Abstract
Rubella is typically a mild viral illness, but it can lead to severe complications when contracted during pregnancy, such as pregnancy loss or developmental defects in the fetus (congenital rubella syndrome). Therefore, it is crucial to develop and maintain protective immunity in women of childbearing age. In this study, we assessed the transcriptional factors associated with rubella-specific immune outcomes (IgG binding antibody and avidity, neutralizing antibody, and memory B cell ELISpot response) following a third MMR vaccine dose in women of reproductive age to identify key factors/signatures impacting the immune response. We identified baseline (Day 0) and differentially expressed (Day 28–Day 0) genes associated with several RV-specific immune outcomes, including the transferrin receptor 2 (TFR2), which is an important factor regulating iron homeostasis and macrophage functional activity, and a close functional homolog of TFR1, the cellular receptor of the New World hemorrhagic fever arenaviruses. We also identified enriched KEGG pathways, “cell adhesion molecules”, “antigen processing and presentation”, “natural killer cell-mediated cytotoxicity”, and “immune network for IgA production”, relevant to immune response priming and immune activation to be associated with RV-specific immune outcomes. This study provides novel insights into potential biomarkers of rubella-specific immunity in women of childbearing age.
Keywords:
rubella; rubella vaccine; MMR; immunity; humoral immunity; gene expression; NGS; RNA; transcriptome; genetic markers 1. Introduction
Rubella is considered a mild viral illness characterized by fever, maculopapular rash, and lymphadenopathy, with or without joint involvement (arthritis and arthralgia are noted mostly in adult women) [1]. However, the disease in pregnant women frequently results in pregnancy loss or has detrimental consequences for fetal development, resulting in deafness and ocular, cardiac, and other congenital abnormalities, known as congenital rubella syndrome (CRS) [1]. While the global burden of CRS decreased 66% (from 105,000 cases in 2010 to 32,000 cases in 2019) [2,3], if MMR vaccination rates drop below the herd immunity threshold (83–85%) [4], it will raise concerns about the re-emergence of the disease [5]. The tropism of rubella virus is not fully elucidated, but the virus infects and replicates in the nasopharynx/upper respiratory tract, spreads to the regional lymph nodes, and during systemic viremia in pregnant women crosses/infects the placenta and fetal tissues, including brain tissue (CRS) [6]. Of the immune cells, it is established that rubella virus preferentially infects macrophages, neutrophils, the microglia (the brain resident macrophage cells), and possibly monocytes and dendritic cells [6,7], which may contribute to virus dissemination and host innate/inflammatory immune response and influence rubella virus-specific adaptive immunity.
Rubella vaccination is considered effective and confers protection in 95% of vaccinated individuals after a single dose [1]. There is substantial inter-individual variability in immune responses to rubella, even among immunocompetent individuals with one or two documented doses of a rubella-containing vaccine. Those with suboptimal responses are more likely, over time, to experience waning immunity and fall below the threshold of 10 IU/mL, the established cutoff for defining rubella immunity [8,9,10,11]. In a large study comprising a highly immunized population from Olmsted County, MN, and surrounding areas (N = 1393 subjects, 20 to 44 years of age, 80.2% females), we have reported 2.2% seronegative individuals, a relatively small but sizable percentage [8]. Such seroepidemiological data suggest a small but potential risk for infection and pregnancy complications for women of reproductive age upon rubella wild-type virus exposure. Evaluation of the relevant genetic and transcriptional factors and immunological characteristics of rubella vaccine hyporesponsiveness/waning immunity in vulnerable populations (and other populations) is warranted to identify individuals at higher risk of acquiring the disease and unveil the mechanisms, which might result in long-lasting and robust vaccine response [12,13,14,15,16].
Here, we report the results of a longitudinal vaccine study in 98 women of reproductive age, considering their rubella-specific baseline immunity and the change in immune response after a third dose of MMR vaccine. The study subjects were chosen from those with residual rubella-specific antibody titers after prior MMR doses in the top and bottom 30th percentile of the baseline antibody response measured in a larger cohort (n = 1117) from the local community [8]. We conducted longitudinal gene expression profiling before and after administration of a third MMR vaccine dose to elucidate the potential factors and mechanisms that may be associated with and/or mediate changes in rubella immune response following a third vaccination.
