Epitranscriptomic Regulation of Hepatitis B Virus by RNA 5-Methylcytosine: Functions, Mechanisms, and Therapeutic Potential
Abstract
1. Introduction
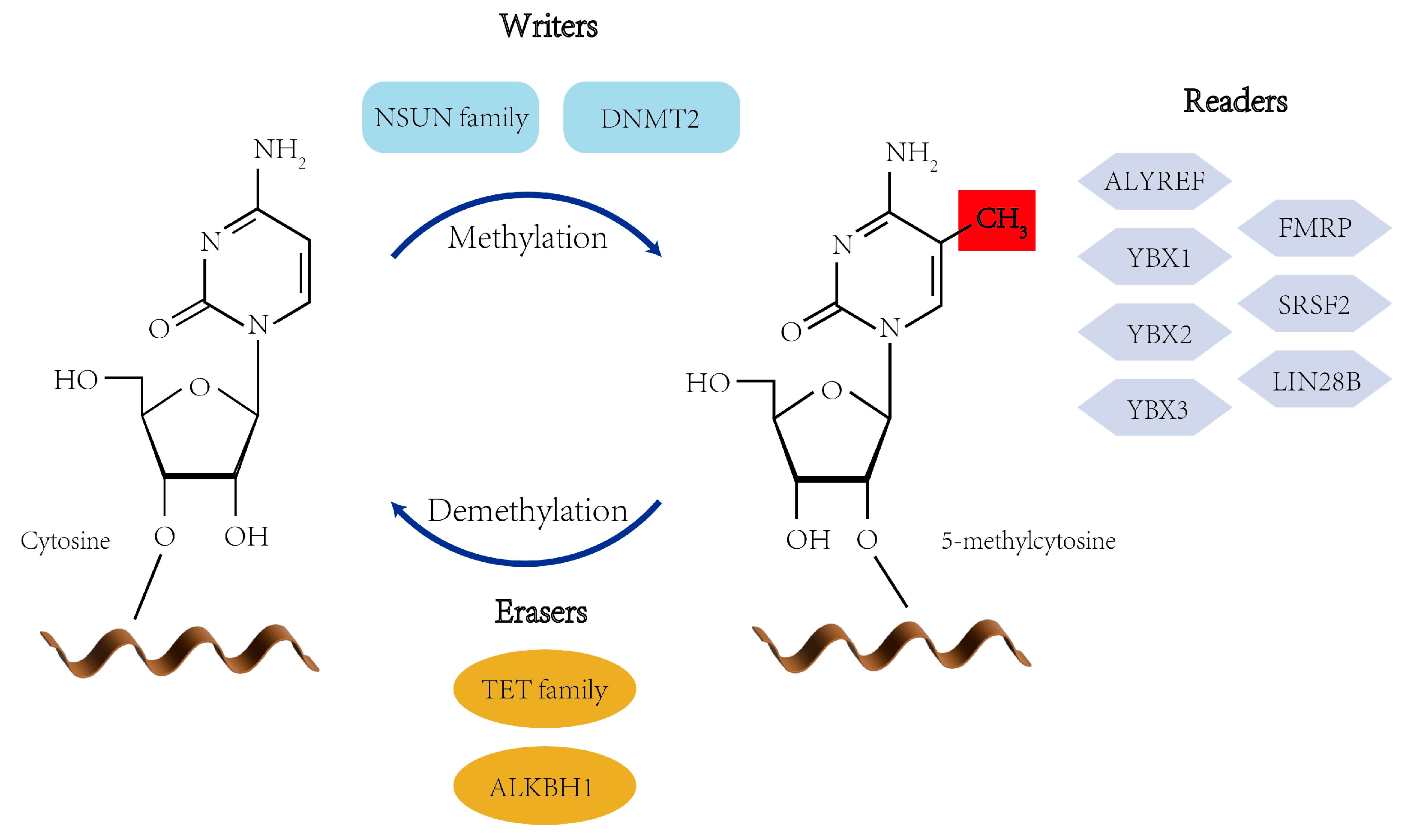
2. RNA m5C: Distribution and Regulatory Mechanisms
2.1. m5C Writers
2.2. m5C Erasers
2.3. m5C Readers
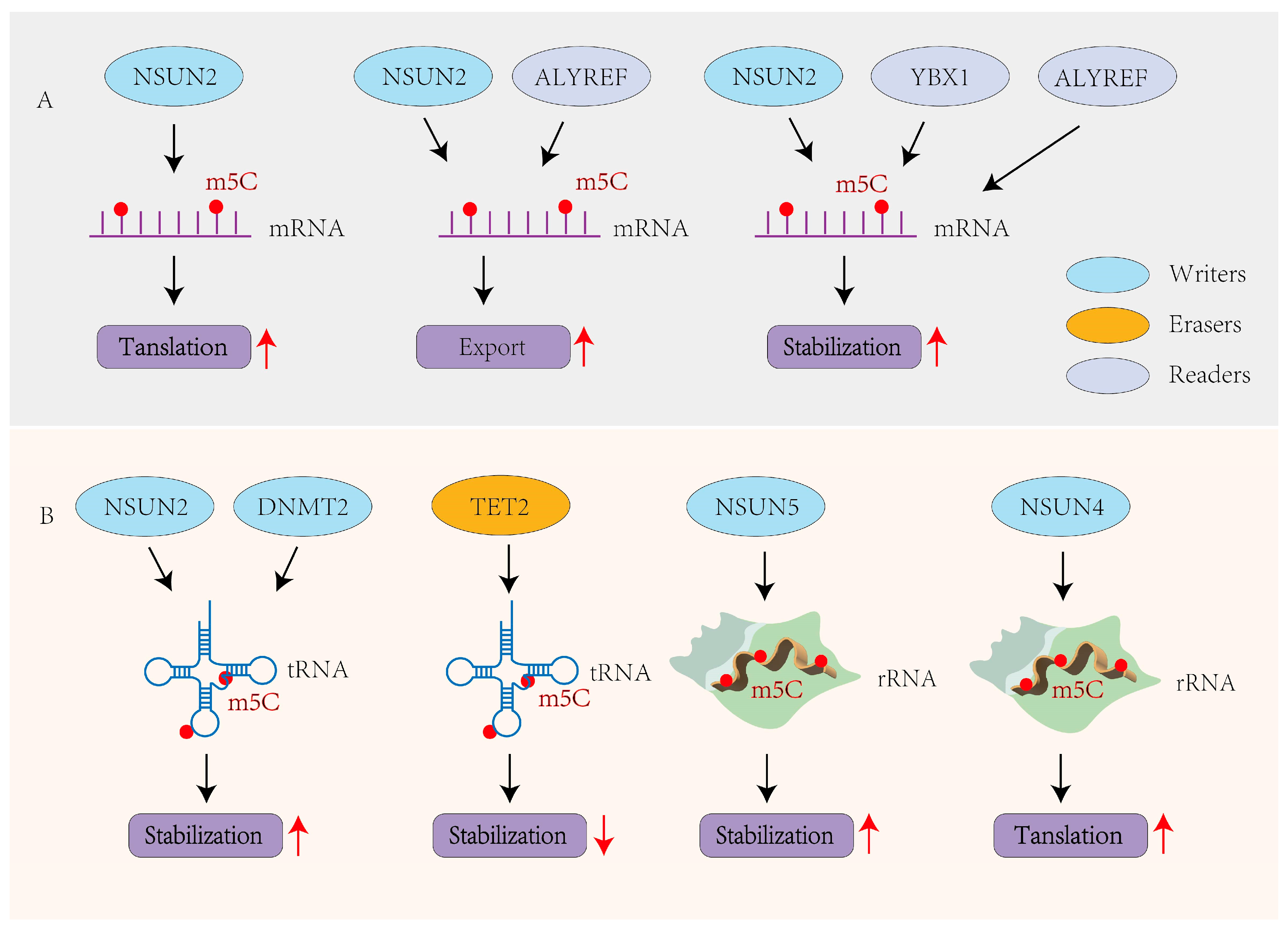
3. Biological Functions of m5C Methylation
3.1. m5C in mRNAs
3.2. m5C in ncRNAs
4. Detection Methods of RNA m5C Methylation
4.1. RNA Bisulfite Sequencing (RNA-BisSeq)
4.2. Immunoprecipitation-Based Techniques
4.3. LC-MS/MS and Nanopore Sequencing
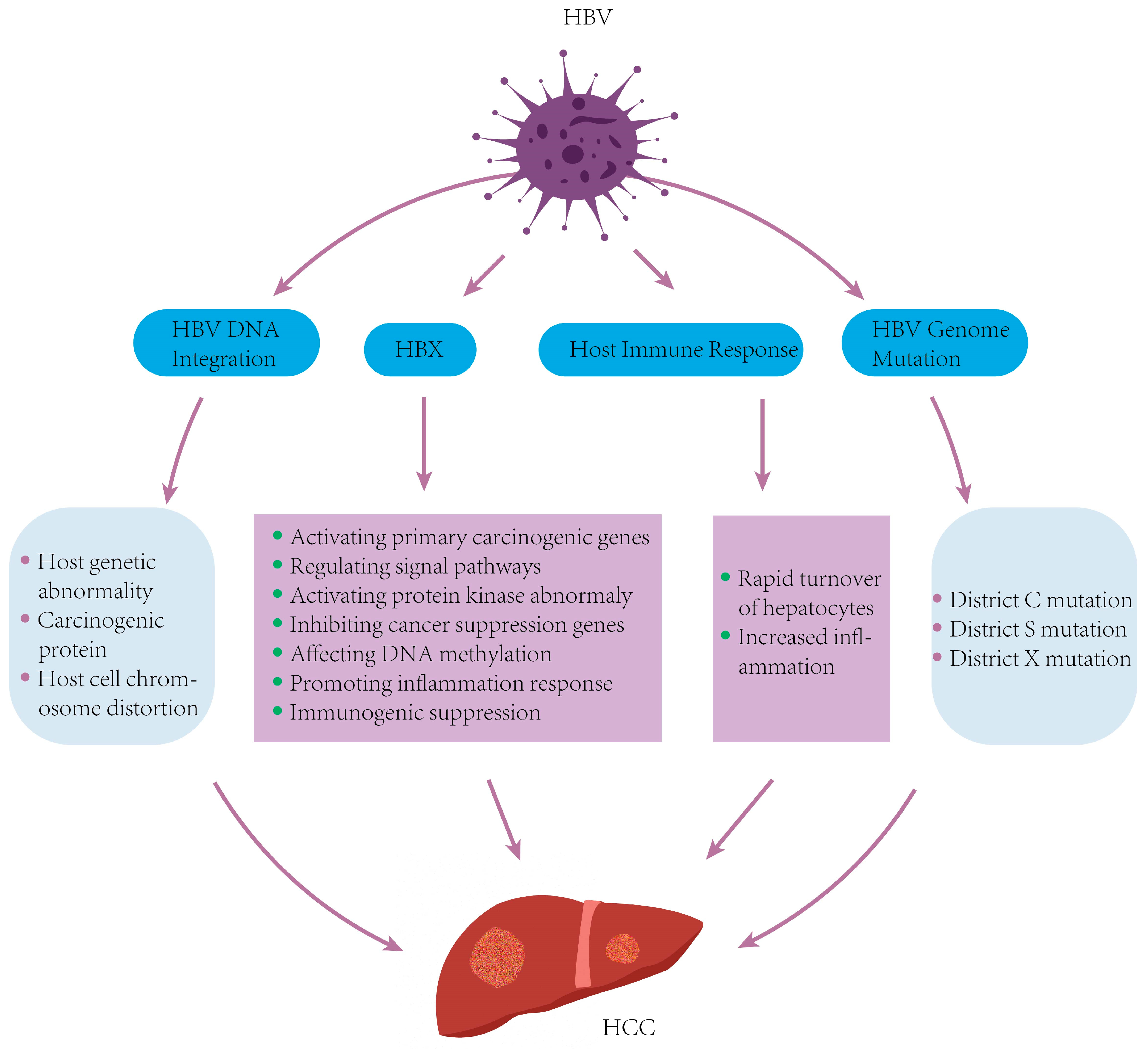
5. Molecular Mechanisms of HBV-Related HCC
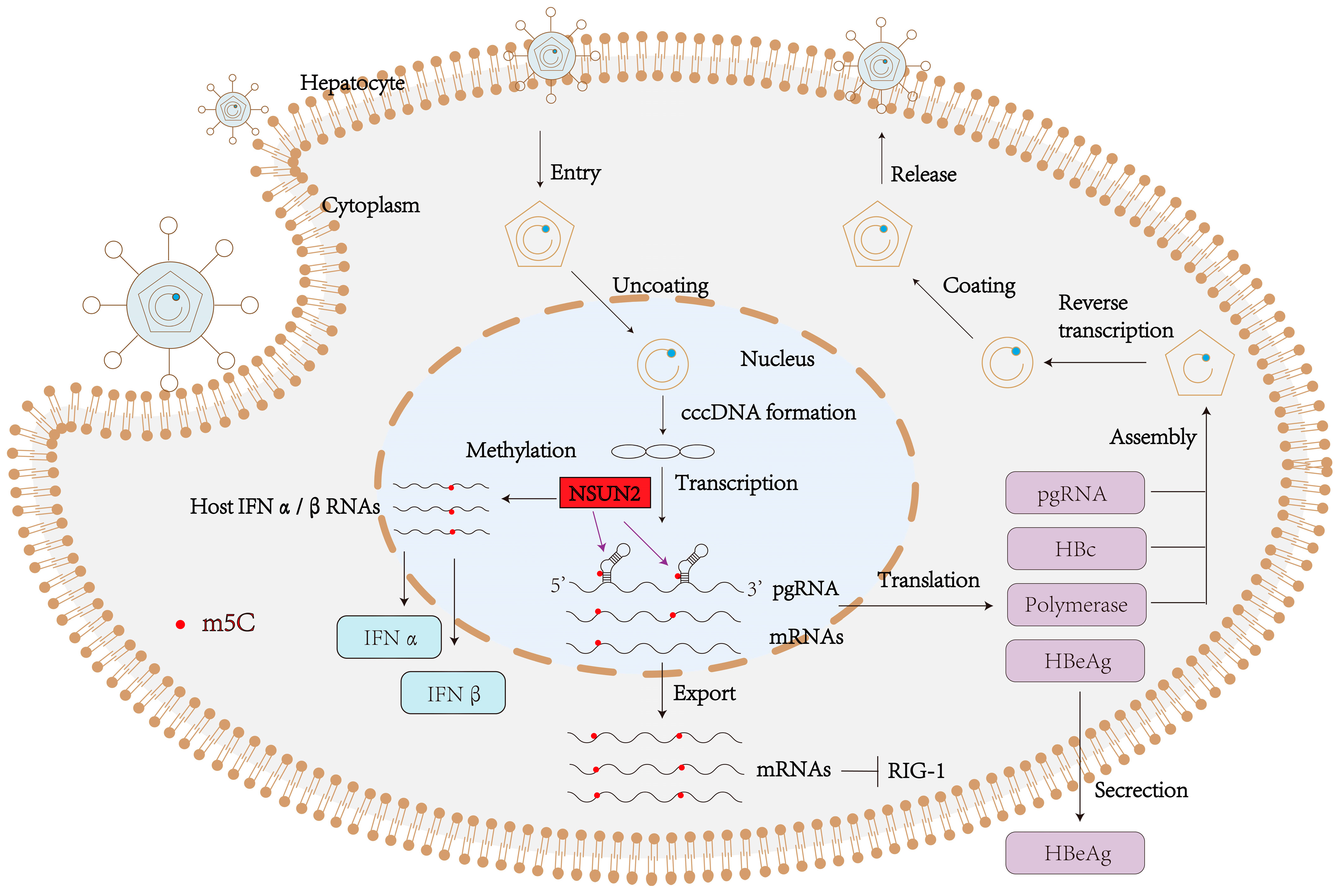
5.1. HBV Replication Cycle
5.2. HBV DNA Integration into the Host Genome
5.3. HBV Genomic Mutations
5.4. Abnormal Expression of the HBx Gene and Its Encoded Protein
6. The Role of m5C in the HBV Life Cycle
6.1. m5C Enhances pgRNA Stability and Encapsidation
6.2. m5C in Element (ε) Regulates HBV Reverse Transcription
6.3. m5C Modulates Innate Immune Recognition and Viral Evasion
7. m5C Methylation in HBV-Related HCC and Associated Liver Diseases
8. Crosstalk Between m5C and Other Modifications in the HBV Life Cycle
9. Comparative Insights: m5C in Other RNA and DNA Viruses
10. Clinical and Therapeutic Perspectives
10.1. Genotype- and Host-Dependent Variation in m5C
10.2. m5C in HBV-Driven Hepatocarcinogenesis
10.3. Targeting the m5C Machinery for Therapy
11. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yardeni, D.; Ghany, M.G. Review article: Hepatitis B-current and emerging therapies. Aliment. Pharmacol. Ther. 2022, 55, 805–819. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yan, L.; Shi, Y.; Lv, D.; Shang, J.; Bai, L.; Tang, H. Hepatitis B Virus Infection: Overview. Adv. Exp. Med. Biol. 2020, 1179, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Protzer, U.; Siddiqui, A. Revisiting Hepatitis B Virus: Challenges of Curative Therapies. J. Virol. 2019, 93, e01032-19. [Google Scholar] [CrossRef] [PubMed]
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; et al. MODOMICS: A database of RNA modification pathways. Nucleic Acids Res. 2022, 50, D231–D235. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Chen, X.; Li, A.; Sun, B.F.; Yang, Y.; Han, Y.N.; Yuan, X.; Chen, R.X.; Wei, W.S.; Liu, Y.; Gao, C.C.; et al. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 2019, 21, 978–990. [Google Scholar] [CrossRef]
- Chen, Z.; Huang, L.; Wang, K.; Zhang, L.; Zhong, X.; Yan, Z.; Liu, B.; Zhu, P. rtcisE2F promotes the self-renewal and metastasis of liver tumor-initiating cells via N6-methyladenosine-dependent E2F3/E2F6 mRNA stability. Sci. China Life Sci. 2022, 65, 1840–1854. [Google Scholar] [CrossRef]
- Zheng, Z.H.; Zhang, G.L.; Jiang, R.F.; Hong, Y.Q.; Zhang, Q.Y.; He, J.P.; Liu, X.R.; Yang, Z.S.; Yang, L.; Jiang, X.; et al. METTL3 is essential for normal progesterone signaling during embryo implantation via m6A-mediated translation control of progesterone receptor. Proc. Natl. Acad. Sci. USA 2023, 120, e2214684120. [Google Scholar] [CrossRef]
- Qiao, Y.; Sun, Q.; Chen, X.; He, L.; Wang, D.; Su, R.; Xue, Y.; Sun, H.; Wang, H. Nuclear m6A reader YTHDC1 promotes muscle stem cell activation/proliferation by regulating mRNA splicing and nuclear export. eLife 2023, 12, e82703. [Google Scholar] [CrossRef]
- Li, J.; Zhang, L. The emerging role of m5C modification in viral infection. Virology 2025, 610, 110606. [Google Scholar] [CrossRef]
- Liu, L.; Chen, Z.; Zhang, K.; Hao, H.; Ma, L.; Liu, H.; Yu, B.; Ding, S.; Zhang, X.; Zhu, M.; et al. NSUN2 mediates distinct pathways to regulate enterovirus 71 replication. Virol. Sin. 2024, 39, 574–586. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.S.; Dai, Q.; Chen, P.; Lu, M.; Kairis, E.L.; Murugaiah, V.; Xu, J.; Shukla, R.K.; Liang, X.; et al. 5-methylcytosine (m5C) RNA modification controls the innate immune response to virus infection by regulating type I interferons. Proc. Natl. Acad. Sci. USA 2022, 119, e2123338119. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Yao, X.; Xu, B.; Chen, W.; Lou, H.; Tong, X.; Fang, S.; Zou, R.; Hu, Y.; Wang, Z.; et al. RNA epigenetic modifications in ovarian cancer: The changes, chances, and challenges. Wiley Interdiscip. Rev. RNA 2023, 14, e1784. [Google Scholar] [CrossRef] [PubMed]
- Winans, S.; Beemon, K. m5C Goes Viral. Cell Host Microbe 2019, 26, 154–155. [Google Scholar] [CrossRef]
- Zhang, X.; Peng, Q.; Wang, L. N6-methyladenosine modification-a key player in viral infection. Cell. Mol. Biol. Lett. 2023, 28, 78. [Google Scholar] [CrossRef]
- Zhu, M.; Huang, F.; Sun, H.; Liu, K.; Chen, Z.; Yu, B.; Hao, H.; Liu, H.; Ding, S.; Zhang, X.; et al. Characterization of ACTN4 as a novel antiviral target against SARS-CoV-2. Signal Transduct. Target. Ther. 2024, 9, 243. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef]
