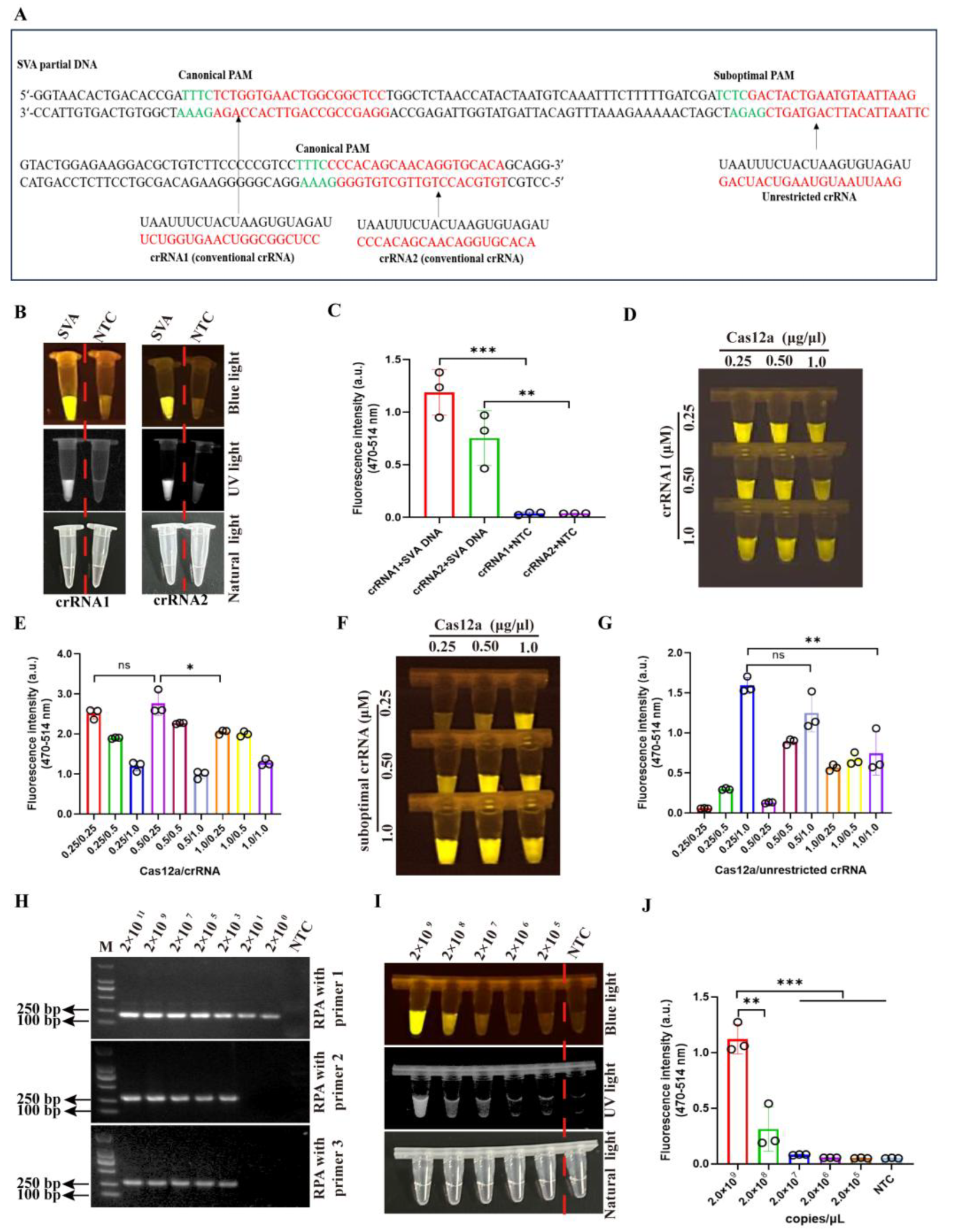
2.4. Design and Synthesization of crRNAs and ssDNA
2.5. Evaluation of Cas12a Endonuclease Activity
2.6. The Optimization of crRNA and Cas12a Concentrations
2.7. The Optimization of Recombinant Polymerase Amplification (RPA)
2.8. The Optimization of Sensitivity and Time in CRISPR/Cas12a-SVA Detection
2.9. Sensitivity and Specificity of Two-Pot RPA-CRISPR/Cas12a-SVA Detection Method
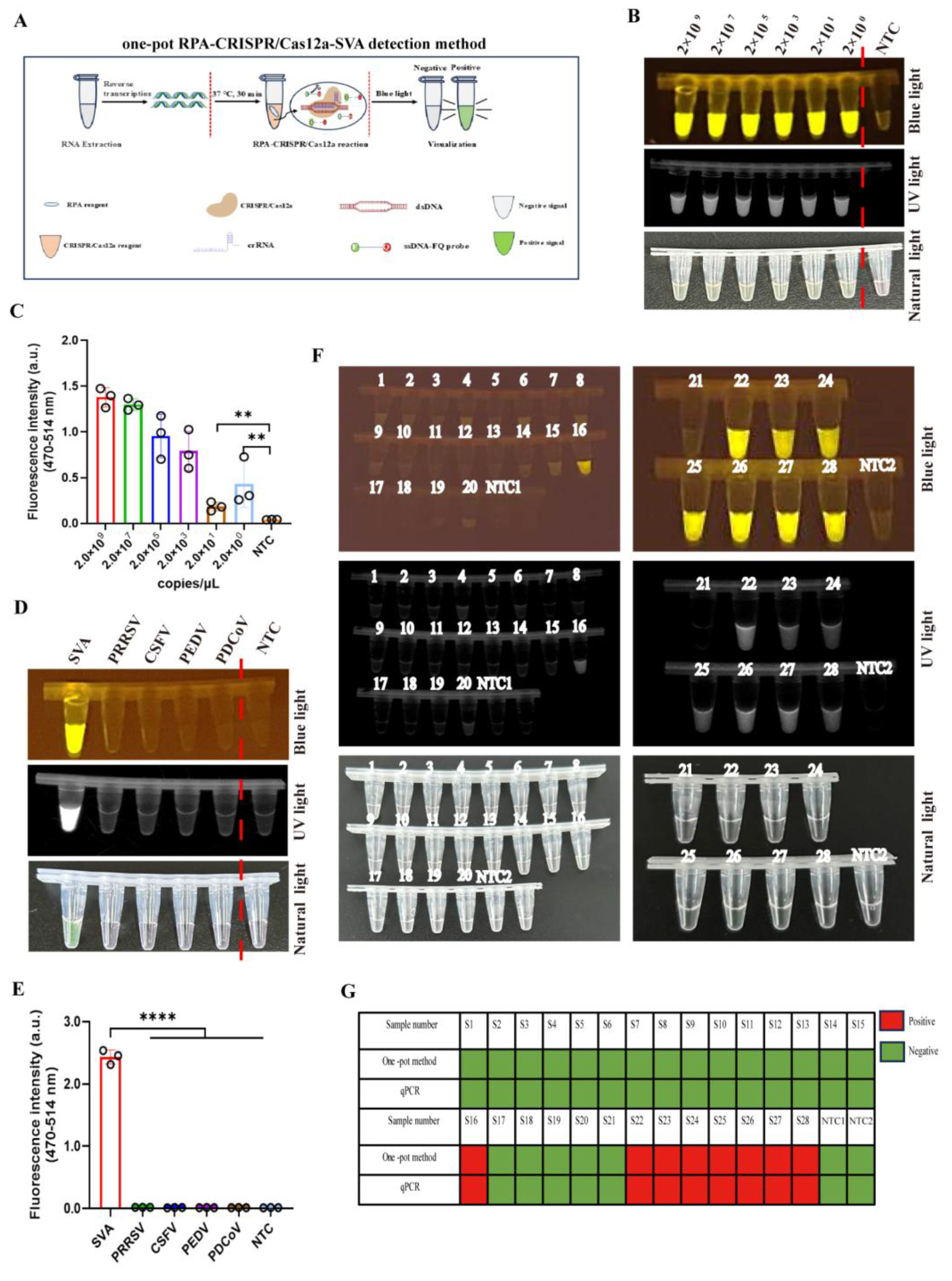
2.10. Establishment and Sensitivity of One-Pot RPA-CRISPR/Cas12a-SVA Detection Method
2.11. The Detection of Clinical Samples
2.12. Ethics Statement
2.13. Statistical Analysis
3. Results
3.1. Endonuclease Activity Assay of Purified Cas12a Protein
3.2. The Optimization of Cas12a and crRNA Concentration and RPA Primers in CRISPR/Cas12a-
SVA Detection Method
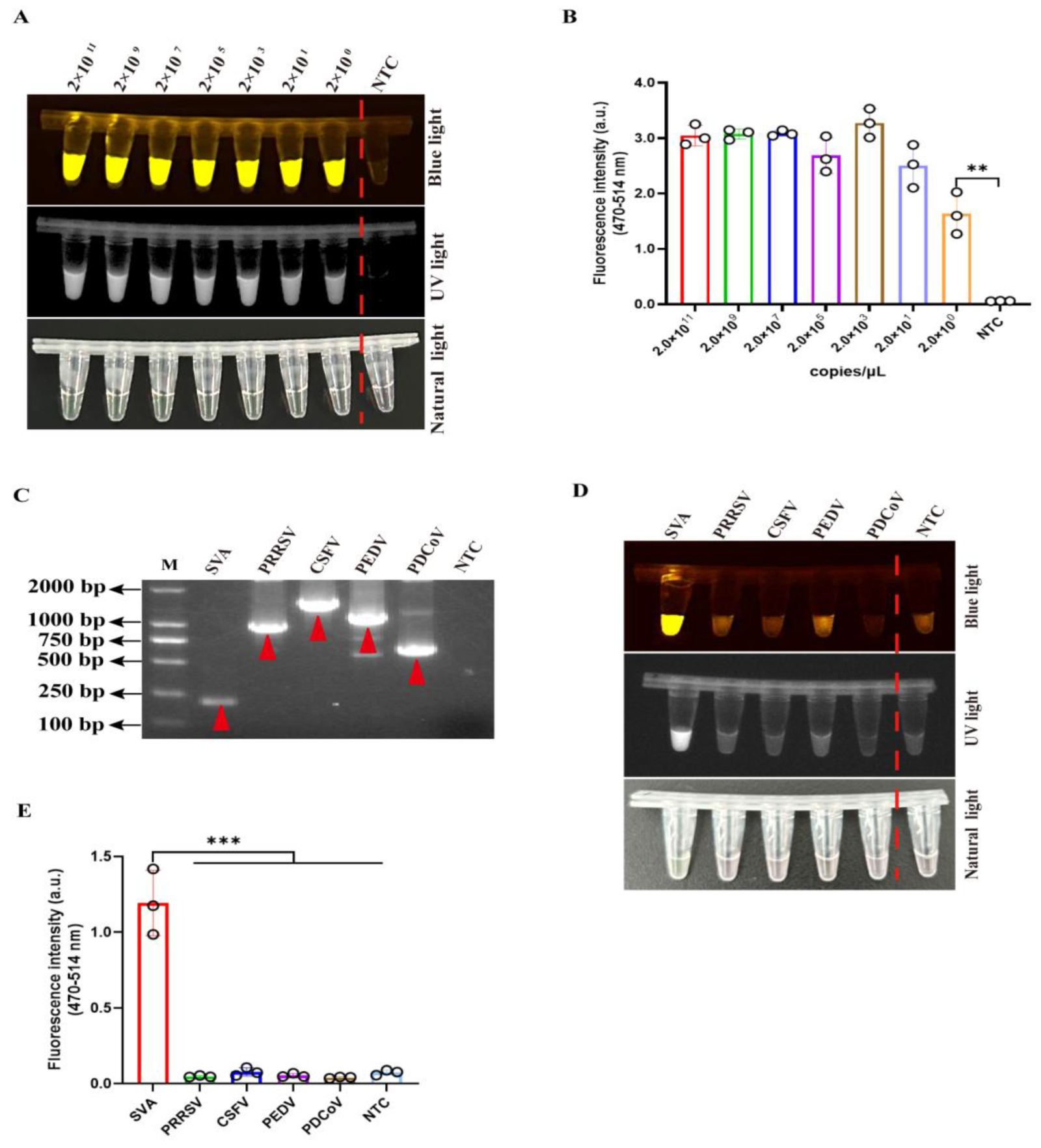
3.3. Development of the Two-Pot RPA-CRISPR/Cas12a Assay for SVA Detection
3.4. Assessment of Detection Sensitivity and Target Specificity in a Two-Pot RPA-CRISPR/Cas12a-
SVA System
3.5. Comparative Evaluation Between RPA-CRISPR/Cas12a-Based and qPCR Methods for SVA
3.6. Clinical Evaluation of a One-Pot RPA-CRISPR/Cas12a Assay for SVA Detection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, X.; Zhu, Z.; Yang, F.; Cao, W.; Tian, H.; Zhang, K.; Zheng, H.; Liu, X. Review of Seneca Valley Virus: A Call for Increased Surveillance and Research. Front. Microbiol. 2018, 9, 940. [Google Scholar] [CrossRef] [PubMed]
- Hales, L.M.; Knowles, N.J.; Reddy, P.S.; Xu, L.; Hay, C.; Hallenbeck, P.L. Complete genome sequence analysis of Seneca Valley virus-001, a novel oncolytic picornavirus. J. Gen. Virol. 2008, 89, 1265–1275. [Google Scholar] [CrossRef]
- Pasma, T.; Davidson, S.; Shaw, S.L. Idiopathic vesicular disease in swine in Manitoba. Can. Vet. J. 2008, 49, 84–85. [Google Scholar]