2. Materials and Methods
The study cohort, rubella IgG and neutralizing antibody assays, memory B cell ELISpot assay, and RNA extraction methods have been thoroughly described in our previous publications [8,10].
2.1. Study Cohort
The study cohort has been described in detail by Haralambieva et al. (2020) [10]. Our analysis included 98 healthy female participants aged 30.7–40.4 years from Olmsted County, MN, USA and nearby areas, who were enrolled at the Mayo Clinic in Rochester, MN, USA and had documented receipt of two previous MMR vaccine doses. Individuals were selected based on rubella virus (RV)-specific antibody IgG titers measured by enzyme-linked immunosorbent assay (ELISA) from a pool of 1117 serum samples collected through the Mayo Clinic Biobank. The cohort represented those in the top and bottom 30% of the baseline IgG titer distribution. Blood samples were obtained at baseline (prior to vaccination) and on Day 8 and Day 28 following administration of a third MMR dose. Written informed consent was obtained, and all study procedures, including those related to the Mayo Clinic Biobank, were approved by the Mayo Clinic Institutional Review Board (IRB #15-007916).
2.2. Rubella Neutralizing Antibody Assay
Neutralizing antibody titers against RV were measured as previously described [10]. Neutralizing titers (NT50) were reported using the Karber method, representing the highest serum dilution at which the viral signal was reduced by at least 50% across the dilution series [10]. The assay demonstrated an intra-class correlation coefficient (ICC) of 0.89 based on log-transformed NT50 values from repeated measurements [10].
2.3. Memory B Cell ELISpot Assay
The frequency of RV-specific memory-like IgG B cells was measured in peripheral blood mononuclear cells (PBMCs) at baseline and Day 28 after administration of a third MMR vaccine dose. Quantification was performed using the Mabtech ELISpotPLUS kit for human IgG (Mabtech Inc.; Cincinnati, OH, USA), following the manufacturer’s instructions. Prior to analysis, PBMCs/B cells underwent non-specific in vitro pre-stimulation for three days with human recombinant IL-2 and Toll-like receptor (TLR) agonist R848. ELISpot plates were coated with RV antigen (HPV77 RV strain) obtained from Meridian Life Science Inc. (Memphis, TN, USA). The frequency of antigen-specific memory B cells was expressed as spot-forming units (SFUs) per 2 × 105 cells, calculated as the median of quadruplicate RV-specific responses after subtracting subject-specific background values (no-antigen control). The assay showed strong reproducibility, with an average intra-class correlation coefficient (ICC) of 0.88 across replicate measurements.
2.4. Gene Expression
Gene expression in response to a third MMR vaccination dose was assessed in PBMCs collected at baseline and Day 28 post-vaccination. Briefly, mRNA was extracted using the Qiagen RNeasy Plus Mini Kit (Qiagen, Valencia, CA, USA) following cell preservation in RNAProtect (Qiagen, Valencia, CA, USA). All RNA and cDNA samples passed a quality control on an Agilent 2100 Bioanalyzer (Agilent; Palo Alto, CA, USA). Libraries were prepared and sequenced on the Illumina HiSeq 4000 platform (101 bp paired-end reads, 9 samples/lane) at Mayo Clinic’s Advanced Genomics Technology Center. Raw paired-end RNA sequencing reads were processed using the MAP-RSEQ version 3.0 pipeline, aligned to the hg38 reference genome using STAR, and quantified with featureCounts [17,18], using Ensembl v78 annotations.
Gene expression data were processed and normalized as previously described [19]. Briefly, conditional quantile normalization was applied, and only genes classified as protein-coding or annotated as processed pseudogenes were retained for analysis. To filter out low-abundance transcripts, genes were excluded if their coefficient of variation fell below the 25th percentile or if the median difference in expression between RV-stimulated and unstimulated cells was under 16. For the remaining 12,925 transcripts, expression values were normalized by centering on the mean and scaling according to standard deviation.
2.5. Statistical Analysis
Demographic and immune response variables were summarized using medians and the first (Q1) and third (Q3) quartiles.