- Meng, Y.; Shu, Z.; Wang, X.; Hong, L.; Wang, B.; Jiang, J.; He, K.; Cao, Q.; Shi, F.; Wang, H.; et al. Hepatitis B Virus-Mediated m6A Demethylation Increases Hepatocellular Carcinoma Stemness and Immune Escape. Mol. Cancer Res. 2024, 22, 642–655. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Mason, C.E. The pivotal regulatory landscape of RNA modifications. Annu. Rev. Genom. Hum. Genet. 2014, 15, 127–150. [Google Scholar] [CrossRef]
- Zhang, X.; Qin, S.; Huang, F.; Liu, H.; Wang, J.; Chen, Z.; Hao, H.; Ding, S.; Liu, L.; Yu, B.; et al. N4-acetylcytidine coordinates with NP1 and CPSF5 to facilitate alternative RNA processing during the replication of minute virus of canines. Nucleic Acids Res. 2025, 53, gkaf229. [Google Scholar] [CrossRef]
- Schiffers, S.; Oberdoerffer, S. ac4C: A fragile modification with stabilizing functions in RNA metabolism. RNA 2024, 30, 583–594. [Google Scholar] [CrossRef]
- Hao, H.; Liu, W.; Miao, Y.; Ma, L.; Yu, B.; Liu, L.; Yang, C.; Zhang, K.; Chen, Z.; Yang, J.; et al. N4-acetylcytidine regulates the replication and pathogenicity of enterovirus 71. Nucleic Acids Res. 2022, 50, 9339–9354. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Gan, H.; Yang, F.; Yao, Y.; Hao, F.; Hong, L.; Jin, L. Cytoplasmic m1A reader YTHDF3 inhibits trophoblast invasion by downregulation of m1A-methylated IGF1R. Cell Discov. 2020, 6, 12. [Google Scholar] [CrossRef]
- Yan, Q.; Zhou, J.; Wang, Z.; Ding, X.; Ma, X.; Li, W.; Jia, X.; Gao, S.J.; Lu, C. NAT10-dependent N4-acetylcytidine modification mediates PAN RNA stability, KSHV reactivation, and IFI16-related inflammasome activation. Nat. Commun. 2023, 14, 6327. [Google Scholar] [CrossRef]
- Tsai, K.; Jaguva Vasudevan, A.A.; Martinez Campos, C.; Emery, A.; Swanstrom, R.; Cullen, B.R. Acetylation of Cytidine Residues Boosts HIV-1 Gene Expression by Increasing Viral RNA Stability. Cell Host Microbe 2020, 28, 306–312.e306. [Google Scholar] [CrossRef]
- Pereira-Montecinos, C.; Valiente-Echeverría, F.; Soto-Rifo, R. Epitranscriptomic regulation of viral replication. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 460–471. [Google Scholar] [CrossRef]
- Levi, O.; Arava, Y.S. Pseudouridine-mediated translation control of mRNA by methionine aminoacyl tRNA synthetase. Nucleic Acids Res. 2021, 49, 432–443. [Google Scholar] [CrossRef]
- Carlile, T.M.; Martinez, N.M.; Schaening, C.; Su, A.; Bell, T.A.; Zinshteyn, B.; Gilbert, W.V. mRNA structure determines modification by pseudouridine synthase 1. Nat. Chem. Biol. 2019, 15, 966–974. [Google Scholar] [CrossRef] [PubMed]
- Martinez, N.M.; Su, A.; Burns, M.C.; Nussbacher, J.K.; Schaening, C.; Sathe, S.; Yeo, G.W.; Gilbert, W.V. Pseudouridine synthases modify human pre-mRNA co-transcriptionally and affect pre-mRNA processing. Mol. Cell 2022, 82, 645–659.e649. [Google Scholar] [CrossRef]
- Li, S.; Feng, T.; Liu, Y.; Yang, Q.; Song, A.; Wang, S.; Xie, J.; Zhang, J.; Yuan, B.; Sun, Z. m1A inhibition fuels oncolytic virus-elicited antitumor immunity via downregulating MYC/PD-L1 signaling. Int. J. Oral Sci. 2024, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- Filipowicz, W. Functions of the 5′-terminal m7G cap in eukaryotic mRNA. FEBS Lett. 1978, 96, 1–11. [Google Scholar] [CrossRef]
- Kiriakidou, M.; Tan, G.S.; Lamprinaki, S.; De Planell-Saguer, M.; Nelson, P.T.; Mourelatos, Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell 2007, 129, 1141–1151. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Han, H.; Huang, Y.; Yang, C.; Zheng, S.; Cai, T.; Bi, J.; Huang, X.; Liu, R.; Huang, L.; et al. METTL1/WDR4-mediated m7G tRNA modifications and m7G codon usage promote mRNA translation and lung cancer progression. Mol. Ther. 2021, 29, 3422–3435. [Google Scholar] [CrossRef] [PubMed]
- Létoquart, J.; Huvelle, E.; Wacheul, L.; Bourgeois, G.; Zorbas, C.; Graille, M.; Heurgué-Hamard, V.; Lafontaine, D.L. Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc. Natl. Acad. Sci. USA 2014, 111, E5518–E5526. [Google Scholar] [CrossRef]
- Werner, M.; Purta, E.; Kaminska, K.H.; Cymerman, I.A.; Campbell, D.A.; Mittra, B.; Zamudio, J.R.; Sturm, N.R.; Jaworski, J.; Bujnicki, J.M. 2′-O-ribose methylation of cap2 in human: Function and evolution in a horizontally mobile family. Nucleic Acids Res. 2011, 39, 4756–4768. [Google Scholar] [CrossRef] [PubMed]
- Belanger, F.; Stepinski, J.; Darzynkiewicz, E.; Pelletier, J. Characterization of hMTr1, a human Cap1 2′-O-ribose methyltransferase. J. Biol. Chem. 2010, 285, 33037–33044. [Google Scholar] [CrossRef]
- Drazkowska, K.; Tomecki, R.; Warminski, M.; Baran, N.; Cysewski, D.; Depaix, A.; Kasprzyk, R.; Kowalska, J.; Jemielity, J.; Sikorski, P.J. 2′-O-Methylation of the second transcribed nucleotide within the mRNA 5′ cap impacts the protein production level in a cell-specific manner and contributes to RNA immune evasion. Nucleic Acids Res. 2022, 50, 9051–9071. [Google Scholar] [CrossRef]
