Human-Derived H3N2 Influenza A Viruses Detected in Pigs in Northern Italy
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Processing Workflow
2.2. Sequence Alignments and Phylogenetic Analyses
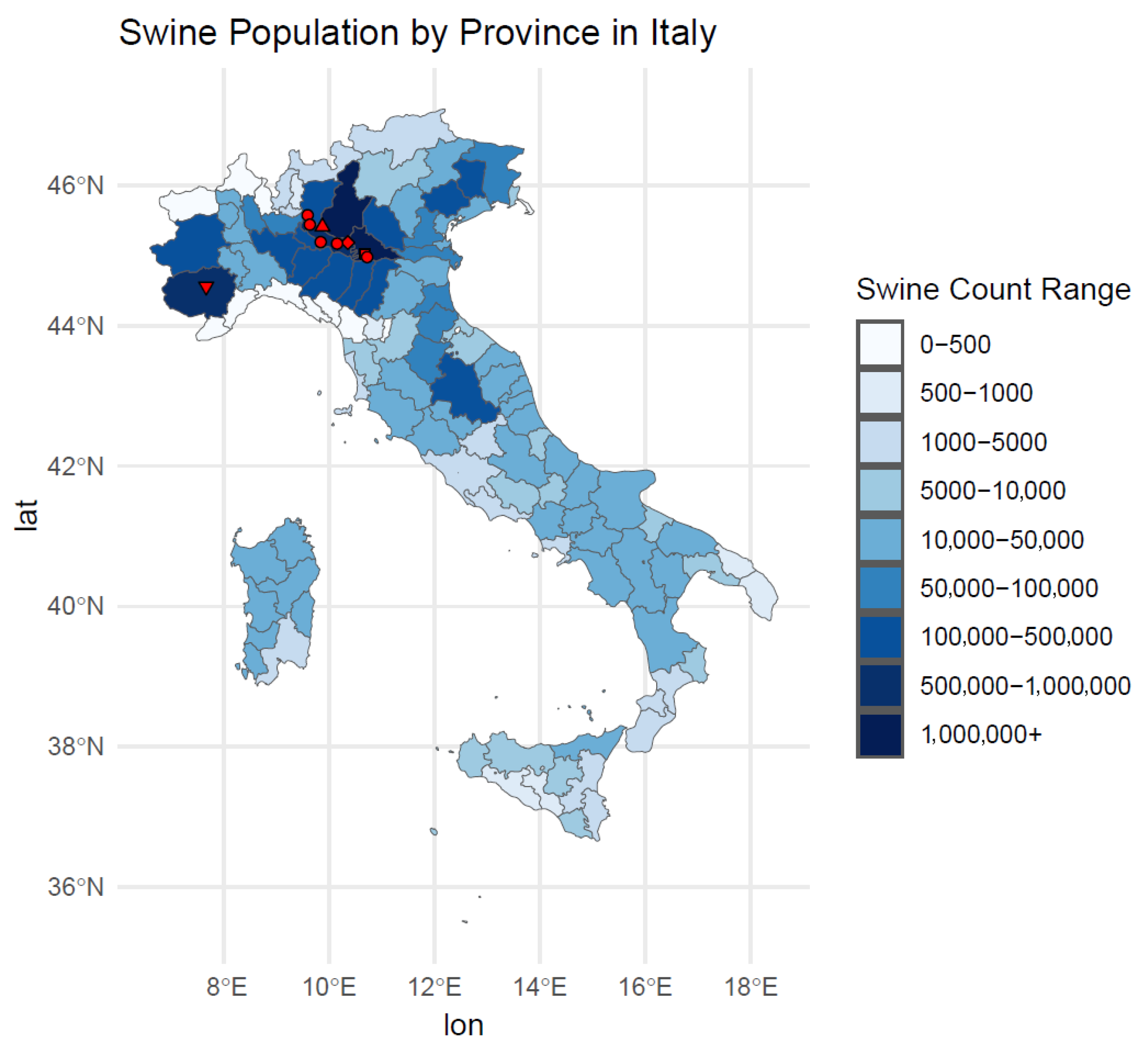
2.3. Map Representing the Swine Population in Italian Provinces
2.4. Hemagglutination Inhibition (HI) Tests
3. Results
3.1. Virological Analysis
3.2. Serological Testing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Henritzi, D.; Zhao, N.; Starick, E.; Simon, G.; Krog, J.S.; Larsen, L.E.; Reid, S.M.; Brown, I.H.; Chiapponi, C.; Foni, E.; et al. Rapid Detection and Subtyping of European Swine Influenza Viruses in Porcine Clinical Samples by Haemagglutinin- and Neuraminidase-Specific Tetra- and Triplex Real-Time RT-PCRs. Influenza Other Respir. Viruses 2016, 10, 504–517. [Google Scholar] [CrossRef] [PubMed]
- Ma, W. Swine Influenza Virus: Current Status and Challenge. Virus Res. 2020, 288, 198118. [Google Scholar] [CrossRef] [PubMed]
- Chiapponi, C.; Prosperi, A.; Moreno, A.; Baioni, L.; Faccini, S.; Manfredi, R.; Zanni, I.; Gabbi, V.; Calanchi, I.; Fusaro, A.; et al. Genetic Variability among Swine Influenza Viruses in Italy: Data Analysis of the Period 2017–2020. Viruses 2022, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Henritzi, D.; Petric, P.P.; Lewis, N.S.; Graaf, A.; Pessia, A.; Starick, E.; Breithaupt, A.; Strebelow, G.; Luttermann, C.; Parker, L.M.K.; et al. Surveillance of European Domestic Pig Populations Identifies an Emerging Reservoir of Potentially Zoonotic Swine Influenza A Viruses. Cell Host Microbe 2020, 28, 614–627.e6. [Google Scholar] [CrossRef]
- Watson, S.J.; Pinky, L.; Reid, S.M.; Lam, T.T.; Cotten, M.; Kelly, M.; Van Reeth, K.; Qiu, Y.; Simon, G.; Bonin, E.; et al. Molecular Epidemiology and Evolution of Influenza Viruses Circulating within European Swine between 2009 and 2013. J. Virol. 2015, 89, 9920–9931. [Google Scholar] [CrossRef]
- Anderson, T.K.; Macken, C.A.; Lewis, N.S.; Scheuermann, R.H.; Van Reeth, K.; Brown, I.H.; Swenson, S.L.; Simon, G.; Saito, T.; Berhane, Y.; et al. A Phylogeny-Based Global Nomenclature System and Automated Annotation Tool for H1 Hemagglutinin Genes from Swine Influenza A Viruses. mSphere 2016, 1, e00275-16. [Google Scholar] [CrossRef]
- Anderson, T.K.; Chang, J.; Arendsee, Z.W.; Venkatesh, D.; Souza, C.K.; Kimble, J.B.; Lewis, N.S.; Davis, C.T.; Vincent, A.L. Swine Influenza A Viruses and the Tangled Relationship with Humans. Cold Spring Harb. Perspect. Med. 2021, 11, a038737. [Google Scholar] [CrossRef]
