Comprehensive Analysis of the Impact of Weight Loss Thresholds on Mouse Models of Fatal Viral Infection
Abstract
1. Introduction
2. Methods
2.1. Institutional Approvals
2.2. Mammalian Cell Lines
2.3. SARS-CoV-2 Viral Stock Preparation and Titration
2.4. Influenza A/Puerto Rico/8/1934 (H1N1) Viral Stock Preparation and Titration
2.5. Powassan Virus (LB Strain) Viral Stock Preparation and Titration
2.6. Mouse Strains
2.7. SARS-CoV-2 and IAV PR8 Infections in Mice
2.8. POWV Infections in Mice
2.9. Clinical Scoring and Monitoring for Survival
2.10. Statistical Analysis
3. Results
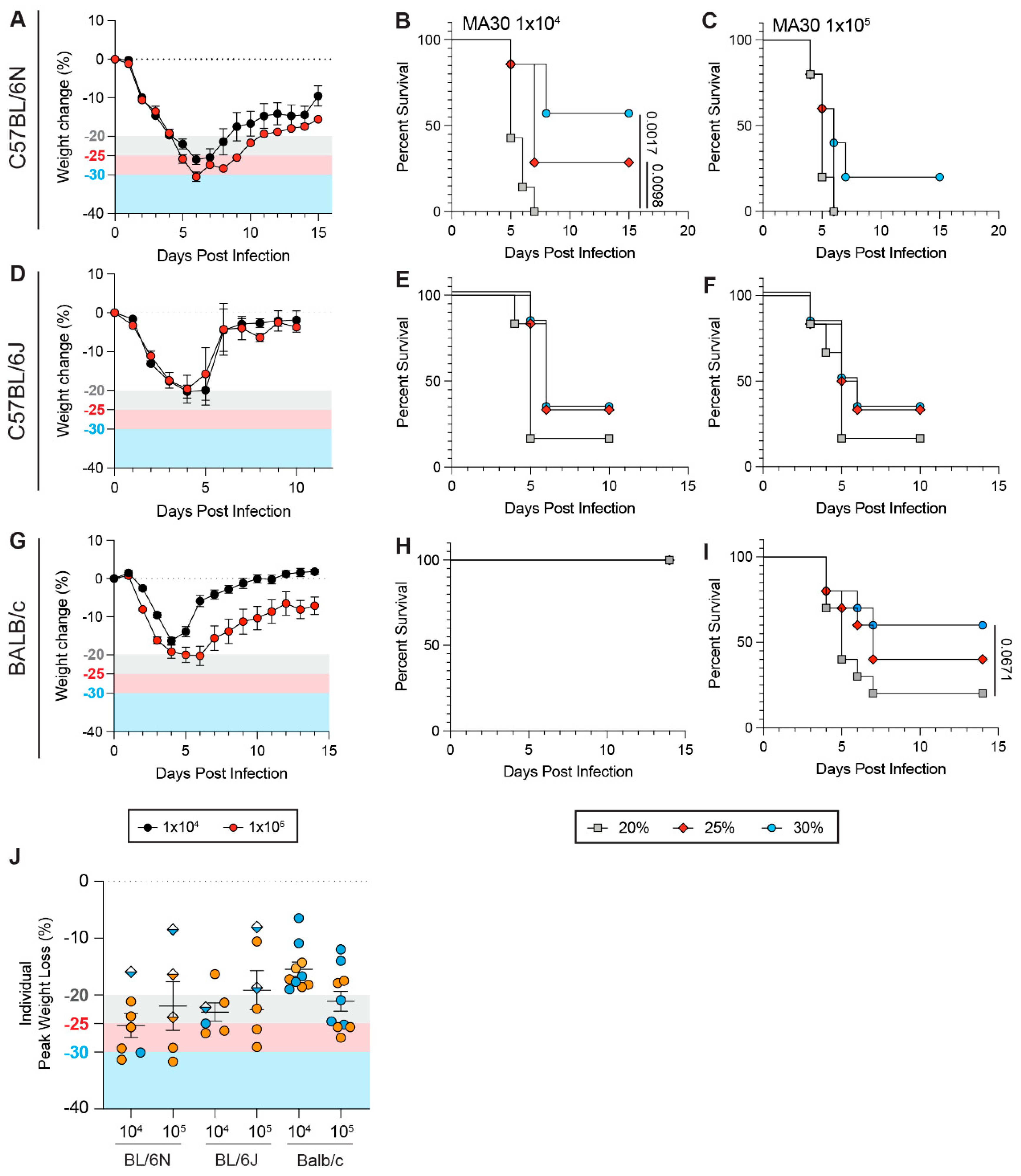
3.1. Weight Loss Threshold Significantly Impacts the Survival Rates of C57BL/6Ntac Mice upon SARS-CoV-2 Challenge
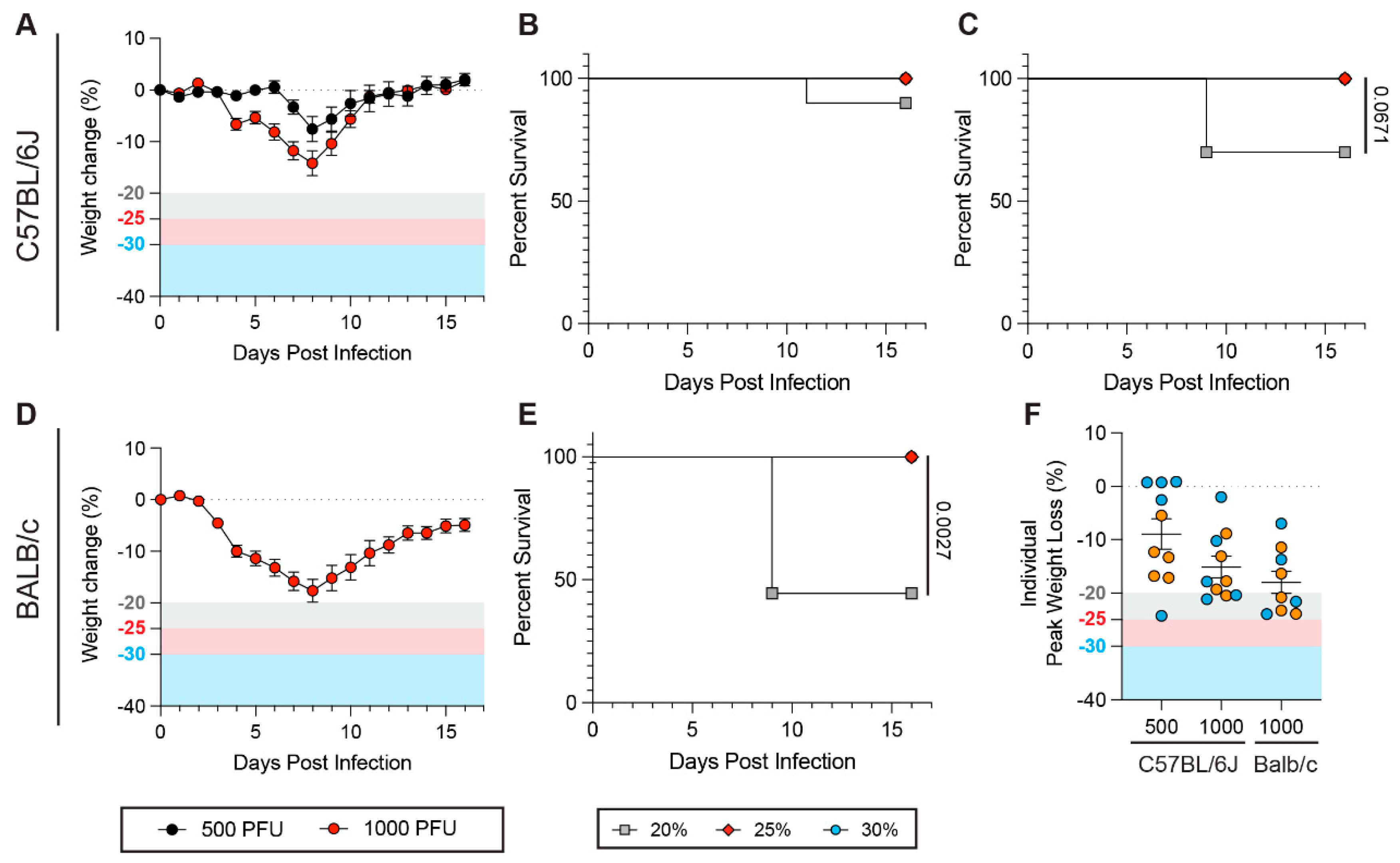
3.2. Genetic Polymorphisms Between Mouse Strains and Sub-Strains Influence How Weight Loss Thresholds Affect Survival Rates During SARS-CoV-2 Infection
3.3. Viral Genetic Variants Influence the Impact of Weight Loss Thresholds on Survival Rates During SARS-CoV-2 Infection
3.4. Biological Sex Is a Confounding Variable When Establishing Weight Loss Thresholds
3.5. Weight Loss Thresholds Significantly Impact Survival Rates upon Influenza A Virus Infection
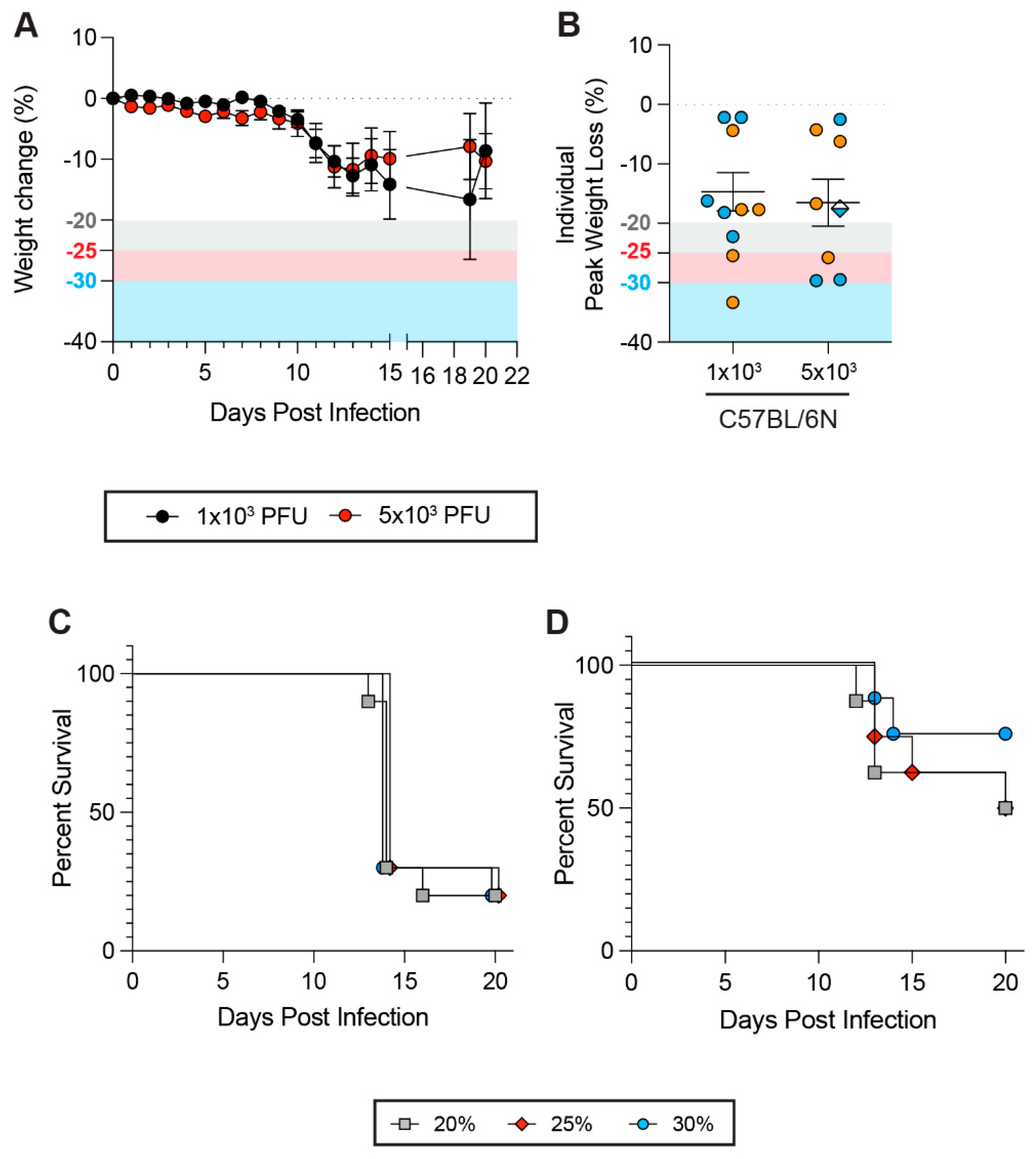
3.6. Impacts of Weight Loss Thresholds on Non-Respiratory Viral Infections
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brochut, M.; Heinonen, T.; Snaka, T.; Gilbert, C.; Le Roy, D.; Roger, T. Using weight loss to predict outcome and define a humane endpoint in preclinical sepsis studies. Sci. Rep. 2024, 14, 21150. [Google Scholar] [CrossRef] [PubMed]
- Rutigliano, J.A.; Sharma, S.; Morris, M.Y.; Oguin, T.H., 3rd; McClaren, J.L.; Doherty, P.C.; Thomas, P.G. Highly pathological influenza A virus infection is associated with augmented expression of PD-1 by functionally compromised virus-specific CD8+ T cells. J. Virol. 2014, 88, 1636–1651. [Google Scholar] [CrossRef] [PubMed]
- Felgenhauer, J.L.; Brune, J.E.; Long, M.E.; Manicone, A.M.; Chang, M.Y.; Brabb, T.L.; Altemeier, W.A.; Frevert, C.W. Evaluation of Nutritional Gel Supplementation in C57BL/6J Mice Infected with Mouse-Adapted Influenza A/PR/8/34 Virus. Comp. Med. 2020, 70, 471–486. [Google Scholar] [CrossRef]
- Gilchuk, I.M.; Bangaru, S.; Gilchuk, P.; Irving, R.P.; Kose, N.; Bombardi, R.G.; Thornburg, N.J.; Creech, C.B.; Edwards, K.M.; Li, S.; et al. Influenza H7N9 Virus Neuraminidase-Specific Human Monoclonal Antibodies Inhibit Viral Egress and Protect from Lethal Influenza Infection in Mice. Cell Host Microbe 2019, 26, 715–728.e8. [Google Scholar] [CrossRef]
- Song, Y.; Wang, X.; Zhang, H.; Tang, X.; Li, M.; Yao, J.; Jin, X.; Ertl, H.C.; Zhou, D. Repeated Low-Dose Influenza Virus Infection Causes Severe Disease in Mice: A Model for Vaccine Evaluation. J. Virol. 2015, 89, 7841–7851. [Google Scholar] [CrossRef]