For analytical purposes, we defined the response to a third dose of MMR as the change in antibody measurement from Day 0 to Day 28 (i.e., the Day 28 value minus the Day 0 value). To explore this relationship between humoral immunity and gene expression, we modeled three response variables: (1) the change in RV-specific IgG titer, (2) the change in neutralizing antibody titer, and (3) the Day 28 memory B cell ELISpot response (estimating RV-specific memory B cell frequencies) at Day 28. Since the ELISpot response is returned as counts, we fit a Poisson regression model, adjusting for batch effects. The residuals from this model were used as the response variable.
Elastic-net linear regression (α = 0.9 with 10-fold cross validation) was used to identify genes associated with each endpoint. Response variables and gene expression values were standardized to allow for within-model comparisons of the regression coefficients. Analysis was conducted using the glmnet package in R version 4.2.2 [20].
Over-representation analysis using KEGG pathways was run using the enrichKEGG function in the R cluster Profiler package [21]. The p-value cutoff was 0.05, and adjusted p-values were calculated using the Benjamini–Hochberg method.
3. Results and Discussion
3.1. Demographic and Immune Response Characterization of Study Subjects
This analysis included 98 females with a median age at enrollment (third MMR dose) of 35.2 years. The median time from the second rubella vaccination to study enrollment was 23.2 years (IQR: 18.7–25.5), as summarized in Appendix A, Table A1. Of the 98 study subjects, 53 were recruited as a low-antibody group and 45 as a high-antibody group according to prior screening of RV-specific IgG antibody titers [8]. Most study participants identified as White/Non-Hispanic or Latino. Demographic and clinical characteristics were balanced between the groups. Immunological outcomes (IgG titer, neutralizing antibody titer, avidity index, and memory B cell frequencies) at baseline and Day 28 post-vaccination have been previously described [10] and are summarized in Appendix A, Table A2. Consistent with other MMR vaccine studies [22,23,24], the subjects with a lower baseline antibody titer responded better to the third MMR vaccine dose, compared to those with higher antibody titer. In our study, the change in RV-specific IgG titers and avidity indexes from baseline to Day 28 was significantly greater in the group with low screening antibody titers (median change in IgG: 95.1 in the low vs. 49.3 in high, p < 0.001; median change in avidity index: 11.2 in the low vs. 5.6 in the high, p < 0.001), whereas in our study the change in neutralizing antibodies and RV-specific memory B cell frequencies did not differ significantly between groups [10].
3.2. The Impact of Baseline Gene Expression on Rubella-Specific Immune Outcomes
We used elastic-net linear regression models to identify baseline and differentially expressed (Day 28–Day 0) genes associated with rubella-specific immune outcomes following a third dose of MMR vaccination. The immune outcome of interest in these analyses was the change (Day 28–Day 0) in the immune response (IgG titer, neutralizing antibody titer, avidity index) after vaccination to account for pre-existing immunity (baseline immunity). The only exception was the RV-specific memory B cell response measured by ELISpot, where the Day 28 measure was included in the model due to the minimal detectable baseline counts.
Interestingly, we observed significant association between many baseline genes (predictors) on the antibody outcomes and/or the Day 28 memory B cell frequencies after vaccination, while the models assessing differentially expressed (Day 28–Day 0) genes produced limited results. We identified 46 baseline genes predictive of IgG response, 35 predictive of neutralizing antibody response, and 19 predictive of Day 28 memory B cell frequency. Baseline predictor genes associated with RV-specific immune outcomes are presented in Table 1, and model coefficients (mc) are illustrated in Figure 1. Notably, nine genes overlapped across at least two of these outcomes, as described in Table 2. No baseline genes were identified as being associated with changes in avidity index.

Table 1.
Elastic-net model results for the effect of baseline gene expression on rubella-specific immune outcomes after MMR vaccination. This table shows baseline genes associated with rubella virus-specific change (Day 28–Day 0) in (A) IgG antibody titer, (B) neutralizing antibody titer (reported as Karber NT), or (C) Day 28 RV-specific memory B cell frequencies (measured in memory B cell ELISpot) following a third dose of MMR vaccine that remained in the model.