- Lu, Y.; Yang, L.; Feng, Q.; Liu, Y.; Sun, X.; Liu, D.; Qiao, L.; Liu, Z. RNA 5-Methylcytosine Modification: Regulatory Molecules, Biological Functions, and Human Diseases. Genom. Proteom. Bioinform. 2024, 22, qzae063. [Google Scholar] [CrossRef]
- Bourgeois, G.; Ney, M.; Gaspar, I.; Aigueperse, C.; Schaefer, M.; Kellner, S.; Helm, M.; Motorin, Y. Eukaryotic rRNA Modification by Yeast 5-Methylcytosine-Methyltransferases and Human Proliferation-Associated Antigen p120. PLoS ONE 2015, 10, e0133321. [Google Scholar] [CrossRef]
- Xu, Y.; Qi, S.; Zhang, G.; Liu, D.; Xu, D.; Qin, T.; Cheng, Q.; Kang, H.; Hu, B.; Huang, Z. One-pot ligation of multiple mRNA fragments on dsDNA splint advancing regional modification and translation. Nucleic Acids Res. 2025, 53, gkae1280. [Google Scholar] [CrossRef]
- Trixl, L.; Lusser, A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. Wiley Interdiscip. reviews. RNA 2019, 10, e1510. [Google Scholar] [CrossRef]
- Goll, M.G.; Kirpekar, F.; Maggert, K.A.; Yoder, J.A.; Hsieh, C.L.; Zhang, X.; Golic, K.G.; Jacobsen, S.E.; Bestor, T.H. Methylation of tRNA Asp by the DNA methyltransferase homolog Dnmt2. Science 2006, 311, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, H.; Zhu, X.; Yadav, T.; Ouyang, J.; Truesdell, S.S.; Tan, J.; Wang, Y.; Duan, M.; Wei, L.; et al. m5C modification of mRNA serves a DNA damage code to promote homologous recombination. Nat. Commun. 2020, 11, 2834. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.; Xu, H.; Sun, Z.; Shen, H.; Chen, S.; Ouyang, J.; Zhou, Q.; Hu, X.; Cui, H. Depletion of TRDMT1 affects 5-methylcytosine modification of mRNA and inhibits HEK293 cell proliferation and migration. Biochem. Biophys. Res. Commun. 2019, 520, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, H.; Long, T.; Dong, H.; Wang, E.D.; Liu, R.J. Archaeal NSUN6 catalyzes m5C72 modification on a wide-range of specific tRNAs. Nucleic Acids Res. 2019, 47, 2041–2055. [Google Scholar] [CrossRef]
- Squires, J.E.; Patel, H.R.; Nousch, M.; Sibbritt, T.; Humphreys, D.T.; Parker, B.J.; Suter, C.M.; Preiss, T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012, 40, 5023–5033. [Google Scholar] [CrossRef]
- Gonskikh, Y.; Tirrito, C.; Bommisetti, P.; Mendoza-Figueroa, M.S.; Stoute, J.; Kim, J.; Wang, Q.; Song, Y.; Liu, K.F. Spatial regulation of NSUN2-mediated tRNA m5C installation in cognitive function. Nucleic Acids Res. 2025, 53, gkae1169. [Google Scholar] [CrossRef]
- Dominissini, D.; Rechavi, G. 5-methylcytosine mediates nuclear export of mRNA. Cell Res. 2017, 27, 717–719. [Google Scholar] [CrossRef]
- Shinoda, S.; Kitagawa, S.; Nakagawa, S.; Wei, F.Y.; Tomizawa, K.; Araki, K.; Araki, M.; Suzuki, T.; Suzuki, T. Mammalian NSUN2 introduces 5-methylcytidines into mitochondrial tRNAs. Nucleic Acids Res. 2019, 47, 8734–8745. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ying, S.; Zou, Y.; Wang, X.; Zhang, R.; Huang, C.; Dai, M.; Xu, K.; Feng, G.; Li, X.; et al. NSun2-Mediated tsRNAs Alleviate Liver Fibrosis via FAK Dephosphorylation. Cell Prolif. 2025, e70058. [Google Scholar] [CrossRef]
- Li, M.; Tao, Z.; Zhao, Y.; Li, L.; Zheng, J.; Li, Z.; Chen, X. 5-methylcytosine RNA methyltransferases and their potential roles in cancer. J. Transl. Med. 2022, 20, 214. [Google Scholar] [CrossRef]
- Chen, Y.S.; Yang, W.L.; Zhao, Y.L.; Yang, Y.G. Dynamic transcriptomic m5C and its regulatory role in RNA processing. Wiley Interdiscip. Rev. RNA 2021, 12, e1639. [Google Scholar] [CrossRef]
- Shen, Q.; Zhang, Q.; Shi, Y.; Shi, Q.; Jiang, Y.; Gu, Y.; Li, Z.; Li, X.; Zhao, K.; Wang, C.; et al. Tet2 promotes pathogen infection-induced myelopoiesis through mRNA oxidation. Nature 2018, 554, 123–127. [Google Scholar] [CrossRef]
- Shen, H.; Ontiveros, R.J.; Owens, M.C.; Liu, M.Y.; Ghanty, U.; Kohli, R.M.; Liu, K.F. TET-mediated 5-methylcytosine oxidation in tRNA promotes translation. J. Biol. Chem. 2021, 296, 100087. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xue, M.; Deng, X.; Dong, L.; Nguyen, L.X.T.; Ren, L.; Han, L.; Li, C.; Xue, J.; Zhao, Z.; et al. TET2-mediated mRNA demethylation regulates leukemia stem cell homing and self-renewal. Cell Stem Cell 2023, 30, 1072–1090.e1010. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Shi, Y.G. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development 2012, 139, 1895–1902. [Google Scholar] [CrossRef]
- Wang, X.; Jin, X.-Y.; Cheng, L. Recent Advance in the Study on 5-Formylcytosine (f5C) RNA Modification. Isr. J. Chem. 2024, 64, e202300178. [Google Scholar] [CrossRef]
