The Role of HBx Mutations in Chronic Hepatitis B with Acute Exacerbation
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Exclusion Criteria
2.3. Serologic Testing
2.4. HBV DNA Quantification
2.5. HBx Gene Amplification
- Forward primer: 5′-ATGGCTGCTAGGCTGTGCTGCCAAC-3′.
- Reverse primer: 5′-TTAGGCAGAGGTGAAAAAGTTGCAT-3′.
- (1)
- Initial denaturation at 95 °C for 5 min.
- (2)
- 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s.
- (3)
- Final extension at 72 °C for 10 min.
2.6. HBx Mutation Analysis
2.7. Statistical Analysis
3. Results
3.1. Baseline Characteristics of Patients
3.1.1. Study Cohort Characteristics
3.1.2. Baseline Comparison Between CHB-AE and HBV-Related Liver Failure
3.2. The HBx Mutations and the Severity of CHB
3.3. HBx Mutations and HBV DNA Viral Load
3.4. The HBx Mutations and Patients’ Age
3.5. HBx Mutations and Antiviral Treatment History
3.6. Risk Factors Related to Prognosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ACLF | Acute-on-chronic liver failure |
| ALT | Alanine aminotransferase |
| cccDNA | Covalently closed circular DNA |
| CHB-AE | Acute exacerbation of chronic hepatitis B |
| HBV | Hepatitis B virus |
| HBx | Hepatitis B virus X protein |
| HCC | Hepatocellular carcinoma |
| NAs | Nucleos(t)ide analogues |
| ORFs | Open reading frames |
References
- WHO. Hepatitis B. 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-b (accessed on 9 April 2024).
- Jeng, W.J.; Papatheodoridis, G.V.; Lok, A.S.F. Hepatitis B. Lancet 2023, 401, 1039–1052. [Google Scholar] [CrossRef]
- Chang, M.L.; Liaw, Y.F. Hepatitis B Flare in Hepatitis B e Antigen-Negative Patients: A Complicated Cascade of Innate and Adaptive Immune Responses. Int. J. Mol. Sci. 2022, 23, 1552. [Google Scholar] [CrossRef]
- Tsai, W.L.; Sun, W.C.; Cheng, J.S. Chronic Hepatitis B with Spontaneous Severe Acute Exacerbation. Int. J. Mol. Sci. 2015, 16, 28126–28145. [Google Scholar] [CrossRef]
- Liaw, Y.F. Acute exacerbation and superinfection in patients with chronic viral hepatitis. J. Formos. Med. Assoc. 1995, 94, 521–528. [Google Scholar]