- Douam, F.; Soto Albrecht, Y.E.; Hrebikova, G.; Sadimin, E.; Davidson, C.; Kotenko, S.V.; Ploss, A. Type III Interferon-Mediated Signaling Is Critical for Controlling Live Attenuated Yellow Fever Virus Infection In Vivo. Mbio 2017, 8. [Google Scholar] [CrossRef]
- Dowall, S.D.; Graham, V.A.; Rayner, E.; Atkinson, B.; Hall, G.; Watson, R.J.; Bosworth, A.; Bonney, L.C.; Kitchen, S.; Hewson, R. A Susceptible Mouse Model for Zika Virus Infection. PLoS Negl. Trop. Dis. 2016, 10, e0004658. [Google Scholar] [CrossRef] [PubMed]
- Winkler, C.W.; Myers, L.M.; Woods, T.A.; Messer, R.J.; Carmody, A.B.; McNally, K.L.; Scott, D.P.; Hasenkrug, K.J.; Best, S.M.; Peterson, K.E. Adaptive Immune Responses to Zika Virus Are Important for Controlling Virus Infection and Preventing Infection in Brain and Testes. J. Immunol. 2017, 198, 3526–3535. [Google Scholar] [CrossRef]
- Aliota, M.T.; Caine, E.A.; Walker, E.C.; Larkin, K.E.; Camacho, E.; Osorio, J.E. Characterization of Lethal Zika Virus Infection in AG129 Mice. PLoS Negl. Trop. Dis. 2016, 10, e0004682. [Google Scholar] [CrossRef]
- Zust, R.; Toh, Y.X.; Valdes, I.; Cerny, D.; Heinrich, J.; Hermida, L.; Marcos, E.; Guillen, G.; Kalinke, U.; Shi, P.Y.; et al. Type I interferon signals in macrophages and dendritic cells control dengue virus infection: Implications for a new mouse model to test dengue vaccines. J. Virol. 2014, 88, 7276–7285. [Google Scholar] [CrossRef]
- Zheng, Z.; Li, M.; Liu, Z.; Jin, X.; Sun, J. Establishment of Murine Infection Models with Biological Clones of Dengue Viruses Derived from a Single Clinical Viral Isolate. Virol. Sin. 2020, 35, 626–636. [Google Scholar] [CrossRef]
- Watson, A.M.; Lam, L.K.; Klimstra, W.B.; Ryman, K.D. The 17D-204 Vaccine Strain-Induced Protection against Virulent Yellow Fever Virus Is Mediated by Humoral Immunity and CD4+ but not CD8+ T Cells. PLoS Pathog. 2016, 12, e1005786. [Google Scholar] [CrossRef]
- Graham, J.B.; Swarts, J.L.; Lund, J.M. A Mouse Model of West Nile Virus Infection. Curr. Protoc. Mouse Biol. 2017, 7, 221–235. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhou, L.; Liu, Q.; Zheng, Y.; Tan, X.; Huang, Z.; Guo, M.; Wang, X.; Chen, X.; Liang, S.; et al. The lethal K18-hACE2 knock-in mouse model mimicking the severe pneumonia of COVID-19 is practicable for antiviral development. Emerg. Microbes Infect. 2024, 13, 2353302. [Google Scholar] [CrossRef] [PubMed]
- Gan, E.S.; Syenina, A.; Linster, M.; Ng, B.; Zhang, S.L.; Watanabe, S.; Rajarethinam, R.; Tan, H.C.; Smith, G.J.; Ooi, E.E. A mouse model of lethal respiratory dysfunction for SARS-CoV-2 infection. Antivir. Res. 2021, 193, 105138. [Google Scholar] [CrossRef] [PubMed]