- Leme, R.A.; Alfieri, A.F.; Alfieri, A.A. Update on Senecavirus Infection in Pigs. Viruses 2017, 9, 170. [Google Scholar] [CrossRef]
- Wu, Q.; Zhao, X.; Bai, Y.; Sun, B.; Xie, Q.; Ma, J. The First Identification and Complete Genome of Senecavirus A Affecting Pig with Idiopathic Vesicular Disease in China. Transbound. Emerg. Dis. 2017, 64, 1633–1640. [Google Scholar] [CrossRef]
- Qian, S.; Fan, W.; Qian, P.; Chen, H.; Li, X. Isolation and full-genome sequencing of Seneca Valley virus in piglets from China, 2016. Virol. J. 2016, 13, 173. [Google Scholar] [CrossRef]
- Zhao, X.; Wu, Q.; Bai, Y.; Chen, G.; Zhou, L.; Wu, Z.; Li, Y.; Zhou, W.; Yang, H.; Ma, J. Phylogenetic and genome analysis of seven senecavirus A isolates in China. Transbound. Emerg. Dis. 2017, 64, 2075–2082. [Google Scholar] [CrossRef]
- Joshi, L.R.; Fernandes, M.H.V.; Clement, T.; Lawson, S.; Pillatzki, A.; Resende, T.P.; Vannucci, F.A.; Kutish, G.F.; Nelson, E.A.; Diel, D.G. Pathogenesis of Senecavirus A infection in finishing pigs. J. Gen. Virol. 2016, 97, 3267–3279. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, C.M.; Akkutay-Yoldar, Z.; Stone, S.R.; Tousignant, S.J.; Vannucci, F.A.; Murtaugh, M.P. An indirect enzyme-linked immunosorbent assay for the identification of antibodies to Senecavirus A in swine. BMC Vet. Res. 2017, 13, 50. [Google Scholar] [CrossRef] [PubMed]
- Feronato, C.; Leme, R.A.; Diniz, J.A.; Agnol, A.M.D.; Alfieri, A.F.; Alfieri, A.A. Development and evaluation of a nested-PCR assay for Senecavirus A diagnosis. Trop. Anim. Health Prod. 2018, 50, 337–344. [Google Scholar] [CrossRef]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef]