- Lall, S.; Agarwala, P.; Kumar, G.; Sharma, M.K.; Gupta, E. The dilemma of differentiating between acute hepatitis B and chronic hepatitis B with acute exacerbation: Is quantitative serology the answer? Clin. Mol. Hepatol. 2020, 26, 187–195. [Google Scholar] [CrossRef]
- Chang, M.L.; Liaw, Y.F. Hepatitis B flares in chronic hepatitis B: Pathogenesis, natural course, and management. J. Hepatol. 2014, 61, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.W.S.; Chan, H.L.Y. Severe acute exacerbation of chronic hepatitis B: A unique presentation of a common disease. J. Gastroenterol. Hepatol. 2009, 24, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Sarin, S.K.; Kumar, M.; Lau, G.K.; Abbas, Z.; Chan, H.L.; Chen, C.J.; Chen, D.S.; Chen, H.L.; Chen, P.J.; Chien, R.N.; et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: A 2015 update. Hepatol. Int. 2016, 10, 1–98. [Google Scholar] [CrossRef]
- Jiang, Y.; Han, Q.J.; Zhao, H.J.; Zhao, J. The Mechanisms of HBV-Induced Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2021, 8, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Nassal, M. HBV cccDNA: Viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 2015, 64, 1972–1984. [Google Scholar] [CrossRef]
- Zhou, B.; He, W.M.; Hou, J.L. HBx mutations emerged during antiviral therapy: A new face of a multifaceted HBV protein? Hepatol. Int. 2020, 14, 944–946. [Google Scholar] [CrossRef]
- Tang, H.; Oishi, N.; Kaneko, S.; Murakami, S. Molecular functions and biological roles of hepatitis B virus x protein. Cancer Sci. 2006, 97, 977–983. [Google Scholar] [CrossRef]
- Hernández, S.; Venegas, M.; Brahm, J.; Villanueva, R.A. The viral transactivator HBx protein exhibits a high potential for regulation via phosphorylation through an evolutionarily conserved mechanism. Infect. Agent. Cancer 2012, 7, 27. [Google Scholar] [CrossRef]