- Wang, X.; Wang, M.; Dai, X.; Han, X.; Zhou, Y.; Lai, W.; Zhang, L.; Yang, Y.; Chen, Y.; Wang, H.; et al. RNA 5-methylcytosine regulates YBX2-dependent liquid-liquid phase separation. Fundam. Res. 2022, 2, 48–55. [Google Scholar] [CrossRef]
- Yang, H.; Wang, Y.; Xiang, Y.; Yadav, T.; Ouyang, J.; Phoon, L.; Zhu, X.; Shi, Y.; Zou, L.; Lan, L. FMRP promotes transcription-coupled homologous recombination via facilitating TET1-mediated m5C RNA modification demethylation. Proc. Natl. Acad. Sci. USA 2022, 119, e2116251119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Li, C.T.; Zhou, Y.J.; Li, H.; Li, J.; Xiong, Q.P.; Zhou, W.; Huang, W.; Zhang, Q.C.; Xiang, Y.; et al. A cohort of mRNAs undergo high-stoichiometry NSUN6-mediated site-specific m5C modification. Nat. Commun. 2025, 16, 6119. [Google Scholar] [CrossRef]
- Yang, X.; Yang, Y.; Sun, B.F.; Chen, Y.S.; Xu, J.W.; Lai, W.Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export—NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Zhang, H.; Wu, X.; He, Z.; Wang, L.; Yin, S.; Tian, B.; Li, G.; Cheng, H. ALYREF mainly binds to the 5′ and the 3′ regions of the mRNA in vivo. Nucleic Acids Res. 2017, 45, 9640–9653. [Google Scholar] [CrossRef]
- Su, J.; Wu, G.; Ye, Y.; Zhang, J.; Zeng, L.; Huang, X.; Zheng, Y.; Bai, R.; Zhuang, L.; Li, M.; et al. NSUN2-mediated RNA 5-methylcytosine promotes esophageal squamous cell carcinoma progression via LIN28B-dependent GRB2 mRNA stabilization. Oncogene 2021, 40, 5814–5828. [Google Scholar] [CrossRef]
- Ma, H.L.; Bizet, M.; Soares Da Costa, C.; Murisier, F.; de Bony, E.J.; Wang, M.K.; Yoshimi, A.; Lin, K.T.; Riching, K.M.; Wang, X.; et al. SRSF2 plays an unexpected role as reader of m5C on mRNA, linking epitranscriptomics to cancer. Mol. Cell 2023, 83, 4239–4254.e4210. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Zhang, J.; Liu, B.; Xu, J.; Cai, B.; Yang, H.; Straube, J.; Yu, X.; Ma, T. Biological roles of RNA m5C modification and its implications in Cancer immunotherapy. Biomark. Res. 2022, 10, 15. [Google Scholar] [CrossRef]
- Huang, T.; Chen, W.; Liu, J.; Gu, N.; Zhang, R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat. Struct. Mol. Biol. 2019, 26, 380–388. [Google Scholar] [CrossRef]
- Schumann, U.; Zhang, H.N.; Sibbritt, T.; Pan, A.; Horvath, A.; Gross, S.; Clark, S.J.; Yang, L.; Preiss, T. Multiple links between 5-methylcytosine content of mRNA and translation. BMC Biol. 2020, 18, 40. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Y.; Wu, R.; Gao, C.C.; Liao, X.; Han, X.; Zeng, B.; Huang, C.; Luo, Y.; Liu, Y.; et al. mRNA m5C inhibits adipogenesis and promotes myogenesis by respectively facilitating YBX2 and SMO mRNA export in ALYREF-m5C manner. Cell. Mol. Life Sci. 2022, 79, 481. [Google Scholar] [CrossRef]
- Chen, Y.; Zuo, X.; Wei, Q.; Xu, J.; Liu, X.; Liu, S.; Wang, H.; Luo, Q.; Wang, Y.; Yang, Y.; et al. Upregulation of LRRC8A by m5C modification-mediated mRNA stability suppresses apoptosis and facilitates tumorigenesis in cervical cancer. Int. J. Biol. Sci. 2023, 19, 691–704. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, L.; Han, X.; Yang, W.L.; Zhang, M.; Ma, H.L.; Sun, B.F.; Li, A.; Xia, J.; Chen, J.; et al. RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay. Mol. Cell 2019, 75, 1188–1202.e1111. [Google Scholar] [CrossRef]
- Wang, J.Z.; Zhu, W.; Han, J.; Yang, X.; Zhou, R.; Lu, H.C.; Yu, H.; Yuan, W.B.; Li, P.C.; Tao, J.; et al. The role of the HIF-1α/ALYREF/PKM2 axis in glycolysis and tumorigenesis of bladder cancer. Cancer Commun. 2021, 41, 560–575. [Google Scholar] [CrossRef]
- Wang, N.; Chen, R.X.; Deng, M.H.; Wei, W.S.; Zhou, Z.H.; Ning, K.; Li, Y.H.; Li, X.D.; Ye, Y.L.; Wen, J.H.; et al. m5C-dependent cross-regulation between nuclear reader ALYREF and writer NSUN2 promotes urothelial bladder cancer malignancy through facilitating RABL6/TK1 mRNAs splicing and stabilization. Cell Death Dis. 2023, 14, 139. [Google Scholar] [CrossRef]
- Navarro-Torné, A.; Anderson, A.; Panwar, K.; Ghys, E.; Benninghoff, B.; Weynants, V.; Beddows, S.; Checchi, M. Corrigendum to How has post-implementation surveillance of high-coverage vaccination with HPV16/18-AS04 vaccine in England added to evidence about its cross-protective effects? Vaccine 2024, 42, 126215, Erratum in Vaccine 2025, 47, 126701. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Yao, J.; Fu, J.; Huang, Q.; Luo, Y.; You, Y.; Zhang, W.; Zhong, Q.; Xia, T.; Xia, L. ALYREF promotes the metastasis of nasopharyngeal carcinoma by increasing the stability of NOTCH1 mRNA. Cell Death Dis. 2024, 15, 578. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Sierzputowska-Gracz, H.; Guenther, R.; Everett, K.; Agris, P.F. 5-Methylcytidine is required for cooperative binding of Mg2+ and a conformational transition at the anticodon stem-loop of yeast phenylalanine tRNA. Biochemistry 1993, 32, 10249–10253. [Google Scholar] [CrossRef]