- Espy, M.J.; Uhl, J.R.; Sloan, L.M.; Buckwalter, S.P.; Jones, M.F.; Vetter, E.A.; Yao, J.D.; Wengenack, N.L.; Rosenblatt, J.E.; Cockerill, F.R., 3rd; et al. Real-time PCR in clinical microbiology: Applications for routine laboratory testing. Clin. Microbiol. Rev. 2006, 19, 165–256. [Google Scholar] [CrossRef]
- Li, S.Y.; Cheng, Q.X.; Wang, J.M.; Li, X.Y.; Zhang, Z.L.; Gao, S.; Cao, R.B.; Zhao, G.P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef]
- Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.; Wang, J. HOLMESv2: A CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. ACS Synth. Biol. 2019, 8, 2228–2237. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ji, P.; Fan, H.; Dang, L.; Wan, W.; Liu, S.; Li, Y.; Yu, W.; Li, X.; Ma, X.; et al. CRISPR/Cas12a technology combined with immunochromatographic strips for portable detection of African swine fever virus. Commun. Biol. 2020, 3, 62. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef]
- Zhang, L.; Sun, R.; Yang, M.; Peng, S.; Cheng, Y.; Chen, C. Conformational Dynamics and Cleavage Sites of Cas12a Are Modulated by Complementarity between crRNA and DNA. iScience 2019, 19, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Mao, S.; Wang, L.; Fu, S.; Dong, Y.; Jaffrezic-Renault, N.; Guo, Z. CRISPR/Cas and Argonaute-Based Biosensors for Pathogen Detection. ACS Sens. 2023, 8, 3623–3642. [Google Scholar] [CrossRef]
- Jiang, G.; Yang, X.; Li, Z.; Mao, J.; Zeng, P.; Wang, D.; Wu, Z.; Liu, C.; Qiu, Y.; Cui, Y.; et al. Recombinant Polymerase Amplification Coupled with CRISPR/Cas12a Detection System for Rapid Visual Detection of Porcine Circovirus 3. Animals 2024, 14, 2527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Jiang, S.; Xia, N.; Zhang, Y.; Zhang, J.; Liu, A.; Zhang, C.; Chen, N.; Meurens, F.; Zheng, W.; et al. Rapid Visual Detection of African Swine Fever Virus with a CRISPR/Cas12a Lateral Flow Strip Based on Structural Protein Gene D117L. Animals 2023, 13, 3712. [Google Scholar] [CrossRef]