- Belloni, L.; Pollicino, T.; Nicola, F.D.; Guerrieri, F.; Raffa, G.; Fanciulli, M.; Raimondo, G.; Levrero, M. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc. Natl. Acad. Sci. USA 2009, 106, 19975–19979. [Google Scholar] [CrossRef]
- Bouchard, M.J.; Schneider, R.J. The enigmatic X gene of hepatitis B virus. J. Virol. 2004, 78, 12725–12734. [Google Scholar] [CrossRef] [PubMed]
- Benhenda, S.; Cougot, D.; Buendia, M.A.; Neuveut, C. Hepatitis B virus X protein molecular functions and its role in virus life cycle and pathogenesis. Adv. Cancer Res. 2009, 103, 75–109. [Google Scholar] [PubMed]
- Schollmeier, A.; Glitscher, M.; Hildt, E. Relevance of HBx for Hepatitis B Virus-Associated Pathogenesis. Int. J. Mol. Sci. 2023, 24, 4964. [Google Scholar] [CrossRef]
- Ma, J.; Sun, T.; Park, S.; Shen, G.; Liu, J. The role of hepatitis B virus X protein is related to its differential intracellular localization. Acta Biochim. Biophys. Sin. 2011, 43, 583–588. [Google Scholar] [CrossRef]
- Slagle, B.L.; Bouchard, M.J. Role of HBx in hepatitis B virus persistence and its therapeutic implications. Curr. Opin. Virol. 2018, 30, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.T.; Shen, C.H.; Tai, D.I.; Chu, C.M.; Liaw, Y.F. Identification and characterization of a prevalent hepatitis B virus X protein mutant in Taiwanese patients with hepatocellular carcinoma. Oncogene 2000, 19, 5213–5220. [Google Scholar] [CrossRef]
- Chen, G.G.; Li, M.Y.; Ho, R.L.K.; Chak, E.C.W.; Lau, W.Y.; Lai, P.B.S. Identification of hepatitis B virus X gene mutation in Hong Kong patients with hepatocellular carcinoma. J. Clin. Virol. 2005, 34, 7–12. [Google Scholar] [CrossRef]
- Salarnia, F.; Besharat, S.; Zhand, S.; Javid, N.; Khodabakhshi, B.; Moradi, A. Mutations in Hepatitis-B X-Gene Region: Chronic Hepatitis-B versus Cirrhosis. J. Clin. Diagn. Res. 2017, 11, OC31–OC34. [Google Scholar] [CrossRef]
- Chaturvedi, V.K.; Singh, A.; Dubey, S.K.; Hetta, H.F.; John, J.; Singh, M.P. Molecular mechanistic insight of hepatitis B virus mediated hepatocellular carcinoma. Microb. Pathog. 2019, 128, 184–194. [Google Scholar] [CrossRef]