- Myeni, S.K.; Leijs, A.A.; Bredenbeek, P.J.; Morales, S.T.; Linger, M.E.; Fougeroux, C.; van Zanen-Gerhardt, S.; Zander, S.A.L.; Sander, A.F.; Kikkert, M. Protection of K18-hACE2 Mice against SARS-CoV-2 Challenge by a Capsid Virus-like Particle-Based Vaccine. Vaccines 2024, 12, 766. [Google Scholar] [CrossRef] [PubMed]
- Tarres-Freixas, F.; Trinite, B.; Pons-Grifols, A.; Romero-Durana, M.; Riveira-Munoz, E.; Avila-Nieto, C.; Perez, M.; Garcia-Vidal, E.; Perez-Zsolt, D.; Munoz-Basagoiti, J.; et al. Heterogeneous Infectivity and Pathogenesis of SARS-CoV-2 Variants Beta, Delta and Omicron in Transgenic K18-hACE2 and Wildtype Mice. Front. Microbiol. 2022, 13, 840757. [Google Scholar] [CrossRef]
- Carossino, M.; Kenney, D.; O’Connell, A.K.; Montanaro, P.; Tseng, A.E.; Gertje, H.P.; Grosz, K.A.; Ericsson, M.; Huber, B.R.; Kurnick, S.A.; et al. Fatal Neurodissemination and SARS-CoV-2 Tropism in K18-hACE2 Mice Is Only Partially Dependent on hACE2 Expression. Viruses 2022, 14, 535. [Google Scholar] [CrossRef]
- Mann, B.J.; Chhabra, P.; Ma, M.; Brovero, S.G.; Hannan, R.T.; Sturek, J.M.; Jones, M.K.; Linden, J.; Brayman, K.L. Improved survival of SARS COV-2-infected K18-hACE2 mice treated with adenosine A(2A)R agonist. Heliyon 2023, 9, e19226. [Google Scholar] [CrossRef]
- Chen, D.Y.; Chin, C.V.; Kenney, D.; Tavares, A.H.; Khan, N.; Conway, H.L.; Liu, G.; Choudhary, M.C.; Gertje, H.P.; O’Connell, A.K.; et al. Spike and nsp6 are key determinants of SARS-CoV-2 Omicron BA.1 attenuation. Nature 2023, 615, 143–150. [Google Scholar] [CrossRef]
- Iwata-Yoshikawa, N.; Shiwa, N.; Sekizuka, T.; Sano, K.; Ainai, A.; Hemmi, T.; Kataoka, M.; Kuroda, M.; Hasegawa, H.; Suzuki, T.; et al. A lethal mouse model for evaluating vaccine-associated enhanced respiratory disease during SARS-CoV-2 infection. Sci. Adv. 2022, 8, eabh3827. [Google Scholar] [CrossRef] [PubMed]
- Silvas, J.A.; Vasquez, D.M.; Park, J.G.; Chiem, K.; Allue-Guardia, A.; Garcia-Vilanova, A.; Platt, R.N.; Miorin, L.; Kehrer, T.; Cupic, A.; et al. Contribution of SARS-CoV-2 Accessory Proteins to Viral Pathogenicity in K18 Human ACE2 Transgenic Mice. J. Virol. 2021, 95, e0040221. [Google Scholar] [CrossRef]
- Oladunni, F.S.; Park, J.G.; Pino, P.A.; Gonzalez, O.; Akhter, A.; Allue-Guardia, A.; Olmo-Fontanez, A.; Gautam, S.; Garcia-Vilanova, A.; Ye, C.; et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020, 11, 6122. [Google Scholar] [CrossRef]
- Davis, M.; Voss, K.; Turnbull, J.B.; Gustin, A.T.; Knoll, M.; Muruato, A.; Hsiang, T.Y.; Dinnon, K.H., 3rd; Leist, S.R.; Nickel, K.; et al. A C57BL/6 Mouse model of SARS-CoV-2 infection recapitulates age- and sex-based differences in human COVID-19 disease and recovery. Res. Sq. 2022. [Google Scholar] [CrossRef]