- Yang, L.; Perrera, V.; Saplaoura, E.; Apelt, F.; Bahin, M.; Kramdi, A.; Olas, J.; Mueller-Roeber, B.; Sokolowska, E.; Zhang, W.; et al. m5C Methylation Guides Systemic Transport of Messenger RNA over Graft Junctions in Plants. Curr. Biol. 2019, 29, 2465–2476.e2465. [Google Scholar] [CrossRef]
- Burgess, A.L.; David, R.; Searle, I.R. Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol. 2015, 15, 199. [Google Scholar] [CrossRef] [PubMed]
- Metodiev, M.D.; Spåhr, H.; Loguercio Polosa, P.; Meharg, C.; Becker, C.; Altmueller, J.; Habermann, B.; Larsson, N.G.; Ruzzenente, B. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLoS Genet. 2014, 10, e1004110. [Google Scholar] [CrossRef] [PubMed]
- Amort, T.; Rieder, D.; Wille, A.; Khokhlova-Cubberley, D.; Riml, C.; Trixl, L.; Jia, X.Y.; Micura, R.; Lusser, A. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017, 18, 1. [Google Scholar] [CrossRef] [PubMed]
- David, R.; Burgess, A.; Parker, B.; Li, J.; Pulsford, K.; Sibbritt, T.; Preiss, T.; Searle, I.R. Transcriptome-Wide Mapping of RNA 5-Methylcytosine in Arabidopsis mRNAs and Noncoding RNAs. Plant Cell 2017, 29, 445–460. [Google Scholar] [CrossRef]
- Dubin, D.T.; Taylor, R.H. The methylation state of poly A-containing messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975, 2, 1653–1668. [Google Scholar] [CrossRef]
- Iwanami, Y.; Brown, G.M. Methylated bases of ribosomal ribonucleic acid from HeLa cells. Arch. Biochem. Biophys. 1968, 126, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Frommer, M.; McDonald, L.E.; Millar, D.S.; Collis, C.M.; Watt, F.; Grigg, G.W.; Molloy, P.L.; Paul, C.L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 1992, 89, 1827–1831. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Hurto, R.L.; Hopper, A.K.; Grayhack, E.J.; Phizicky, E.M. Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m5C. Mol. Cell. Biol. 2005, 25, 8191–8201. [Google Scholar] [CrossRef]
- Guo, G.; Pan, K.; Fang, S.; Ye, L.; Tong, X.; Wang, Z.; Xue, X.; Zhang, H. Advances in mRNA 5-methylcytosine modifications: Detection, effectors, biological functions, and clinical relevance. Mol. Ther. Nucleic Acids 2021, 26, 575–593. [Google Scholar] [CrossRef]
- Zhang, L.S.; Dai, Q.; He, C. Base-Resolution Sequencing Methods for Whole-Transcriptome Quantification of mRNA Modifications. Acc. Chem. Res. 2024, 57, 47–58. [Google Scholar] [CrossRef] [PubMed]
- Khoddami, V.; Cairns, B.R. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 2013, 31, 458–464. [Google Scholar] [CrossRef]
- George, H.; Ule, J.; Hussain, S. Illustrating the Epitranscriptome at Nucleotide Resolution Using Methylation-iCLIP (miCLIP). Methods Mol. Biol. 2017, 1562, 91–106. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, H.; Kohnen, M.V.; Prasad, K.; Gu, L.; Reddy, A.S.N. Analysis of Transcriptome and Epitranscriptome in Plants Using PacBio Iso-Seq and Nanopore-Based Direct RNA Sequencing. Front. Genet. 2019, 10, 253. [Google Scholar] [CrossRef]
- Chan, S.L.; Wong, V.W.; Qin, S.; Chan, H.L. Infection and Cancer: The Case of Hepatitis B. J. Clin. Oncol. 2016, 34, 83–90. [Google Scholar] [CrossRef]
- Rizzo, G.E.M.; Cabibbo, G.; Craxì, A. Hepatitis B Virus-Associated Hepatocellular Carcinoma. Viruses 2022, 14, 986. [Google Scholar] [CrossRef]
- D’Souza, S.; Lau, K.C.; Coffin, C.S.; Patel, T.R. Molecular mechanisms of viral hepatitis induced hepatocellular carcinoma. World J. Gastroenterol. 2020, 26, 5759–5783. [Google Scholar] [CrossRef]
- Herrscher, C.; Roingeard, P.; Blanchard, E. Hepatitis B Virus Entry into Cells. Cells 2020, 9, 1486. [Google Scholar] [CrossRef]
- Tsukuda, S.; Watashi, K. Hepatitis B virus biology and life cycle. Antivir. Res. 2020, 182, 104925. [Google Scholar] [CrossRef] [PubMed]
- Pollicino, T.; Caminiti, G. HBV-Integration Studies in the Clinic: Role in the Natural History of Infection. Viruses 2021, 13, 368. [Google Scholar] [CrossRef]
- Mason, W.S.; Liu, C.; Aldrich, C.E.; Litwin, S.; Yeh, M.M. Clonal expansion of normal-appearing human hepatocytes during chronic hepatitis B virus infection. J. Virol. 2010, 84, 8308–8315. [Google Scholar] [CrossRef]
- Péneau, C.; Imbeaud, S.; La Bella, T.; Hirsch, T.Z.; Caruso, S.; Calderaro, J.; Paradis, V.; Blanc, J.F.; Letouzé, E.; Nault, J.C.; et al. Hepatitis B virus integrations promote local and distant oncogenic driver alterations in hepatocellular carcinoma. Gut 2022, 71, 616–626. [Google Scholar] [CrossRef]
- Wang, J.; Zindy, F.; Chenivesse, X.; Lamas, E.; Henglein, B.; Bréchot, C. Modification of cyclin A expression by hepatitis B virus DNA integration in a hepatocellular carcinoma. Oncogene 1992, 7, 1653–1656. [Google Scholar]