- Rajoriya, N.; Combet, C.; Zoulim, F.; Janssen, H.L.A. How viral genetic variants and genotypes influence disease and treatment outcome of chronic hepatitis B. Time for an individualised approach? J. Hepatol. 2017, 67, 1281–1297. [Google Scholar] [CrossRef]
- Akrami, H.; Monjezi, M.R.; Ilbeigi, S.; Amiri, F.; Fattahi, M.R. The association between hepatitis B Virus mutations and the risk of liver disease and hepatocellular carcinoma. Curr. Mol. Med. 2022, 22, 514–523. [Google Scholar] [CrossRef]
- Fujiyama, A.; Miyanohara, A.; Nozaki, C.; Yoneyama, T.; Ohtomo, N.; Matsubara, K. Cloning and structural analyses of hepatitis B virus DNAs, subtype adr. Nucleic Acids Res. 1983, 11, 4601–4610. [Google Scholar] [CrossRef]
- Hou, J.; Wang, G.; Wang, F.; Cheng, J.; Ren, H.; Zhuang, H.; Sun, J.; Li, L.; Li, J.; Meng, Q.; et al. Guideline of Prevention and Treatment for Chronic Hepatitis B (2015 Update). J. Clin. Transl. Hepatol. 2017, 5, 297–318. [Google Scholar] [CrossRef]
- Liver Failure and Artificial Liver Group; Chinese Society of Infectious Diseases; Chinese Medical Association; Severe Liver Diseases and Artificial Liver Group; Chinese Society of Hepatology; Chinese Medical Association. Diagnostic and treatment guidelines for liver failure (2012 version). Zhonghua Gan Zang Bing Za Zhi 2013, 21, 177–183. [Google Scholar]
- Bouchard, M.J.; Wang, L.; Schneider, R.J. Activation of focal adhesion kinase by hepatitis B virus HBx protein: Multiple functions in viral replication. J. Virol. 2006, 80, 4406–4414. [Google Scholar] [CrossRef] [PubMed]
- Al-Qahtani, A.A.; Al-Anazi, M.R.; Nazir, N.; Ghai, R.; Abdo, A.A.; Sanai, F.M.; Al-Hamoudi, W.K.; Alswat, K.A.; Al-Ashgar, H.I.; Khan, M.Q.; et al. Hepatitis B virus (HBV) X gene mutations and their association with liver disease progression in HBV-infected patients. Oncotarget 2017, 8, 105115–105125. [Google Scholar] [CrossRef] [PubMed]
- Luber, B.; Lauer, U.; Weiss, L.; Hohne, M.; Hofschneider, P.H.; Kekule, A.S. The hepatitis B virus transactivator HBx causes elevation of diacylglycerol and activation of protein kinase C. Res. Virol. 1993, 144, 311–321. [Google Scholar] [CrossRef]