- Nelson, M.I.; Detmer, S.E.; Wentworth, D.E.; Tan, Y.; Schwartzbard, A.; Halpin, R.A.; Stockwell, T.B.; Lin, X.; Vincent, A.L.; Gramer, M.R.; et al. Genomic Reassortment of Influenza A Virus in North American Swine, 1998–2011. J. Gen. Virol. 2012, 93, 2584–2589. [Google Scholar] [CrossRef]
- Krumbholz, A.; Lange, J.; Sauerbrei, A.; Groth, M.; Platzer, M.; Kanrai, P.; Pleschka, S.; Scholtissek, C.; Büttner, M.; Dürrwald, R.; et al. Origin of the European Avian-like Swine Influenza Viruses. J. Gen. Virol. 2014, 95, 2372–2376. [Google Scholar] [CrossRef]
- Dunham, E.J.; Dugan, V.G.; Kaser, E.K.; Perkins, S.E.; Brown, I.H.; Holmes, E.C.; Taubenberger, J.K. Different Evolutionary Trajectories of European Avian-Like and Classical Swine H1N1 Influenza A Viruses. J. Virol. 2009, 83, 5485–5494. [Google Scholar] [CrossRef]
- Smith, G.J.D.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and Evolutionary Genomics of the 2009 Swine-Origin H1N1 Influenza A Epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef]
- Welsh, M.D.; Baird, P.M.; Guelbenzu-Gonzalo, M.P.; Hanna, A.; Reid, S.M.; Essen, S.; Russell, C.; Thomas, S.; Barrass, L.; McNeilly, F.; et al. Initial Incursion of Pandemic (H1N1) 2009 Influenza A Virus into European Pigs. Vet. Rec. 2010, 166, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Howard, W.A.; Essen, S.C.; Strugnell, B.W.; Russell, C.; Barrass, L.; Reid, S.M.; Brown, I.H. Reassortant Pandemic (H1N1) 2009 Virus in Pigs, United Kingdom. Emerg. Infect. Dis. J. 2011, 17, 1049. [Google Scholar] [CrossRef]
- Zell, R.; Groth, M.; Krumbholz, A.; Lange, J.; Philipps, A.; Dürrwald, R. Displacement of the Gent/1999 Human-like Swine H1N2 Influenza A Virus Lineage by Novel H1N2 Reassortants in Germany. Arch. Virol. 2020, 165, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.I.; Wentworth, D.E.; Culhane, M.R.; Vincent, A.L.; Viboud, C.; LaPointe, M.P.; Lin, X.; Holmes, E.C.; Detmer, S.E. Introductions and Evolution of Human-Origin Seasonal Influenza A Viruses in Multinational Swine Populations. J. Virol. 2014, 88, 10110–10119. [Google Scholar] [CrossRef]
- Simon, G.; Larsen, L.E.; Dürrwald, R.; Foni, E.; Harder, T.; Van Reeth, K.; Markowska-Daniel, I.; Reid, S.M.; Dan, A.; Maldonado, J.; et al. European Surveillance Network for Influenza in Pigs: Surveillance Programs, Diagnostic Tools and Swine Influenza Virus Subtypes Identified in 14 European Countries from 2010 to 2013. PLoS ONE 2014, 9, e115815. [Google Scholar] [CrossRef]