- Lu, S.; Tong, X.; Han, Y.; Zhang, K.; Zhang, Y.; Chen, Q.; Duan, J.; Lei, X.; Huang, M.; Qiu, Y.; et al. Fast and sensitive detection of SARS-CoV-2 RNA using suboptimal protospacer adjacent motifs for Cas12a. Nat. Biomed. Eng. 2022, 6, 286–297. [Google Scholar] [CrossRef]
- Kanitchinda, S.; Srisala, J.; Suebsing, R.; Prachumwat, A.; Chaijarasphong, T. CRISPR-Cas fluorescent cleavage assay coupled with recombinase polymerase amplification for sensitive and specific detection of Enterocytozoon hepatopenaei. Biotechnol. Rep. 2020, 27, e00485. [Google Scholar] [CrossRef]
- Low, S.J.; O’Neill, M.T.; Kerry, W.J.; Krysiak, M.; Papadakis, G.; Whitehead, L.W.; Savic, I.; Prestedge, J.; Williams, L.; Cooney, J.P.; et al. Rapid detection of monkeypox virus using a CRISPR-Cas12a mediated assay: A laboratory validation and evaluation study. Lancet Microbe 2023, 4, e800–e810. [Google Scholar] [CrossRef]
- Ma, L.; Zhu, M.; Meng, Q.; Wang, Y.; Wang, X. Real-time detection of Seneca Valley virus by one-tube RPA-CRISPR/Cas12a assay. Front Cell Infect Microbiol. 2023, 13, 1305222. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Liao, K.; Li, R.; Peng, W.; Qian, B.; Zeng, D.; Tang, F.; Xue, F.; Jung, Y.S.; Dai, J. Development of a CRISPR/Cas12a-based fluorescent detection method of Senecavirus A. BMC Vet Res. 2024, 20, 258. [Google Scholar] [CrossRef] [PubMed]
- Fowler, V.L.; Ransburgh, R.H.; Poulsen, E.G.; Wadsworth, J.; King, D.P.; Mioulet, V.; Knowles, N.J.; Williamson, S.; Liu, X.; Anderson, G.A.; et al. Development of a novel real-time RT-PCR assay to detect Seneca Valley virus-1 associated with emerging cases of vesicular disease in pigs. J. Virol. Methods 2017, 239, 34–37. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J.; von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2018, 144, 31–67. [Google Scholar] [CrossRef]