- Robertson, S.J.; Bedard, O.; McNally, K.L.; Shaia, C.; Clancy, C.S.; Lewis, M.; Broeckel, R.M.; Chiramel, A.I.; Shannon, J.G.; Sturdevant, G.L.; et al. Genetically diverse mouse models of SARS-CoV-2 infection reproduce clinical variation in type I interferon and cytokine responses in COVID-19. Nat. Commun. 2023, 14, 4481. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, J.; Jang, J.Y.; Noh, H.; Park, J.; Jeong, H.; Jeon, D.; Uhm, C.; Oh, H.; Cho, K.; et al. Mouse models of lung-specific SARS-CoV-2 infection with moderate pathological traits. Front. Immunol. 2022, 13, 1055811. [Google Scholar] [CrossRef]
- Yinda, C.K.; Port, J.R.; Bushmaker, T.; Offei Owusu, I.; Purushotham, J.N.; Avanzato, V.A.; Fischer, R.J.; Schulz, J.E.; Holbrook, M.G.; Hebner, M.J.; et al. K18-hACE2 mice develop respiratory disease resembling severe COVID-19. PLoS Pathog. 2021, 17, e1009195. [Google Scholar] [CrossRef] [PubMed]
- Winkler, E.S.; Bailey, A.L.; Kafai, N.M.; Nair, S.; McCune, B.T.; Yu, J.; Fox, J.M.; Chen, R.E.; Earnest, J.T.; Keeler, S.P.; et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020, 21, 1327–1335. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Wong, L.R.; Li, K.; Verma, A.K.; Ortiz, M.E.; Wohlford-Lenane, C.; Leidinger, M.R.; Knudson, C.M.; Meyerholz, D.K.; McCray, P.B., Jr.; et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 2021, 589, 603–607. [Google Scholar] [CrossRef]
- Ellsworth, C.R.; Wang, C.; Katz, A.R.; Chen, Z.; Islamuddin, M.; Yang, H.; Scheuermann, S.E.; Goff, K.A.; Maness, N.J.; Blair, R.V.; et al. Natural Killer Cells Do Not Attenuate a Mouse-Adapted SARS-CoV-2-Induced Disease in Rag2(-/-) Mice. Viruses 2024, 16, 611. [Google Scholar] [CrossRef]
- Montoya, B.; Melo-Silva, C.R.; Tang, L.; Kafle, S.; Lidskiy, P.; Bajusz, C.; Vadovics, M.; Muramatsu, H.; Abraham, E.; Lipinszki, Z.; et al. mRNA-LNP vaccine-induced CD8(+) T cells protect mice from lethal SARS-CoV-2 infection in the absence of specific antibodies. Mol. Ther. 2024, 32, 1790–1804. [Google Scholar] [CrossRef]
- Dong, W.; Mead, H.; Tian, L.; Park, J.G.; Garcia, J.I.; Jaramillo, S.; Barr, T.; Kollath, D.S.; Coyne, V.K.; Stone, N.E.; et al. The K18-Human ACE2 Transgenic Mouse Model Recapitulates Non-severe and Severe COVID-19 in Response to an Infectious Dose of the SARS-CoV-2 Virus. J. Virol. 2022, 96, e0096421. [Google Scholar] [CrossRef]
- Rosenfeld, R.; Noy-Porat, T.; Mechaly, A.; Makdasi, E.; Levy, Y.; Alcalay, R.; Falach, R.; Aftalion, M.; Epstein, E.; Gur, D.; et al. Post-exposure protection of SARS-CoV-2 lethal infected K18-hACE2 transgenic mice by neutralizing human monoclonal antibody. Nat. Commun. 2021, 12, 944. [Google Scholar] [CrossRef]
- Bayarri-Olmos, R.; Johnsen, L.B.; Idorn, M.; Reinert, L.S.; Rosbjerg, A.; Vang, S.; Hansen, C.B.; Helgstrand, C.; Bjelke, J.R.; Bak-Thomsen, T.; et al. The alpha/B.1.1.7 SARS-CoV-2 variant exhibits significantly higher affinity for ACE-2 and requires lower inoculation doses to cause disease in K18-hACE2 mice. Elife 2021, 10. [Google Scholar] [CrossRef]