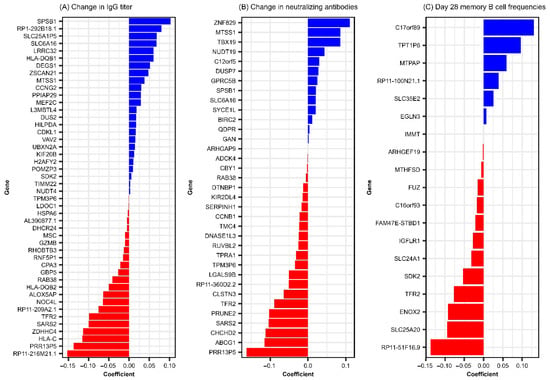
Figure 1.
Elastic net linear model coefficients for baseline genes predictive of rubella-specific humoral response following MMR vaccination. This figure illustrates linear regression coefficients results from GLMNET modeling and summarizes the relative contribution of genes to RV-specific immune outcomes. (A) baseline genes predictive of change (Day 28–Day 0) in rubella IgG titer. (B) baseline genes predictive of change (Day 28–Day 0) in neutralizing antibodies. (C) Baseline genes predictive of Day 28 rubella-specific memory B cell frequencies.

Table 2.
Genes predictive of two or more outcomes.
Of the identified predictors, we consider TFR2 (transferrin receptor 2) to be the most consistent finding, since it emerged as a baseline predictor of rubella-specific immune response outcomes across both antibody immune response (RV-specific IgG titers, mc = −0.099 and neutralizing antibody titers, mc = −0.089, Figure 1 and Table 1) and rubella virus-specific memory B cell ELISPOT response (mc = −0.077, Figure 1 and Table 1). Interestingly, TFR2 was also identified in the model of (Day 28–Day 0) gene expression, exhibiting a negative association with Day 28 memory B cell frequencies, albeit with a smaller effect (mc = 0.0031) on the immune outcome. TFR2 encodes a protein involved in iron homeostasis, erythrocyte differentiation, and inflammatory response. It is preferentially expressed in the liver, muscle, lung, spleen, bone marrow, erythroid progenitors, neurons, microglia, and macrophages, regulating their polarization and functional activity (including cytokine and chemotactic factor production, reactive oxygen species/ROS production, antigen presentation, Toll-like receptor, and interferon signaling) [25,26]. Importantly, TFR2 has close structural similarity and is functionally related to TFR1. TFR1 has been identified as a bona fide cellular receptor, mediating the binding and entry of New World arenaviruses linked to hemorrhagic fever [27]. It is also suggested to facilitate the entry of other viruses, including influenza A, hepatitis C, and rabies [28,29,30]. Given this background, it is tempting to speculate that TFR2 may play a role in rubella virus binding and entry, potentially influencing viral cell entry, replication, antigen abundance, and immune response priming. We recognize that this is a working hypothesis that will require experimental validation. Our results point to a negative association between baseline gene expression and vaccine-induced rubella immune outcomes. This is counterintuitive but can be explained by the viral direct targeting of immune cells (e.g., macrophages, neutrophils, DCs) [6,7], which may directly impact innate/inflammatory response and skew/suppress cell immune function (e.g., antigen presentation, cytokine production, adaptive immune response priming), leading to blunting of adaptive immune response [26,31]. It has been demonstrated that lower CCR5 expression on CD4+ T cells is associated with improved immune responses to HIV [32]. This hypothesis is also supported by evidence that conserved linear structural motifs in viral proteins of different viruses can enable receptor binding and exploitation of host cellular machinery. For example, rubella virus E1 has been reported to contain epitope regions that align with those in measles virus [33,34,35], and its main antigen/target of neutralizing antibody response has structural similarity with the class II fusion proteins of alphaviruses, flaviviruses, and paleoviruses, despite minimal sequence homology [36,37]. These structural parallels suggest that shared binding motifs across unrelated viruses may enable attachment to similar cellular receptors, such as those in the transferrin receptor family, and promote viral entry. Further investigation and in-depth functional studies are necessary to reveal the role of TFR2 in regulating rubella virus-specific humoral outcomes.