- Ma, M.R.; Yi, L.; Pei, Y.F.; Zhang, Q.M.; Tong, C.; Zhao, M.Y.; Chen, Y.H.; Zhu, J.H.; Zhang, W.G.; Yao, F.; et al. USP26 as a hepatitis B virus-induced deubiquitinase primes hepatocellular carcinogenesis by epigenetic remodeling. Nat. Commun. 2024, 15, 7856. [Google Scholar] [CrossRef]
- Damme, E.V.; Vanhove, J.; Severyn, B.; Verschueren, L.; Pauwels, F. The Hepatitis B Virus Interactome: A Comprehensive Overview. Front. Microbiol. 2021, 12, 724877. [Google Scholar] [CrossRef]
- He, T.; Zhang, N.; Wang, L.; Wan, B.S.; Wang, X.Q.; Zhang, L. GPR43 regulates HBV X protein (HBx)-induced inflammatory response in human LO2 hepatocytes. Biomed. Pharmacother. 2020, 123, 109737. [Google Scholar] [CrossRef]
- Zhang, D.; Guo, S.C.; Schrodi, S.J. Mechanisms of DNA Methylation in Virus-Host Interaction in Hepatitis B Infection: Pathogenesis and Oncogenetic Properties. Int. J. Mol. Sci. 2021, 22, 9858. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, G.E.M.; Cabibbo, G.; Craxì, A. Hepatitis B Virus-Associated Hepatocellular Carcinoma. Viruses 2022, 14, 986. [Google Scholar] [CrossRef]
- Waris, G.; Huh, K.W.; Siddiqui, A. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-kappa B via oxidative stress. Mol. Cell. Biol. 2001, 21, 7721–7730. [Google Scholar] [CrossRef]
- Lucifora, J.; Arzberger, S.; Durantel, D.; Belloni, L.; Strubin, M.; Levrero, M.; Zoulim, F.; Hantz, O.; Protzer, U. Hepatitis B Virus X protein is essential to initiate and maintain virus replication after infection. J. Hepatol. 2011, 55, 996–1003. [Google Scholar] [CrossRef]
- Tsuge, M.; Hiraga, N.; Akiyama, R.; Tanaka, S.; Matsushita, M.; Mitsui, F.; Abe, H.; Kitamura, S.; Hatakeyama, T.; Kimura, T.; et al. HBx protein is indispensable for development of viraemia in human hepatocyte chimeric mice. J. Gen. Virol. 2010, 91, 1854–1864. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.S.; Kaneko, S.; Girones, R.; Anderson, R.W.; Hornbuckle, W.E.; Tennant, B.C.; Cote, P.J.; Gerin, J.L.; Purcell, R.H.; Miller, R.H. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J. Virol. 1993, 67, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- Zoulim, F.; Saputelli, J.; Seeger, C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 1994, 68, 2026–2030. [Google Scholar] [CrossRef]
- Yang, H.I.; Yeh, S.H.; Chen, P.J.; Iloeje, U.H.; Jen, C.L.; Su, J.; Wang, L.Y.; Lu, S.N.; You, S.L.; Chen, D.S.; et al. REVEAL-HBV Study Group: Associations between hepatitis B virus genotype and mutations and the risk of hepatocellular carcinoma. J. Nat Cancer Inst. 2008, 100, 1134–1143. [Google Scholar] [CrossRef]
- Yuen, L.; Revill, P.A.; Rosenberg, G.; Wagner, J.; Littlejohn, M.; Bayliss, J.; Jackson, K.; Tan, S.K.; Gaggar, A.; Kitrinos, K.; et al. HBV variants are common in the ‘immune-tolerant’ phase of chronic hepatitis B. J. Viral. Hepat. 2020, 27, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.L.; Zeng, L.I.; Chen, W.Q. HBV X gene point mutations are associated with the risk of hepatocellular carcinoma: A systematic review and meta-analysis. Mol. Clin. Oncol. 2016, 4, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.L.; Chien, R.N.; Chu, Y.D.; Liang, H.K.; Huang, Y.H.; Ke, P.Y.; Lin, K.H.; Lin, Y.H.; Yeh, C.T. Hepatitis B virus X gene mutants emerge during antiviral therapy and increase cccDNA levels to compensate for replication suppression. Hepatol. Int. 2020, 14, 973–984. [Google Scholar] [CrossRef]
- Tang, H.; Delgermaa, L.; Huang, F.; Oishi, N.; Liu, L.; He, F.; Zhao, L.S.; Murakami, S. The transcriptional transactivation function of HBx protein is important for its augmentation role in hepatitis B virus replication. J. Virol. 2005, 79, 5548–5556. [Google Scholar] [CrossRef]