- Leist, S.R.; Dinnon, K.H., 3rd; Schafer, A.; Tse, L.V.; Okuda, K.; Hou, Y.J.; West, A.; Edwards, C.E.; Sanders, W.; Fritch, E.J.; et al. A Mouse-Adapted SARS-CoV-2 Induces Acute Lung Injury and Mortality in Standard Laboratory Mice. Cell 2020, 183, 1070–1085.e12. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.R.; Zheng, J.; Wilhelmsen, K.; Li, K.; Ortiz, M.E.; Schnicker, N.J.; Thurman, A.; Pezzulo, A.A.; Szachowicz, P.J.; Li, P.; et al. Eicosanoid signalling blockade protects middle-aged mice from severe COVID-19. Nature 2022, 605, 146–151. [Google Scholar] [CrossRef] [PubMed]
- Fumagalli, V.; Rava, M.; Marotta, D.; Di Lucia, P.; Laura, C.; Sala, E.; Grillo, M.; Bono, E.; Giustini, L.; Perucchini, C.; et al. Administration of aerosolized SARS-CoV-2 to K18-hACE2 mice uncouples respiratory infection from fatal neuroinvasion. Sci. Immunol. 2022, 7, eabl9929. [Google Scholar] [CrossRef]
- Drelich, A.K.; Rayavara, K.; Hsu, J.; Saenkham-Huntsinger, P.; Judy, B.M.; Tat, V.; Ksiazek, T.G.; Peng, B.H.; Tseng, C.K. Characterization of Unique Pathological Features of COVID-Associated Coagulopathy: Studies with AC70 hACE2 Transgenic Mice Highly Permissive to SARS-CoV-2 Infection. PLoS Pathog. 2024, 20, e1011777. [Google Scholar] [CrossRef] [PubMed]
- Dinnon, K.H., 3rd; Leist, S.R.; Schafer, A.; Edwards, C.E.; Martinez, D.R.; Montgomery, S.A.; West, A.; Yount, B.L., Jr.; Hou, Y.J.; Adams, L.E.; et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 2020, 586, 560–566. [Google Scholar] [CrossRef]
- de Wit, E.; Spronken, M.I.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Osterhaus, A.D.; Fouchier, R.A. Efficient generation and growth of influenza virus A/PR/8/34 from eight cDNA fragments. Virus Res. 2004, 103, 155–161. [Google Scholar] [CrossRef]
- Matrosovich, M.; Matrosovich, T.; Garten, W.; Klenk, H.D. New low-viscosity overlay medium for viral plaque assays. Virol. J. 2006, 3, 63. [Google Scholar] [CrossRef]
- Mekada, K.; Yoshiki, A. Substrains matter in phenotyping of C57BL/6 mice. Exp. Anim. 2021, 70, 145–160. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.M.; Greenaway, S.; White, J.K.; Fuchs, H.; Gailus-Durner, V.; Wells, S.; Sorg, T.; Wong, K.; Bedu, E.; Cartwright, E.J.; et al. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 2013, 14, R82. [Google Scholar] [CrossRef]
- Malm Tillgren, S.; Nieto-Fontarigo, J.J.; Cerps, S.; Ramu, S.; Menzel, M.; Mahmutovic Persson, I.; Meissner, A.; Akbarshahi, H.; Uller, L. C57Bl/6N mice have an attenuated lung inflammatory response to dsRNA compared to C57Bl/6J and BALB/c mice. J. Inflamm. 2023, 20, 6. [Google Scholar] [CrossRef]
- Watanabe, H.; Numata, K.; Ito, T.; Takagi, K.; Matsukawa, A. Innate immune response in Th1- and Th2-dominant mouse strains. Shock 2004, 22, 460–466. [Google Scholar] [CrossRef]