Among the 46 baseline genes predictive of change (Day 28–Day 0) in rubella IgG titer following a third MMR dose, 8 (MTSS1, RAB38, PRR13P5, SARS2, SPSB1, TPM3P6, SLC6A16, and TFR2) were also predictive of change in neutralizing antibody titer, manifesting positive (MTSS1, SPSB1, SLC6A16) or negative (RAB38, PRR13P5, SARS2, TMP3P6, TFR2) associations with immune outcome (Table 2) Moreover, these genes exhibited consistent coefficient directions across both models, as demonstrated in Table 2 and Figure 1A,B, suggesting shared transcriptional determinants of binding and functional/neutralizing antibody responses and highlighting potential baseline biomarkers of rubella antibody responsiveness following a third dose of MMR. Interestingly, a recent study analyzing RNA sequencing data from identical twins discordant for autism spectrum disorder identified PRR13P5 as one of the differentially expressed genes and found enrichment in pathways related to immune cell signaling and immune response, suggesting a potential link between this gene’s expression and humoral immunity function [38]. Due to the limited understanding of PRR13P5 (a gene negatively associated with rubella virus-specific humoral immunity in our study) function, further research is necessary to elucidate its role in immune regulation and potential impact on humoral response.
We performed KEGG pathway over-enrichment analysis to identify relevant pathways and immune function-related cellular activities impacting the RV-specific antibody response following MMR vaccination. This analysis revealed 12 pathways that were significantly enriched and associated with antibody titer, illustrated in Figure 2. Four enriched pathways, “antigen processing and presentation,” “natural killer cell-mediated cytotoxicity,” “intestinal immune network for IgA production,” and, importantly, “cell adhesion molecules,” reflected pertinent biological processes/immune activity related to the viral attachment/cell entry and initial priming of immune response upon vaccination.
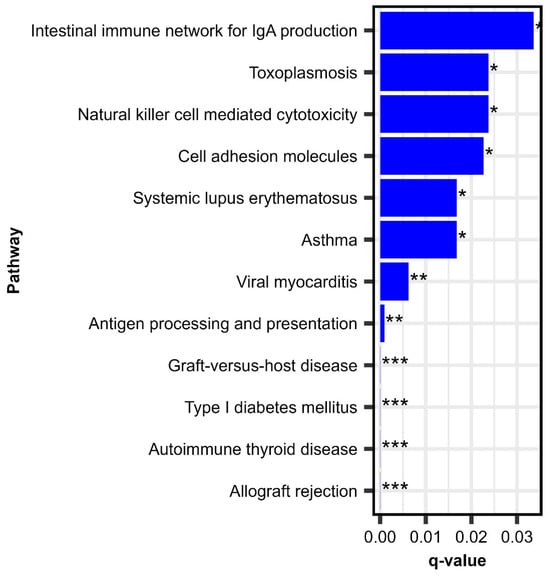
Figure 2.
Enriched pathways of baseline gene expression associated with change (Day 28–Day 0) in rubella IgG antibody titer. This figure depicts q-values from KEGG pathway enrichment analysis on baseline genes associated with change (Day 28–Day 0) in RV-specific IgG antibody titer following a third dose MMR. * Adjusted p-value < 0.05; ** adjusted p-value < 0.01; *** adjusted p-value < 0.001.
Of these, “antigen processing and presentation” is fundamental to the activation of CD4+ and CD8+ T cells, key factors in adaptive immunity and B cell help [39]. Simultaneously, natural killer (NK) cell-mediated cytotoxicity represents a crucial early defense, targeting infected cells and producing cytokines such as IFN-γ, which augment T cell activation/function [40]. Cell adhesion molecules can mediate attachment of viruses and facilitate the migration and interaction of immune cells, ensuring efficient trafficking to lymphoid tissues and stable contact between T cells and antigen-presenting cells [41]. Finally, the intestinal immune network for IgA production reflects mucosal immune activation and/or immunoglobulin production in general, potentially contributing to viral neutralization and the prevention of viral dissemination [42]. Collectively, these pathways highlight the coordinated activation of innate and adaptive immunity in response to MMR vaccination, underlining the biological mechanisms that drive effective priming and long-term protection against rubella.