- Zell, R.; Groth, M.; Krumbholz, A.; Lange, J.; Philipps, A.; Dürrwald, R. Novel Reassortant Swine H3N2 Influenza A Viruses in Germany. Sci. Rep. 2020, 10, 14296. [Google Scholar] [CrossRef]
- Lewis, N.S.; Russell, C.A.; Langat, P.; Anderson, T.K.; Berger, K.; Bielejec, F.; Burke, D.F.; Dudas, G.; Fonville, J.M.; Fouchier, R.A.M.; et al. The Global Antigenic Diversity of Swine Influenza A Viruses. Elife 2016, 5, e12217. [Google Scholar] [CrossRef]
- Nelson, M.I.; Vincent, A.L. Reverse Zoonosis of Influenza to Swine: New Perspectives on the Human–Animal Interface. Trends Microbiol. 2015, 23, 142–153. [Google Scholar] [CrossRef]
- Busch, M.G.; Bateman, A.C.; Landolt, G.A.; Karasin, A.I.; Brockman-Schneider, R.A.; Gern, J.E.; Suresh, M.; Olsen, C.W. Identification of Amino Acids in the HA of H3 Influenza Viruses That Determine Infectivity Levels in Primary Swine Respiratory Epithelial Cells. Virus Res. 2008, 133, 269–279. [Google Scholar] [CrossRef]
- Rajão, D.S.; Gauger, P.C.; Anderson, T.K.; Lewis, N.S.; Abente, E.J.; Killian, M.L.; Perez, D.R.; Sutton, T.C.; Zhang, J.; Vincent, A.L. Novel Reassortant Human-Like H3N2 and H3N1 Influenza A Viruses Detected in Pigs Are Virulent and Antigenically Distinct from Swine Viruses Endemic to the United States. J. Virol. 2015, 89, 11213–11222. [Google Scholar] [CrossRef]
- Dibárbora, M.; Cappuccio, J.; Olivera, V.; Quiroga, M.; Machuca, M.; Perfumo, C.; Pérez, D.; Pereda, A. Swine Influenza: Clinical, Serological, Pathological, and Virological Cross-Sectional Studies in Nine Farms in Argentina. Influenza Other Respir. Viruses 2013, 7, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Ngo, L.T.; Hiromoto, Y.; Pham, V.P.; Le, H.T.H.; Nguyen, H.T.; Le, V.T.; Takemae, N.; Saito, T. Isolation of Novel Triple-Reassortant Swine H3N2 Influenza Viruses Possessing the Hemagglutinin and Neuraminidase Genes of a Seasonal Influenza Virus in Vietnam in 2010. Influenza Other Respir. Viruses 2012, 6, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Krog, J.S.; Hjulsager, C.K.; Larsen, M.A.; Larsen, L.E. Triple-Reassortant Influenza A Virus with H3 of Human Seasonal Origin, NA of Swine Origin, and Internal A(H1N1) Pandemic 2009 Genes Is Established in Danish Pigs. Influenza Other Respir. Viruses 2017, 11, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Zeller, M.A.; Li, G.; Harmon, K.M.; Zhang, J.; Vincent, A.L.; Anderson, T.K.; Gauger, P.C. Complete Genome Sequences of Two Novel Human-Like H3N2 Influenza A Viruses, A/Swine/Oklahoma/65980/2017 (H3N2) and A/Swine/Oklahoma/65260/2017 (H3N2), Detected in Swine in the United States. Microbiol. Resour. Announc. 2018, 7, e01203-18. [Google Scholar] [CrossRef]