- Tan, M.; Liao, C.; Liang, L.; Yi, X.; Zhou, Z.; Wei, G. Recent advances in recombinase polymerase amplification: Principle, advantages, disadvantages and applications. Front. Cell Infect. Microbiol. 2022, 12, 1019071. [Google Scholar] [CrossRef]
- Rolando, J.C.; Jue, E.; Barlow, J.T.; Ismagilov, R.F. Real-time kinetics and high-resolution melt curves in single-molecule digital LAMP to differentiate and study specific and non-specific amplification. Nucleic Acids Res. 2020, 48, e42. [Google Scholar] [CrossRef]
- Li, Y.; Li, S.; Wang, J.; Liu, G. CRISPR/Cas Systems towards Next-Generation Biosensing. Trends Biotechnol. 2019, 37, 730–743. [Google Scholar] [CrossRef]
- Li, Y.; Liu, L.; Liu, G. CRISPR/Cas Multiplexed Biosensing: A Challenge or an Insurmountable Obstacle? Trends Biotechnol. 2019, 37, 792–795. [Google Scholar] [CrossRef] [PubMed]
- Ren, M.; Mei, H.; Zhou, M.; Fu, Z.F.; Han, H.; Bi, D.; Peng, F.; Zhao, L. Development of A Super-Sensitive Diagnostic Method for African Swine Fever Using CRISPR Techniques. Virol. Sin. 2021, 36, 220–230. [Google Scholar] [CrossRef]
- Sun, Y.; Yu, L.; Liu, C.; Ye, S.; Chen, W.; Li, D.; Huang, W. One-tube SARS-CoV-2 detection platform based on RT-RPA and CRISPR/Cas12a. J. Transl. Med. 2021, 19, 74. [Google Scholar] [CrossRef]
- Yin, K.; Ding, X.; Li, Z.; Zhao, H.; Cooper, K.; Liu, C. Dynamic Aqueous Multiphase Reaction System for One-Pot CRISPR-Cas12a-Based Ultrasensitive and Quantitative Molecular Diagnosis. Anal. Chem. 2020, 92, 8561–8568. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Guo, J.; Guo, X.; Li, Q.; Li, X.; Sun, Z.; Zhao, Z.; Weng, J.; Wu, J.; Zhang, R.; et al. Fast and visual detection of nucleic acids using a one-step RPA-CRISPR detection (ORCD) system unrestricted by the PAM. Anal. Chim. Acta. 2023, 1248, 340938. [Google Scholar] [CrossRef] [PubMed]