It is interesting to compare our rubella findings with the findings from other vaccine and/or transcriptional studies, in particular with measles and mumps. Comparative gene expression studies of dendritic cells infected with MV vs. other pathogens (but not rubella virus) have demonstrated an MV-specific pronounced effect on the regulation of antigen presentation and innate antiviral immunity [43], consistent with our rubella study findings. Transcriptomic studies in measles vaccine recipients have emphasized the importance of plasma cell survival factors (e.g., CD93 expression), chemokine and cytokine activity, cell adhesion, and cell migration for neutralizing antibody response to vaccination [44] and have identified early B cell transcriptomic signatures (IL20RB, PMAIP1, BEX2, FAIM, and IL16 contributing to the selection of high-affinity B cells and the control of apoptotic pathways) that impact MV-specific antibody response after MMR vaccination [45]. Interestingly, studies exploring the influence of host genetic factors on immune response to measles and mumps vaccine have highlighted the critical roles of polymorphisms/genes linked to viral entry and innate/inflammatory pathways in governing antiviral immunity, as in our rubella vaccine study. A genome-wide association study demonstrated that polymorphisms in the measles virus receptor-encoding CD46 gene and in IFI44L contribute to inter-individual differences in neutralizing antibody response following live measles vaccination [46]. Several studies have established that variance in the 19q13 genomic region (including the glycosyltransferase FUT2 gene involved in host glycosylation and the sialic acid-recognizing receptors SIGLEC5/SIGLEC14) may influence mumps virus susceptibility/entry and contribute to cellular and inflammatory responses following mumps vaccination [47,48,49]. Thus, research on measles and mumps (both members of the Paramyxoviridae family) has identified both overlapping and distinct factors, genes, biological processes, and pathways regulating antiviral immunity in comparison to our findings with rubella.
3.3. The Impact of (Day 28–Day 0) Gene Expression Change on Rubella-Specific Immune Outcomes
These modeling efforts identified 18 differentially expressed (Day 28–Day 0) genes associated with Day 28 memory B cell ELISpot frequencies (RV-specific), depicted in Table 3 and Figure 3.

Table 3.
Elastic-net model results for the effect of (Day 28–Day 0) gene expression associated with Day 28 rubella virus-specific memory B cell frequencies. This table shows (Day 28–Day 0) genes associated with Day 28 rubella virus-specific memory B cell frequencies (measured in memory B cell ELISpot) following a third dose of MMR vaccine.
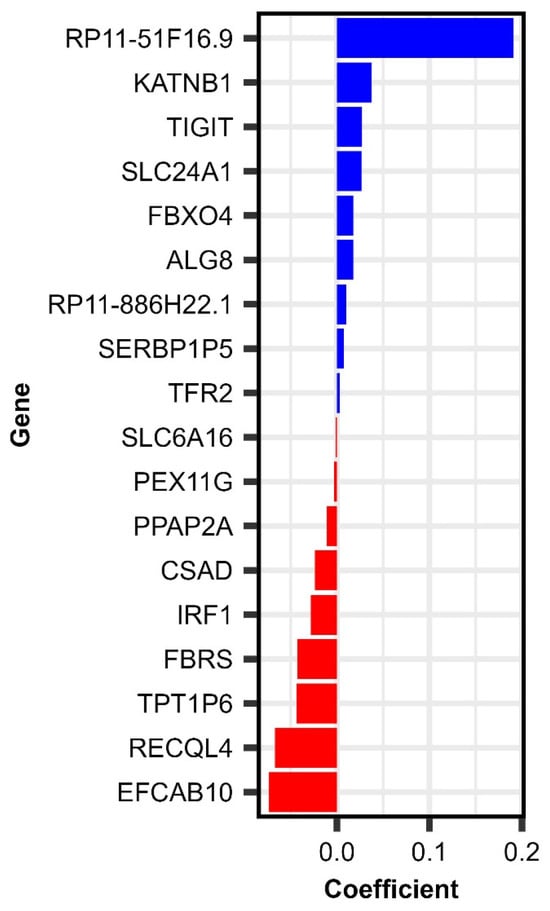
Figure 3.