- Wong, F.Y.K.; Donato, C.; Deng, Y.; Teng, D.; Komadina, N.; Baas, C.; Modak, J.; O’Dea, M.; Smith, D.W.; Effler, P.V.; et al. Divergent Human-Origin Influenza Viruses Detected in Australian Swine Populations. J. Virol. 2018, 92, e00316-18. [Google Scholar] [CrossRef]
- Chiapponi, C.; Ebranati, E.; Pariani, E.; Faccini, S.; Luppi, A.; Baioni, L.; Manfredi, R.; Carta, V.; Merenda, M.; Affanni, P.; et al. Genetic Analysis of Human and Swine Influenza A Viruses Isolated in Northern Italy during 2010–2015. Zoonoses Public Health 2018, 65, 114–123. [Google Scholar] [CrossRef]
- Slomka, M.J.; Densham, A.L.E.; Coward, V.J.; Essen, S.; Brookes, S.M.; Irvine, R.M.; Spackman, E.; Ridgeon, J.; Gardner, R.; Hanna, A.; et al. Original Article: Real Time Reverse Transcription (RRT)-Polymerase Chain Reaction (PCR) Methods for Detection of Pandemic (H1N1) 2009 Influenza Virus and European Swine Influenza A Virus Infections in Pigs. Influenza Other Respir. Viruses 2010, 4, 277–293. [Google Scholar] [CrossRef]
- Chiapponi, C.; Zanni, I.; Garbarino, C.; Barigazzi, G.; Foni, E. Comparison of the Usefulness of the CACO-2 Cell Line with Standard Substrates for Isolation of Swine Influenza A Viruses. J. Virol. Methods 2010, 163, 162–165. [Google Scholar] [CrossRef]
- Lycett, S.J.; Baillie, G.; Coulter, E.; Bhatt, S.; Kellam, P.; McCauley, J.W.; Wood, J.L.N.; Brown, I.H.; Pybus, O.G.; Leigh Brown, A.J.; et al. Estimating Reassortment Rates in Co-Circulating Eurasian Swine Influenza Viruses. J. Gen. Virol. 2012, 93, 2326–2336. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Larsson, A. AliView: A Fast and Lightweight Alignment Viewer and Editor for Large Datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v6: Recent Updates to the Phylogenetic Tree Display and Annotation Tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Olson, R.D.; Assaf, R.; Brettin, T.; Conrad, N.; Cucinell, C.; Davis, J.J.; Dempsey, D.M.; Dickerman, A.; Dietrich, E.M.; Kenyon, R.W.; et al. Introducing the Bacterial and Viral Bioinformatics Resource Center (BV-BRC): A Resource Combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023, 51, D678–D689. [Google Scholar] [CrossRef]
- Statistiche—BDN. Available online: https://www.vetinfo.it/j6_statistiche/#/report-pbi/31 (accessed on 24 June 2024).