| Sample Types | Sample Numbers | RPA-CRISPR/Cas12a | qPCR |
|---|---|---|---|
| Spleen | 3 | 1/3 | 1/3 |
| Lung | 4 | 0/4 | 0/4 |
| Kidney | 2 | 1/2 | 1/2 |
| colorectal | 4 | 0/4 | 0/4 |
| tonsil | 3 | 1/3 | 1/3 |
| heart | 2 | 0/2 | 0/2 |
| liver | 4 | 1/4 | 1/4 |
| bronchial lymph node | 3 | 2/3 | 2/3 |
| Submandibular lymph node | 1 | 1/1 | 1/1 |
| Inguinal lymph node | 2 | 1/2 | 1/2 |
| Positive rates | 28.5% | 28.5% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Jiang, G.; Ruan, Q.; Qu, Y.; Yang, X.; Shi, Y.; Wang, D.; Zhou, J.; Liu, J.; Hou, L. Rapid Visual Detection of Senecavirus A Based on RPA-CRISPR/Cas12a System with Canonical or Suboptimal PAM. Viruses 2025, 17, 1264. https://doi.org/10.3390/v17091264
Zhao X, Jiang G, Ruan Q, Qu Y, Yang X, Shi Y, Wang D, Zhou J, Liu J, Hou L. Rapid Visual Detection of Senecavirus A Based on RPA-CRISPR/Cas12a System with Canonical or Suboptimal PAM. Viruses. 2025; 17(9):1264. https://doi.org/10.3390/v17091264
Chicago/Turabian StyleZhao, Xinrui, Genghong Jiang, Qinyi Ruan, Yunjie Qu, Xiaoyu Yang, Yongyan Shi, Dedong Wang, Jianwei Zhou, Jue Liu, and Lei Hou. 2025. "Rapid Visual Detection of Senecavirus A Based on RPA-CRISPR/Cas12a System with Canonical or Suboptimal PAM" Viruses 17, no. 9: 1264. https://doi.org/10.3390/v17091264
APA StyleZhao, X., Jiang, G., Ruan, Q., Qu, Y., Yang, X., Shi, Y., Wang, D., Zhou, J., Liu, J., & Hou, L. (2025). Rapid Visual Detection of Senecavirus A Based on RPA-CRISPR/Cas12a System with Canonical or Suboptimal PAM. Viruses, 17(9), 1264. https://doi.org/10.3390/v17091264