- Zoulim, F.; Chen, P.J.; Dandri, M.; Kennedy, P.T.; Seeger, C. Hepatitis B virus DNA integration: Implications for diagnostics, therapy, and outcome. J. Hepatol. 2024, 81, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.G.; Mahmud, M.M.; Nazir, K.H.M.N.H.; Ueda, K. PreS1 Mutations Alter the Large HBsAg Antigenicity of a Hepatitis B Virus Strain Isolated in Bangladesh. Int. J. Mol. Sci. 2020, 21, 546. [Google Scholar] [CrossRef] [PubMed]
- Chevaliez, S.; Rodriguez, C.; Poiteau, L.; Soulier, A.; Donati, F.; Darty-Mercier, M.; Pioche, C.; Leroy, V.; Brodard, V.; Zoulim, F.; et al. Primary resistance of hepatitis B virus to nucleoside and nucleotide analogues. J. Viral Hepat. 2019, 26, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Podlaha, O.; Gane, E.; Brunetto, M.; Fung, S.; Chuang, W.L.; Pan, C.Q.; Jiang, Z.; Liu, Y.; Bhardwaj, N.; Mukherjee, P.; et al. Large-scale viral genome analysis identifies novel clinical associations between hepatitis B virus and chronically infected patients. Sci. Rep. 2019, 9, 10529. [Google Scholar] [CrossRef]
- Lin, Y.-T.; Jeng, L.-B.; Chan, W.-L.; Su, I.-J.; Teng, C.-F. Hepatitis B Virus Pre-S Gene Deletions and Pre-S Deleted Proteins: Clinical and Molecular Implications in Hepatocellular Carcinoma. Viruses 2021, 13, 862. [Google Scholar] [CrossRef]
- Panasiuk, Y.V.; Vlasenko, N.V.; Churilova, N.S.; Klushkina, V.V.; Dubodelov, D.V.; Kudryavtseva, E.N.; Korabelnikova, M.I.; Rodionova, Z.S.; Semenenko, T.A.; Kuzin, S.N.; et al. Modern views on the role of X gene of the hepatitis B virus (Hepadnaviridae: Orthohepadnavirus: Hepatitis B virus) in the pathogenesis of the infection it causes. Probl. Virol. 2022, 67, 7–17. [Google Scholar] [CrossRef]
- Ren, F.; Li, W.; Xiang, A.; Wang, L.; Li, M.; Guo, Y. Distribution and difference of APOBEC-induced mutations in the TpCpW context of HBV DNA between HCC and non-HCC. J. Med. Virol. 2020, 92, 53–61. [Google Scholar] [CrossRef]
- Yan, W.; Rao, D.; Fan, F.; Liang, H.; Zhang, Z.; Dong, H. Hepatitis B virus X protein and TGF-β: Partners in the carcinogenic journey of hepatocellular carcinoma. Front. Oncol. 2024, 14, 1407434. [Google Scholar] [CrossRef] [PubMed]
- Salerno, D.; Chiodo, L.; Alfano, V.; Floriot, O.; Cottone, G.; Paturel, A.; Pallocca, M.; Plissonnier, M.L.; Jeddari, S.; Belloni, L.; et al. Hepatitis B protein HBx binds the DLEU2 lncRNA to sustain cccDNA and host cancer-related gene transcription. Gut 2020, 69, 2016–2024. [Google Scholar] [CrossRef]
- An, P.; Xu, J.; Yu, Y.; Winkler, C.A. Host and Viral Genetic Variation in HBV-Related Hepatocellular Carcinoma. Front. Genet. 2018, 9, 261. [Google Scholar] [CrossRef] [PubMed]
- Heukers, R.; Altintas, I.; Raghoenath, S.; De Zan, E.; Pepermans, R.; Roovers, R.C.; Haselberg, R.; Hennink, W.E.; Schiffelers, R.M.; Kok, R.J.; et al. Targeting hepatocyte growth factor receptor (Met) positive tumor cells using internalizing nanobody-decorated albumin nanoparticles. Biomaterials 2014, 35, 601–610. [Google Scholar] [CrossRef]
- Grohmann, M.; Wiede, F.; Dodd, G.T.; Gurzov, E.N.; Ooi, G.J.; Butt, T.; Rasmiena, A.A.; Kaur, S.; Gulati, T.; Goh, P.K.; et al. Obesity Drives STAT-1-Dependent NASH and STAT-3-Dependent HCC. Cell 2018, 175, 1289–1306.e1220. [Google Scholar] [CrossRef]
- Zhou, S.J.; Deng, Y.L.; Liang, H.F.; Jaoude, J.C.; Liu, F.Y. Hepatitis B virus X protein promotes CREB-mediated activation of miR-3188 and Notch signaling in hepatocellular carcinoma. Cell Death Differ. 2017, 24, 1577–1587. [Google Scholar] [CrossRef]
- Tu, W.; Gong, J.; Tian, D.; Wang, Z. Hepatitis B Virus X Protein Induces SATB1 Expression Through Activation of ERK and p38MAPK Pathways to Suppress Anoikis. Dig. Dis. Sci. 2019, 64, 3203–3214. [Google Scholar] [CrossRef]
- Yang, W.Y.; Rao, P.S.; Luo, Y.C.; Lin, H.K.; Huang, S.H.; Yang, J.M.; Yuh, C.H. Omics-based Investigation of Diet-induced Obesity Synergized with HBx, Src, and p53 Mutation Accelerating Hepatocarcinogenesis in Zebrafish Model. Cancers 2019, 11, 1899. [Google Scholar] [CrossRef]
- Wei, X.; Xiang, T.; Ren, G.; Tan, C.; Liu, R.; Xu, X.; Wu, Z. miR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell Signal. 2013, 25, 439–446. [Google Scholar] [CrossRef]
- Ji, L.; Lin, Z.; Wan, Z.; Xia, S.; Jiang, S.; Cen, D.; Cai, L.; Xu, J.; Cai, X. miR-486-3p mediates hepatocellular carcinoma sorafenib resistance by targeting FGFR4 and EGFR. Cell Death Dis. 2020, 11, 250. [Google Scholar] [CrossRef]
- Imam, H.; Kim, G.W.; Mir, S.A.; Khan, M.; Siddiqui, A. Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified Hepatitis B Virus transcripts. PLoS Pathog. 2020, 16, e1008338. [Google Scholar] [CrossRef]
- Kim, G.W.; Siddiqui, A. Hepatitis B Virus X Protein Expression Is Tightly Regulated by N6-Methyladenosine Modification of Its mRNA. J. Virol. 2022, 96, e0165521. [Google Scholar] [CrossRef]