- Choi, W.M.; Yip, T.C.F.; Kim, W.R.; Yee, L.J.; Rooney, C.B.; Curteis, T.; Clark, L.J.; Jafry, Z.; Chen, C.H.; Chen, C.Y.; et al. Chronic hepatitis B baseline viral load and on-treatment liver cancer risk: A multinational cohort study of HBeAg-positive patients. Hepatology 2024, 80, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Chinese Society of Hepatology; Chinese Medical Association. Expert opinion on expanding anti-HBV treatment for chronic hepatitis B. Zhonghua Gan Zang Bing Za Zhi 2022, 30, 131–136. [Google Scholar]
- You, H.; Wang, F.S.; Li, T.S.; Xu, X.Y.; Sun, Y.M.; Nan, Y.M.; Wang, G.Q.; Hou, J.L.; Duan, Z.P.; Wei, L.; et al. Guidelines for the Prevention and Treatment of Chronic Hepatitis B (version 2022). J. Clin. Transl. Hepatol. 2023, 11, 1425–1442. [Google Scholar] [CrossRef]
- Lok, J.; Dusheiko, G. Editorial: Reassessing antiviral treatment criteria for chronic hepatitis B. Hepatology 2023, 78, 1332–1333. [Google Scholar] [CrossRef]
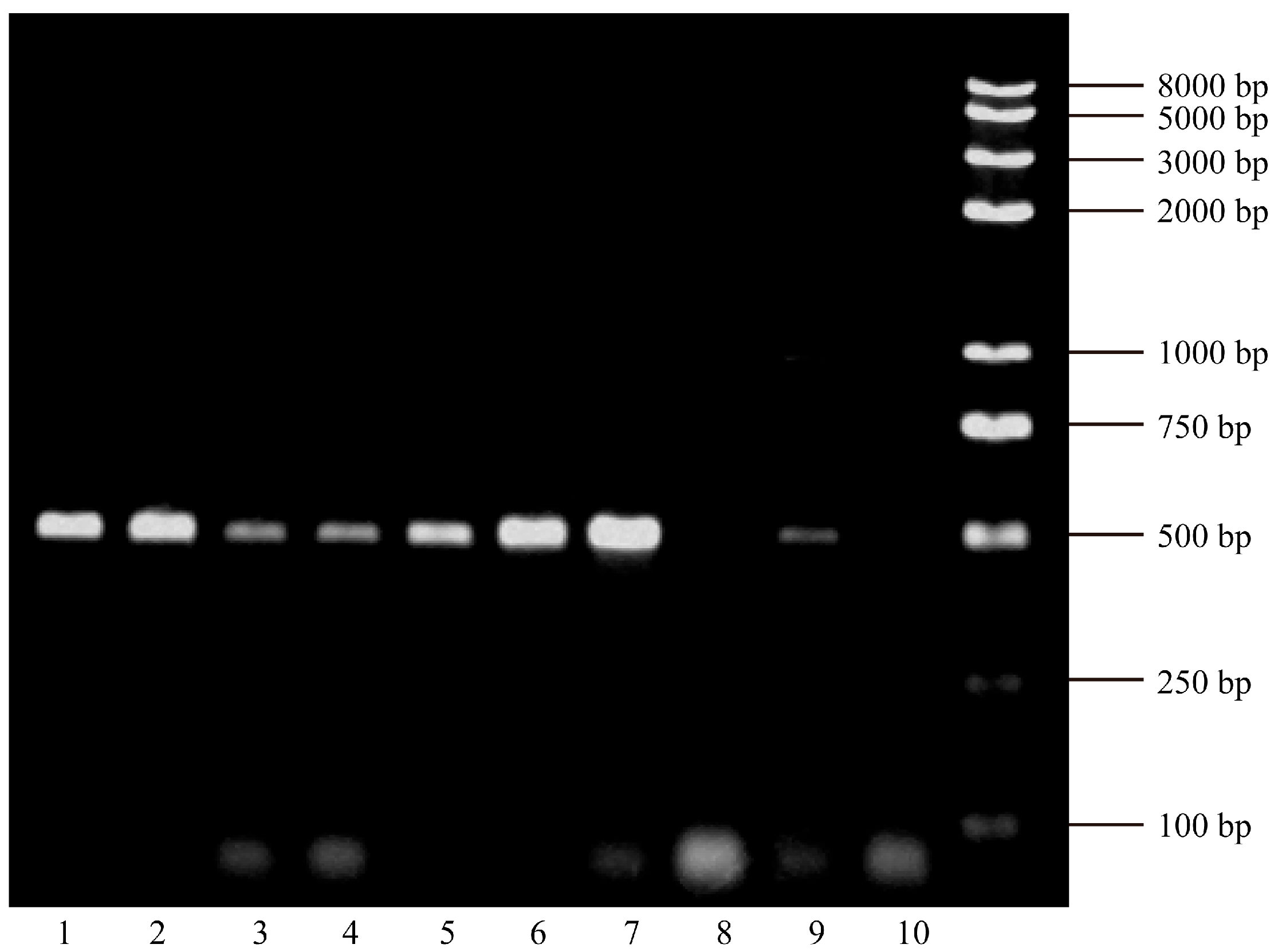
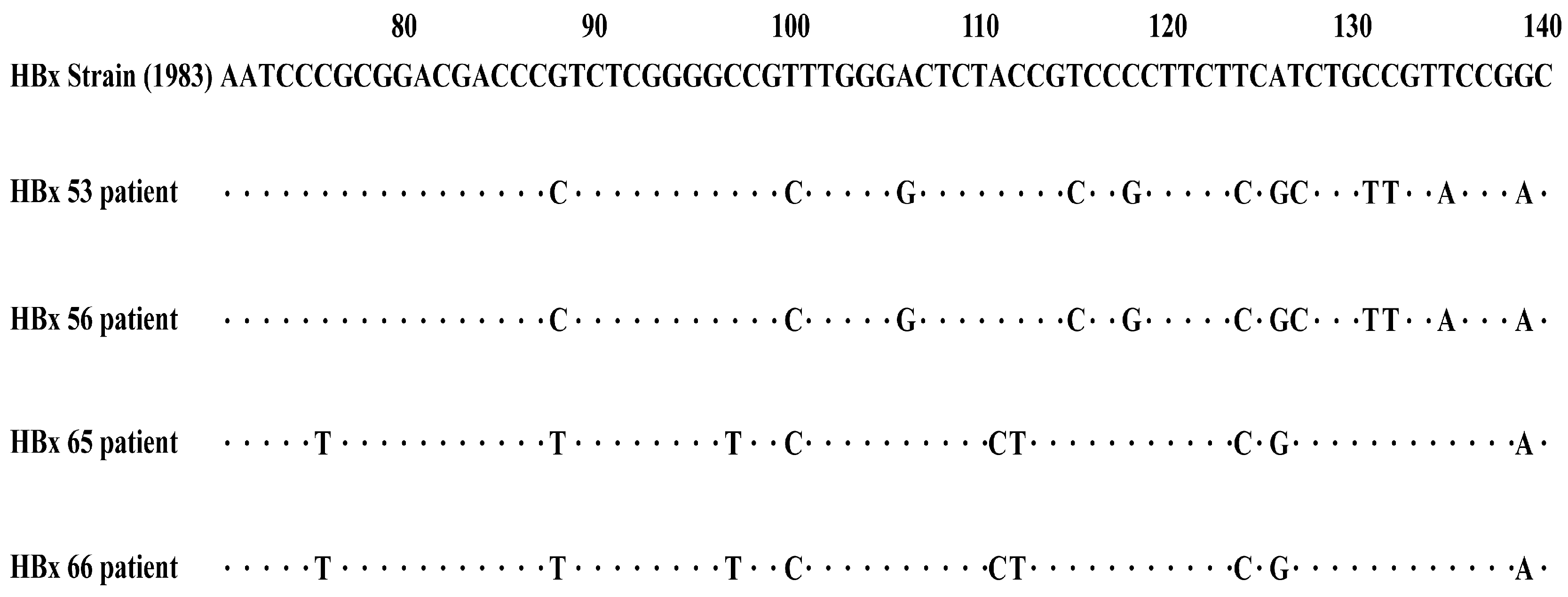
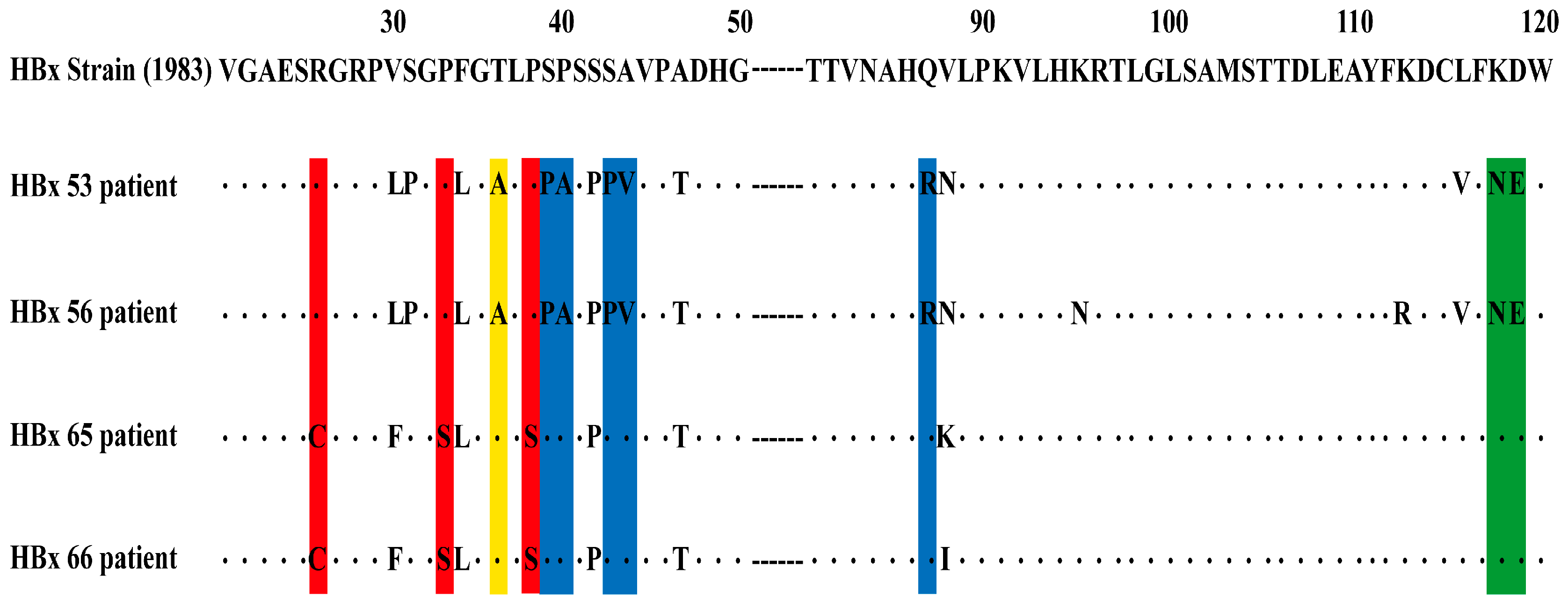
| Chronic Hepatitis B with Acute Exacerbation (n = 33) (%) | HBV-Related Liver Failure (n = 31) (%) | p-Value | ||
|---|---|---|---|---|
| Gender | Male | 26 (78.79) | 28 (90.32) | 0.3546 |
| Female | 7 (21.21) | 3 (9.68) | ||
| Age (years) | ≥35 | 19 (57.58) | 22 (70.97) | 0.2645 |
| <35 | 14 (42.42) | 9 (29.03) | ||
| HBV DNA Log level (copies/mL) | ≥5 | 29 (87.88) | 20 (64.52) | 0.0275 |
| <5 | 4 (12.12) | 11 (35.48) | ||
| HBeAg (0–1 COI) | positive | 21 (63.64) | 10 (32.26) | 0.0121 |
| negative | 12 (36.36) | 21 (67.74) |
| HBx Mutations | Chronic Hepatitis B with Acute Exacerbation (n = 33) (%) | HBV-Related Liver Failure (n = 31) (%) | p-Value |
|---|---|---|---|
| Single Mutation 36 (T36A/S) | 13 (39.39) | 5 (16.13) | 0.0386 |
| Joint Mutation 1 (R26C + P33S + P38S) | 19 (57.58) | 25 (80.65) | 0.0466 |
| Joint Mutation 2 (S39P + P40A + S43P + A44L/V/F + Q87R/W/K/G) | 12 (36.36) | 5 (16.13) | 0.0670 |
| Joint Mutation 3 (K118N/T + D119E) | 11 (33.33) | 5 (16.13) | 0.1122 |
| HBx Mutations | High HBV DNA Log Level ≥ 5 (n = 49) (%) | Low HBV DNA Log Level < 5 (n = 15) (%) | p-Value |
|---|---|---|---|
| Single Mutation 36 (T36A/S) | 18 (36.73) | 0 (0) | 0.0147 |