- McCray, P.B., Jr.; Pewe, L.; Wohlford-Lenane, C.; Hickey, M.; Manzel, L.; Shi, L.; Netland, J.; Jia, H.P.; Halabi, C.; Sigmund, C.D.; et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007, 81, 813–821. [Google Scholar] [CrossRef]
- Yang, J.H.; Yang, M.S.; Kim, D.M.; Kim, B.; Tark, D.; Kang, S.M.; Lee, G.H. Delta (B1.617.2) variant of SARS-CoV-2 induces severe neurotropic patterns in K18-hACE2 mice. Sci. Rep. 2023, 13, 3303. [Google Scholar] [CrossRef]
- Imbiakha, B.; Ezzatpour, S.; Buchholz, D.W.; Sahler, J.; Ye, C.; Olarte-Castillo, X.A.; Zou, A.; Kwas, C.; O’Hare, K.; Choi, A.; et al. Age-dependent acquisition of pathogenicity by SARS-CoV-2 Omicron BA.5. Sci. Adv. 2023, 9, eadj1736. [Google Scholar] [CrossRef] [PubMed]
- Kuruppuarachchi, K.; Jang, Y.; Seo, S.H. Comparison of the Pathogenicity of SARS-CoV-2 Delta and Omicron Variants by Analyzing the Expression Patterns of Immune Response Genes in K18-hACE2 Transgenic Mice. Front. Biosci. 2022, 27, 316. [Google Scholar] [CrossRef]
- Halfmann, P.J.; Iida, S.; Iwatsuki-Horimoto, K.; Maemura, T.; Kiso, M.; Scheaffer, S.M.; Darling, T.L.; Joshi, A.; Loeber, S.; Singh, G.; et al. SARS-CoV-2 Omicron virus causes attenuated disease in mice and hamsters. Nature 2022, 603, 687–692. [Google Scholar] [CrossRef]
- Reed, D.R.; Bachmanov, A.A.; Tordoff, M.G. Forty mouse strain survey of body composition. Physiol. Behav. 2007, 91, 593–600. [Google Scholar] [CrossRef]
- Twitchell, D.K.; Christensen, M.B.; Hackett, G.; Morgentaler, A.; Saad, F.; Pastuszak, A.W. Examining Male Predominance of Severe COVID-19 Outcomes: A Systematic Review. Androg. Clin. Res. Ther. 2022, 3, 41–53. [Google Scholar] [CrossRef]
- Fabiao, J.; Sassi, B.; Pedrollo, E.F.; Gerchman, F.; Kramer, C.K.; Leitao, C.B.; Pinto, L.C. Why do men have worse COVID-19-related outcomes? A systematic review and meta-analysis with sex adjusted for age. Braz. J. Med. Biol. Res. 2022, 55, e11711. [Google Scholar] [CrossRef]
- Jin, J.M.; Bai, P.; He, W.; Wu, F.; Liu, X.F.; Han, D.M.; Liu, S.; Yang, J.K. Gender Differences in Patients With COVID-19: Focus on Severity and Mortality. Front. Public Health 2020, 8, 152. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Chinn, J.; De Ferrante, M.; Kirby, K.A.; Hohmann, S.F.; Amin, A. Male gender is a predictor of higher mortality in hospitalized adults with COVID-19. PLoS ONE 2021, 16, e0254066. [Google Scholar] [CrossRef]
- Giacomelli, A.; De Falco, T.; Oreni, L.; Pedroli, A.; Ridolfo, A.L.; Calabro, E.; Carrozzo, G.; Bonazzetti, C.; Antinori, S.; Brucato, A. Impact of gender on patients hospitalized for SARS-COV-2 infection: A prospective observational study. J. Med. Virol. 2021, 93, 4597–4602. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Adam, M.; Mladinich, M.C. Pathogenicity and virulence of Powassan virus. Virulence 2025, 16, 2523887. [Google Scholar] [CrossRef] [PubMed]