- Kim, G.W.; Imam, H.; Khan, M.; Siddiqui, A. N6-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J. Biol. Chem. 2020, 295, 13123–13133. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Xu, T.; He, M.; Li, J.; Yao, P.; Ma, C.; Yang, S.; Xu, Z.; Yan, K.; Chen, X.; et al. NSUN2-mediated m5C modification of HBV RNA positively regulates HBV replication. PLoS Pathog. 2023, 19, e1011808. [Google Scholar] [CrossRef] [PubMed]
- Su, P.A.; Chang, C.H.; Yen, S.B.; Wu, H.Y.; Tung, W.J.; Hu, Y.P.; Chen, Y.I.; Lin, M.H.; Shih, C.; Chen, P.J.; et al. Epitranscriptomic cytidine methylation of the hepatitis B viral RNA is essential for viral reverse transcription and particle production. Proc. Natl. Acad. Sci. USA 2024, 121, e2400378121. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Li, K.; Kameyama, T.; Hayashi, T.; Ishida, Y.; Murakami, S.; Watanabe, T.; Iijima, S.; Sakurai, Y.; Watashi, K.; et al. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity 2015, 42, 123–132. [Google Scholar] [CrossRef]
- Ding, S.; Liu, H.; Liu, L.; Ma, L.; Chen, Z.; Zhu, M.; Liu, L.; Zhang, X.; Hao, H.; Zuo, L.; et al. Epigenetic addition of m5C to HBV transcripts promotes viral replication and evasion of innate antiviral responses. Cell Death Dis. 2024, 15, 39. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhu, B.; Yang, H.; Li, F.; Qu, Y.; Lu, L.; Zhang, Q. Exploration of YBX1 role in the prognostic value and immune characteristics by single-cell and bulk sequencing analysis for liver hepatocellular carcinoma. J. Gene Med. 2024, 26, e3680. [Google Scholar] [CrossRef]
- Zhang, Q.; Zheng, Q.; Yu, X.; He, Y.; Guo, W. Overview of distinct 5-methylcytosine profiles of messenger RNA in human hepatocellular carcinoma and paired adjacent non-tumor tissues. J. Transl. Med. 2020, 18, 245. [Google Scholar] [CrossRef]
- Xue, C.; Gu, X.; Zheng, Q.; Shi, Q.; Yuan, X.; Su, Y.; Jia, J.; Jiang, J.; Lu, J.; Li, L. ALYREF mediates RNA m5C modification to promote hepatocellular carcinoma progression. Signal Transduct. Target. Ther. 2023, 8, 130. [Google Scholar] [CrossRef]
- Nulali, J.; Zhang, K.; Long, M.; Wan, Y.; Liu, Y.; Zhang, Q.; Yang, L.; Hao, J.; Yang, L.; Song, H. ALYREF-mediated RNA 5-Methylcytosine modification Promotes Hepatocellular Carcinoma Progression Via Stabilizing EGFR mRNA and pSTAT3 activation. Int. J. Biol. Sci. 2024, 20, 331–346. [Google Scholar] [CrossRef]
- Sun, Z.; Xue, S.; Zhang, M.; Xu, H.; Hu, X.; Chen, S.; Liu, Y.; Guo, M.; Cui, H. Aberrant NSUN2-mediated m5C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene 2020, 39, 6906–6919. [Google Scholar] [CrossRef]
- Li, B.; Li, F.; Gu, T.; Guo, Y.; Shen, B.; Xu, X.; Shen, Z.; Chen, L.; Zhang, Q.; Dong, H.; et al. Specific knockdown of Y-box binding protein 1 in hepatic progenitor cells inhibits proliferation and alleviates liver fibrosis. Eur. J. Pharmacol. 2022, 921, 174866. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Pan, Y.; Zhang, J.; Hu, B.; Qin, D.; Liu, S.; Chen, N.; Zhang, L. TET2 regulation of alcoholic fatty liver via Srebp1 mRNA in paraspeckles. iScience 2024, 27, 109278. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhao, Y.; Wu, R.; Chen, Y.; Chen, W.; Liu, Y.; Luo, Y.; Huang, C.; Zeng, B.; Liao, X.; et al. mRNA m5C controls adipogenesis by promoting CDKN1A mRNA export and translation. RNA Biol. 2021, 18, 711–721. [Google Scholar] [CrossRef]
- Zhang, H.; Zhai, X.; Liu, Y.; Xia, Z.; Xia, T.; Du, G.; Zhou, H.; Franziska Strohmer, D.; Bazhin, A.V.; Li, Z.; et al. NOP2-mediated m5C Modification of c-Myc in an EIF3A-Dependent Manner to Reprogram Glucose Metabolism and Promote Hepatocellular Carcinoma Progression. Research 2023, 6, 0184. [Google Scholar] [CrossRef]
- Qu, S.; Jin, L.; Huang, H.; Lin, J.; Gao, W.; Zeng, Z. A positive-feedback loop between HBx and ALKBH5 promotes hepatocellular carcinogenesis. BMC Cancer 2021, 21, 686. [Google Scholar] [CrossRef]
- Kim, G.W.; Imam, H.; Khan, M.; Mir, S.A.; Kim, S.J.; Yoon, S.K.; Hur, W.; Siddiqui, A. HBV-Induced Increased N6 Methyladenosine Modification of PTEN RNA Affects Innate Immunity and Contributes to HCC. Hepatology 2021, 73, 533–547. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.W.; Moon, J.S.; Gudima, S.O.; Siddiqui, A. N6-Methyladenine Modification of Hepatitis Delta Virus Regulates Its Virion Assembly by Recruiting YTHDF1. J. Virol. 2022, 96, e0112422. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Li, J.; Lei, X.; Lin, R.; Wu, Q.; Zhang, Z.; Shuai, S.; Tian, R. Multimodal CRISPR screens uncover DDX39B as a global repressor of A-to-I RNA editing. Cell Rep. 2025, 44, 116009. [Google Scholar] [CrossRef]
- Courtney, D.G.; Tsai, K.; Bogerd, H.P.; Kennedy, E.M.; Law, B.A.; Emery, A.; Swanstrom, R.; Holley, C.L.; Cullen, B.R. Epitranscriptomic Addition of m5C to HIV-1 Transcripts Regulates Viral Gene Expression. Cell Host Microbe 2019, 26, 217–227. [Google Scholar] [CrossRef]