Elastic net linear model coefficients for (Day 28–Day 0) genes associated with rubella-specific Day 28 memory B cell frequencies following MMR vaccination.
Summarizing our results from the (Day 28–Day 0) gene expression and baseline gene expression modeling by relevance, we identified four genes (TFR2, SLC24A1, TPT1P6, and RP11-51F16.9) consistently impacting several rubella-specific immune outcomes following vaccination, including the memory B cell frequencies, which are crucial for recall immune response upon subsequent viral exposure [50]. These genes, including TFR2, may serve as transcriptional markers of recall immune response, with potential implications for long-term humoral immunity and RV-specific memory B cell frequencies following MMR vaccination.
SLC24A1 expression (Day 28–Day 0) change was positively associated with the immune outcome (mc = 0.0265, Table 3). This gene encodes a sodium/potassium/calcium exchanger (NCKX1), known primarily for its role in retinal photoreceptor function and calcium homeostasis. This gene has been reported as INF-stimulated in bats [51], highlighting its potential involvement in antiviral responses. While its specific function in immune cells remains to be fully elucidated, the induction of SLC24A1 expression in response to interferon signaling indicates that it may participate in the modulation of immune responses, including those elicited by rubella vaccination. Perhaps its involvement in calcium homeostasis may influence B cell response, since calcium flux is crucial for B cell activation and differentiation. This highlights SLC24A1 as a potentially significant biomarker of immune response, requiring further investigation into its role and effects in human immunity.
TPT1P6 expression (Day 28–Day 0) change exhibited a negative association with the immune outcome (mc = −0.0435, Table 3). TPT1P6 is a pseudogene (non-coding RNA) related to the translationally controlled tumor protein (TPT1, also known as TCTP). TPT1/TCTP has demonstrated cytokine-like functions in humans and enhancement of B cell proliferation in mouse models, suggesting it may influence B cell functions and response to stimuli [52,53]. Although pseudogenes like TPT1P6 do not encode proteins, they may regulate gene expression post-transcriptionally, and TPT1P6 could, therefore, potentially serve as a modulator of processes/pathways critical to B cell fate in the context of vaccination.
RP11-51F16.9 expression (Day 28–Day 0) change exhibited a strong positive association with the immune outcome (mc = 0.1909, Table 3). It is a long non-coding RNA (lncRNA) that remains uncharacterized in the literature. Given the increasing recognition of lncRNAs in fine-tuning adaptive immune responses [54,55], the consistent association of RP11-51F16.9 with rubella-specific memory B cell outcomes suggests it may be involved in transcriptional programs supporting the establishment of immunological memory.
Together, these genes play diverse roles, such as ionic regulation (SLC24A1), iron metabolism and viral entry (TFR2), and non-coding RNA-mediated gene expression control (TPT1P6 and RP11-51F16.9), including potential modulation of immune responses (TPT1P6). This suggests that effective vaccine responses depend on a complex interplay of systems-level cellular regulation. The fact that these genes were significant at baseline and differentially expressed (for the Day 28–Day 0 gene expression modeling with memory B cell ELISpot response) following vaccination may indicate that pre-vaccination transcriptional profiles could serve as biomarkers for the subsequent RV-specific memory B cell response. Further investigation is required to elucidate the functional roles of these genes in B cell memory response, including whether their expression is directly or indirectly involved in B cell differentiation after antigen exposure.
3.4. Strengths and Limitations
The use of elastic-net linear regression enabled us to effectively handle such a high-dimensional dataset with multicollinearity-dependent data. The elastic-net penalty allowed for the identification of key predictor genes while avoiding overfitting by shrinking the coefficients, thus increasing the likelihood of identifying relevant genes when we have more predictors than samples.
While this study provides novel insights into transcriptional correlates of RV-specific humoral immunity following a third MMR dose, further functional characterization is needed. Future work should focus on validating the roles of candidate genes such as TFR2, SLC24A1, TPT1P6, and RP11-51F16.9, using in vitro models (i.e., siRNA knockdown or CRISPR-Cas9 approaches) and in vivo systems to confirm their contribution to B cell function and antibody responses.