- Telford, S.R.; Piantadosi, A.L. Powassan virus persistence after acute infection. mBio 2023, 14, e0071223. [Google Scholar] [CrossRef]
- Stone, E.T.; Hassert, M.; Geerling, E.; Wagner, C.; Brien, J.D.; Ebel, G.D.; Hirsch, A.J.; German, C.; Smith, J.L.; Pinto, A.K. Balanced T and B cell responses are required for immune protection against Powassan virus in virus-like particle vaccination. Cell Rep. 2022, 38, 110388. [Google Scholar] [CrossRef] [PubMed]
- Bryant, C.D. The blessings and curses of C57BL/6 substrains in mouse genetic studies. Ann. N. Y. Acad. Sci. 2011, 1245, 31–33. [Google Scholar] [CrossRef]
- Song, H.K.; Hwang, D.Y. Use of C57BL/6N mice on the variety of immunological researches. Lab. Anim. Res. 2017, 33, 119–123. [Google Scholar] [CrossRef]
- Bothe, G.W.; Bolivar, V.J.; Vedder, M.J.; Geistfeld, J.G. Genetic and behavioral differences among five inbred mouse strains commonly used in the production of transgenic and knockout mice. Genes Brain Behav. 2004, 3, 149–157. [Google Scholar] [CrossRef]
- Nemoto, S.; Kubota, T.; Ohno, H. Metabolic differences and differentially expressed genes between C57BL/6J and C57BL/6N mice substrains. PLoS ONE 2022, 17, e0271651. [Google Scholar] [CrossRef]
- Threadgill, D.W.; Miller, D.R.; Churchill, G.A.; de Villena, F.P. The collaborative cross: A recombinant inbred mouse population for the systems genetic era. ILAR J. 2011, 52, 24–31. [Google Scholar] [CrossRef]
- Hackett, J.; Gibson, H.; Frelinger, J.; Buntzman, A. Using the Collaborative Cross and Diversity Outbred Mice in Immunology. Curr. Protoc. 2022, 2, e547. [Google Scholar] [CrossRef] [PubMed]
- Noll, K.E.; Ferris, M.T.; Heise, M.T. The Collaborative Cross: A Systems Genetics Resource for Studying Host-Pathogen Interactions. Cell Host Microbe 2019, 25, 484–498. [Google Scholar] [CrossRef]
- Baker, C.N.; Duso, D.; Kothapalli, N.; Hart, T.; Casey, S.; Cookenham, T.; Kummer, L.; Hvizdos, J.; Lanzer, K.; Vats, P.; et al. Characterization of Collaborative Cross mouse founder strain CAST/EiJ as a novel model for lethal COVID-19. Res. Sq. 2024. [Google Scholar] [CrossRef]
- Mattenberger, F.; Vila-Nistal, M.; Geller, R. Increased RNA virus population diversity improves adaptability. Sci. Rep. 2021, 11, 6824. [Google Scholar] [CrossRef]
- Markov, P.V.; Ghafari, M.; Beer, M.; Lythgoe, K.; Simmonds, P.; Stilianakis, N.I.; Katzourakis, A. The evolution of SARS-CoV-2. Nat. Rev. Microbiol. 2023, 21, 361–379. [Google Scholar] [CrossRef] [PubMed]
- Nair, M.; Gettins, L.; Fuller, M.; Kirtley, S.; Hemelaar, J. Global and regional genetic diversity of HIV-1 in 2010-21: Systematic review and analysis of prevalence. Lancet Microbe 2024, 5, 100912. [Google Scholar] [CrossRef] [PubMed]