- Wang, H.; Feng, J.; Fu, Z.; Xu, T.; Liu, J.; Yang, S.; Li, Y.; Deng, J.; Zhang, Y.; Guo, M.; et al. Epitranscriptomic m5C methylation of SARS-CoV-2 RNA regulates viral replication and the virulence of progeny viruses in the new infection. Sci. Adv. 2024, 10, eadn9519. [Google Scholar] [CrossRef]
- Courtney, D.G.; Chalem, A.; Bogerd, H.P.; Law, B.A.; Kennedy, E.M.; Holley, C.L.; Cullen, B.R. Extensive Epitranscriptomic Methylation of A and C Residues on Murine Leukemia Virus Transcripts Enhances Viral Gene Expression. mBio 2019, 10, e01209–e01219. [Google Scholar] [CrossRef]
- Li, Z.L.; Xie, Y.; Xie, Y.; Chen, H.; Zhou, X.; Liu, M.; Zhang, X.L. HCV 5-Methylcytosine Enhances Viral RNA Replication through Interaction with m5C Reader YBX1. ACS Chem. Biol. 2024, 19, 1648–1660. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Feng, J.; Zeng, C.; Liu, J.; Fu, Z.; Wang, D.; Wang, Y.; Zhang, L.; Li, J.; Jiang, A.; et al. NSUN2-mediated M5c methylation of IRF3 mRNA negatively regulates type I interferon responses during various viral infections. Emerg. Microbes Infect. 2023, 12, 2178238. [Google Scholar] [CrossRef]
- Li, Z.L.; Xie, Y.; Wang, Y.; Wang, J.; Zhou, X.; Zhang, X.L. NSUN2-mediated HCV RNA m5C Methylation Facilitates Viral RNA Stability and Replication. Genom. Proteom. Bioinform. 2025, 23, qzaf008. [Google Scholar] [CrossRef]
- Courtney, D.G. Post-Transcriptional Regulation of Viral RNA through Epitranscriptional Modification. Cells 2021, 10, 1129. [Google Scholar] [CrossRef] [PubMed]
- Henry, B.A.; Kanarek, J.P.; Kotter, A.; Helm, M.; Lee, N. 5-methylcytosine modification of an Epstein-Barr virus noncoding RNA decreases its stability. RNA 2020, 26, 1038–1048. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; An, K.; Zhai, W.; Feng, L.; Xu, Y.; Sun, R.; Wang, Y.; Yang, Y.G.; Kan, Q.; Tian, X. NSUN2-mediated mRNA m5C Modification Regulates the Progression of Hepatocellular Carcinoma. Genom. Proteom. Bioinform. 2023, 21, 823–833. [Google Scholar] [CrossRef] [PubMed]

| Modifications | RNA Substrates | Typical Location | Key Roles in Viral Infections | References |
|---|---|---|---|---|
| m5C | mRNA, tRNA, rRNA, viral RNA | CDS/UTR | RNA stability, translation efficiency, immune evasion, nuclear export, and splicing | [10,11,12,13,14] |
| m6A | mRNA, lncRNA, circRNA, rRNA, viral RNA | 3′ UTR, near stop codon | RNA stability, translation efficiency, immune evasion, nuclear export, splicing, and secondary structure | [15,16,17,18,19] |
| ac4C | mRNA, tRNA, rRNA, viral RNA | Coding region | RNA stability, translation efficiency, splicing, and secondary structure | [15,20,21,22,23,24,25] |
| Ψ | tRNA, rRNA, viral RNA | Structural regions | RNA stability, translation efficiency, and pre-mRNA processing | [26,27,28,29] |
| m1A | mRNA, tRNA, viral RNA | 5′ UTR and coding regions | RNA stability and splicing | [22,23,30] |
| m7G | mRNA, rRNA, viral RNA | 5′ cap, internal sites | RNA stability, translation efficiency, and ribosome biogenesis | [31,32,33,34] |
| Nm | rRNA, snRNA, mRNA, viral RNA | Ribose 2′-hydroxyl | RNA stability, translation efficiency, and pre-mRNA processing | [35,36,37] |
| Virus | Writer | Notable m5C Sites | Key Functions | References |
|---|---|---|---|---|
| HIV-1 | NSUN2 | Gag-pol overlap, splice sites | Export, translation, splicing regulation | [136,142] |
| HCV | NSUN2 | C7525 (NS5A) | RNA stability, replication, release | [141] |
| EV71 | NSUN2 | 5′ UTR nt 584, CDS nt 1460 | IRES-mediated translation, RNA stability | [11] |
| SARS-CoV-2 | NSUN2 | 3′ UTR, ORF dynamics | RNA turnover, replication regulation | [140] |
| EBV | NSUN2 | EBER1 C145 | ncRNA stability modulation | [143] |
| MLV | NSUN2 | 5′ UTR, 3′ UTR | Genome packaging, RNA stability, immune evasion | [138] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Huang, Y.; Zhang, X.; Guan, W.; Zhang, F.; Hao, H. Epitranscriptomic Regulation of Hepatitis B Virus by RNA 5-Methylcytosine: Functions, Mechanisms, and Therapeutic Potential. Viruses 2025, 17, 1159. https://doi.org/10.3390/v17091159
Zhou X, Huang Y, Zhang X, Guan W, Zhang F, Hao H. Epitranscriptomic Regulation of Hepatitis B Virus by RNA 5-Methylcytosine: Functions, Mechanisms, and Therapeutic Potential. Viruses. 2025; 17(9):1159. https://doi.org/10.3390/v17091159
Chicago/Turabian StyleZhou, Xuliu, Yanling Huang, Xueyan Zhang, Wuxiang Guan, Fang Zhang, and Haojie Hao. 2025. "Epitranscriptomic Regulation of Hepatitis B Virus by RNA 5-Methylcytosine: Functions, Mechanisms, and Therapeutic Potential" Viruses 17, no. 9: 1159. https://doi.org/10.3390/v17091159
APA StyleZhou, X., Huang, Y., Zhang, X., Guan, W., Zhang, F., & Hao, H. (2025). Epitranscriptomic Regulation of Hepatitis B Virus by RNA 5-Methylcytosine: Functions, Mechanisms, and Therapeutic Potential. Viruses, 17(9), 1159. https://doi.org/10.3390/v17091159