- World Organisation for Animal Health (WOAH). Influenza A Viruses of Swine. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; WOAH: Paris, France, 2023. [Google Scholar]
- Lewis, N.S.; Anderson, T.K.; Kitikoon, P.; Skepner, E.; Burke, D.F.; Vincent, A.L. Substitutions near the Hemagglutinin Receptor-Binding Site Determine the Antigenic Evolution of Influenza A H3N2 Viruses in U.S. Swine. J. Virol. 2014, 88, 4752–4763. [Google Scholar] [CrossRef]
- McHardy, A.C.; Adams, B. The Role of Genomics in Tracking the Evolution of Influenza A Virus. PLoS Pathog. 2009, 5, e1000566. [Google Scholar] [CrossRef]


| Sampling Year | NT | H1Nx | H1N1 | H1N2 | H3Nx | H3N2 | HxN1 | HxN2 | Outbreaks Analyzed n. |
|---|---|---|---|---|---|---|---|---|---|
| 2021 | 15% | 12.4% | 31.1% | 31.1% | 1.6% | 3.1% | 2.1% | 4.1% | 193 |
| 2022 | 15% | 11.4% | 34.3% | 25.9% | 1.2% | 6% | 3% | 3% | 166 |
| 2023 | 8% | 6.3% | 25% | 50% | 0% | 7.1% | 0.9% | 2.7% | 112 |
| 2024 | 11% | 10.6% | 31.7% | 42.3% | 0.7% | 1.4% | 0.7% | 1.4% | 142 |
| 2025 (March) | 5.1% | 5.1% | 38.5% | 28.2% | 0% | 12.8% | 2.6% | 2.6% | 39 |
| Year | H3 (1970.1) | Other-Human-2010 | Total Analyzed | ||
|---|---|---|---|---|---|
| 2021 | 87.5% | 7 | 12.5% | 1 | 8 |
| 2022 | 89% | 8 | 11% | 1 | 9 |
| 2023 | 50% | 1 | 50% | 1 | 2 |
| 2024 | 75% | 3 | 25% | 1 | 4 |
| 2025 | 0% | 0 | 100% | 5 | 5 |
| ID | Sampling Date | Italian Region | H3 HA | NA | PB2 | PB1 | PA | NP | M | NS | Sampling Year |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A/swine/Italy/220316/2021 | 25 June 2021 | Piemonte | Other-Human-2010 | It-N2 | EA | EA | EA | EA | EA | EA | 2021 |
| A/swine/Italy/368357-01/2022 | 9 November 2022 | Lombardia | Other-Human-2010 | It-N2 | EA | EA | EA | EA | EA | EA | 2022 |
| A/swine/Italy/27326-07/2023 | 25 January 2023 | Lombardia | Other-Human-2010 | It-N2 | EA | EA | EA | EA | EA | EA | 2023 |
| A/swine/Italy/111522-01/2024 | 9 April 2024 | Lombardia | Other-Human-2010 | It-N2 | pdm | pdm | pdm | pdm | EA | EA | 2024 |
| A/swine/Italy/21672-01/2025 | 23 January 2025 | Emilia-Romagna | Other-Human-2010 | N2g | pdm | pdm | pdm | pdm | pdm | EA | 2025 |
| A/swine/Italy/44792-01/2025 | 14 February 2025 | Lombardia | Other-Human-2010 | It-N2 | EA | EA | EA | EA | EA | EA | 2025 |
| A/swine/Italy/31869-01/2025 | 3 February 2025 | Lombardia | Other-Human-2010 | It-N2 | EA | EA | EA | EA | EA | EA | 2025 |
| A/swine/Italy/31894-01/2025 | 4 February 2025 | Lombardia | Other-Human-2010 | It-N2 | EA | EA | EA | EA | EA | EA | 2025 |
| A/swine/Italy/53139-01/2025 | 24 February 2025 | Lombardia | Other-Human-2010 | It-N2 | EA | EA | EA | EA | EA | EA | 2025 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soliani, L.; Mescoli, A.; Zanni, I.; Baioni, L.; Alborali, G.; Moreno, A.; Faccini, S.; Rosignoli, C.; Lorenzi, G.D.; Fiorentini, L.; et al. Human-Derived H3N2 Influenza A Viruses Detected in Pigs in Northern Italy. Viruses 2025, 17, 1171. https://doi.org/10.3390/v17091171
Soliani L, Mescoli A, Zanni I, Baioni L, Alborali G, Moreno A, Faccini S, Rosignoli C, Lorenzi GD, Fiorentini L, et al. Human-Derived H3N2 Influenza A Viruses Detected in Pigs in Northern Italy. Viruses. 2025; 17(9):1171. https://doi.org/10.3390/v17091171
Chicago/Turabian StyleSoliani, Laura, Ada Mescoli, Irene Zanni, Laura Baioni, Giovanni Alborali, Ana Moreno, Silvia Faccini, Carlo Rosignoli, Giorgia De Lorenzi, Laura Fiorentini, and et al. 2025. "Human-Derived H3N2 Influenza A Viruses Detected in Pigs in Northern Italy" Viruses 17, no. 9: 1171. https://doi.org/10.3390/v17091171
APA StyleSoliani, L., Mescoli, A., Zanni, I., Baioni, L., Alborali, G., Moreno, A., Faccini, S., Rosignoli, C., Lorenzi, G. D., Fiorentini, L., Torreggiani, C., Cordioli, B., Prosperi, A., Luppi, A., & Chiapponi, C. (2025). Human-Derived H3N2 Influenza A Viruses Detected in Pigs in Northern Italy. Viruses, 17(9), 1171. https://doi.org/10.3390/v17091171