- Le Guillou-Guillemette, H.; Vallet, S.; Gaudy-Graffin, C.; Payan, C.; Pivert, A.; Goudeau, A.; Lunel-Fabiani, F. Genetic diversity of the hepatitis C virus: Impact and issues in the antiviral therapy. World J. Gastroenterol. 2007, 13, 2416–2426. [Google Scholar] [CrossRef]
- Yu, J.M.; Fu, Y.H.; Peng, X.L.; Zheng, Y.P.; He, J.S. Genetic diversity and molecular evolution of human respiratory syncytial virus A and B. Sci. Rep. 2021, 11, 12941. [Google Scholar] [CrossRef]
- Klein, S.L.; Flanagan, K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016, 16, 626–638. [Google Scholar] [CrossRef]
- Scotland, R.S.; Stables, M.J.; Madalli, S.; Watson, P.; Gilroy, D.W. Sex differences in resident immune cell phenotype underlie more efficient acute inflammatory responses in female mice. Blood 2011, 118, 5918–5927. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.E.; McCorkell, L.; Vogel, J.M.; Topol, E.J. Long COVID: Major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023, 21, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Gheorghita, R.; Soldanescu, I.; Lobiuc, A.; Caliman Sturdza, O.A.; Filip, R.; Constantinescu-Bercu, A.; Dimian, M.; Mangul, S.; Covasa, M. The knowns and unknowns of long COVID-19: From mechanisms to therapeutical approaches. Front. Immunol. 2024, 15, 1344086. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Cai, C.; Adamo, S.; Biteus, E.; Kamal, H.; Dager, L.; Miners, K.L.; Llewellyn-Lacey, S.; Ladell, K.; Amratia, P.S.; et al. Identification of soluble biomarkers that associate with distinct manifestations of long COVID. Nat. Immunol. 2025, 26, 692–705. [Google Scholar] [CrossRef]
- O’Mahoney, L.L.; Routen, A.; Gillies, C.; Jenkins, S.A.; Almaqhawi, A.; Ayoubkhani, D.; Banerjee, A.; Brightling, C.; Calvert, M.; Cassambai, S.; et al. The risk of Long Covid symptoms: A systematic review and meta-analysis of controlled studies. Nat. Commun. 2025, 16, 4249. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kenney, D.; Matsuo, M.; Unali, G.; Wacquiez, A.; Saeed, M.; Douam, F. Comprehensive Analysis of the Impact of Weight Loss Thresholds on Mouse Models of Fatal Viral Infection. Viruses 2025, 17, 1225. https://doi.org/10.3390/v17091225
Kenney D, Matsuo M, Unali G, Wacquiez A, Saeed M, Douam F. Comprehensive Analysis of the Impact of Weight Loss Thresholds on Mouse Models of Fatal Viral Infection. Viruses. 2025; 17(9):1225. https://doi.org/10.3390/v17091225
Chicago/Turabian StyleKenney, Devin, Mao Matsuo, Giulia Unali, Alan Wacquiez, Mohsan Saeed, and Florian Douam. 2025. "Comprehensive Analysis of the Impact of Weight Loss Thresholds on Mouse Models of Fatal Viral Infection" Viruses 17, no. 9: 1225. https://doi.org/10.3390/v17091225
APA StyleKenney, D., Matsuo, M., Unali, G., Wacquiez, A., Saeed, M., & Douam, F. (2025). Comprehensive Analysis of the Impact of Weight Loss Thresholds on Mouse Models of Fatal Viral Infection. Viruses, 17(9), 1225. https://doi.org/10.3390/v17091225