| Joint Mutation 1 (R26C + P33S + P38S) | 29 (59.18) | 15 (100) | 0.0077 |
| Joint Mutation 2 (S39P + P40A + S43P + A44L/V/F + Q87R/W/K/G) | 17 (34.69) | 0 (0) | 0.0199 |
| Joint Mutation 3 (K118N/T + D119E) | 16 (32.65) | 0 (0) | 0.0268 |
| HBx Mutations | Age ≥ 35 (n = 41) (%) | Age < 35 (n = 23) (%) | p-Value |
|---|---|---|---|
| Single Mutation 36 (T36A/S) | 8 (19.51) | 10 (43.48) | 0.0407 |
| Joint Mutation 1 (R26C + P33S + P38S) | 32 (78.05) | 12 (52.17) | 0.0321 |
| Joint Mutation 2 (S39P + P40A + S43P + A44L/V/F + Q87R/W/K/G) | 7 (17.07) | 10 (43.48) | 0.0217 |
| Joint Mutation 3 (K118N/T + D119E) | 7 (17.07) | 9 (39.13%) | 0.0505 |
| HBx Mutations | Treatment-Naïve (n = 47) (%) | Treatment-Experienced (n = 17) (%) | p-Value |
|---|---|---|---|
| Single Mutation 36 (T36A/S) | 14 (29.79) | 4 (23.53) | 0.8595 |
| Joint Mutation 1 (R26C + P33S + P38S) | 39 (82.98) | 5 (29.41) | <0.0001 |
| Joint Mutation 2 (S39P + P40A + S43P + A44L/V/F + Q87R/W/K/G) | 14 (29.79) | 3 (17.65) | 0.5152 |
| Joint Mutation 3 (K118N/T + D119E) | 13 (27.66) | 3 (17.65) | 0.6240 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Shi, J.; Zhou, P.; Tian, Y.; Zheng, Y.; Liu, T.; Li, Y.; Zhu, F. The Role of HBx Mutations in Chronic Hepatitis B with Acute Exacerbation. Viruses 2025, 17, 1223. https://doi.org/10.3390/v17091223
Chen X, Shi J, Zhou P, Tian Y, Zheng Y, Liu T, Li Y, Zhu F. The Role of HBx Mutations in Chronic Hepatitis B with Acute Exacerbation. Viruses. 2025; 17(9):1223. https://doi.org/10.3390/v17091223
Chicago/Turabian StyleChen, Xiaobei, Jinzhi Shi, Ping Zhou, Yunyun Tian, Yajing Zheng, Tingting Liu, Yan Li, and Fan Zhu. 2025. "The Role of HBx Mutations in Chronic Hepatitis B with Acute Exacerbation" Viruses 17, no. 9: 1223. https://doi.org/10.3390/v17091223
APA StyleChen, X., Shi, J., Zhou, P., Tian, Y., Zheng, Y., Liu, T., Li, Y., & Zhu, F. (2025). The Role of HBx Mutations in Chronic Hepatitis B with Acute Exacerbation. Viruses, 17(9), 1223. https://doi.org/10.3390/v17091223