A limitation of our study is the use of peripheral blood mononuclear cells (PBMCs), which represent a heterogeneous mixture of immune cell types, including B cells, T cells, monocytes, and natural killer cells. This cellular diversity may dilute transcriptomic signals and hinder the ability to attribute the identified gene expression signatures to specific cell populations. More specifically, PBMC heterogeneity may obscure cell-specific signals by averaging gene expression across multiple lineages, making it difficult to determine whether observed associations are primarily driven by B cells, T cells, or other subsets. Future studies employing single-cell transcriptomics or cell-type-specific gene expression, along with functional studies, could help identify the immune subsets responsible for the observed responses and refine the transcriptional factors and their associated specific cellular pathways critically modulating rubella vaccine-specific immunity.
4. Conclusions
This study provides new insights into transcriptional predictors of rubella-specific humoral immunity after a third MMR vaccine dose in women of childbearing age. We identified both baseline genes associated with key adaptive immune outcomes (IgG titer, neutralizing antibodies, and Day 28 memory B cell frequencies) and Day 28–Day 0 genes associated with Day 28 memory B cell frequencies. While the precise functional roles and mechanisms of action for each specific gene/factor remain to be elucidated, our findings suggest that transcriptional landscape at baseline (involved in cell adhesion, antigen processing and presentation, natural killer cell-mediated cytotoxicity, immunoglobulin production, and immune regulation) can shape immune responses to a third dose of MMR. Notably, transferrin receptor 2 (TFR2), a regulator of iron homeostasis and macrophage function, emerged as key determinant associated with several RV-specific immune outcomes, supporting its potential relevance in modulating rubella vaccine-induced immunity. Given its structural and functional similarity to other viral receptors, we speculate that TFR2 may be involved in rubella virus binding and/or cell entry. Overall, this work underscores the potential for identifying transcriptional biomarkers and lays the groundwork for future investigations to inform personalized vaccination strategies, optimize booster schedules, and enhance vaccine efficiency in populations with variable immune responsiveness.
Author Contributions
Conceptualization, R.B.K., G.A.P., L.I.T., I.H.H.; methodology, I.H.H., K.M.G., D.E.G. and R.B.K.; software, K.M.G. and D.E.G.; validation, I.H.H., R.B.K., K.M.G. and D.E.G.; formal analysis, K.M.G. and D.E.G.; investigation, I.H.H., I.G.O. and R.B.K.; resources, G.A.P. and R.B.K.; data curation, K.M.G. and D.E.G.; writing—original draft preparation, L.I.T. and I.H.H.; writing—review and editing, L.I.T., I.H.H., I.G.O., K.M.G., D.E.G., G.A.P. and R.B.K.; visualization, L.I.T.; supervision, I.G.O., G.A.P. and R.B.K.; project administration, I.G.O., G.A.P. and R.B.K.; funding acquisition, G.A.P. and R.B.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health, grant numbers R37AI048793, R01AI033144, and R01AI138965. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Mayo Clinic Institutional Review Board (IRB #15-007916, 15 August 2016).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets presented in this article are not readily available because the data are part of an ongoing study. Requests to access the datasets should be directed to Richard B. Kennedy (kennedy.rick@mayo.edu).
Acknowledgments
We thank Katherine G. Eberhard and Marguerite Riggenbach for their technical assistance in preparing the samples for NGS. Research reported in this review was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R37AI048793, R01AI033144, and R01AI138965. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflicts of Interest
The authors declare the following financial interests/personal relationships, which may be considered as potential competing interests: Poland is the chair of a Safety Evaluation Committee for novel non-rubella investigational vaccine trials being conducted by Merck Research Laboratories. Poland provides consultative advice to AiZtech; GlaxoSmithKline; Merck & Co., Inc.; Moderna; and Syneos Health. Poland and Ovsyannikova hold patents related to vaccinia and measles peptide vaccines. Kennedy, Poland, and Ovsyannikova hold a patent related to vaccinia peptide vaccines. Poland, Kennedy, and Ovsyannikova have received grant funding and royalties from ICW Ventures for pre-clinical studies on a peptide-based COVID-19 vaccine. Poland, Kennedy, Ovsyannikova and Haralambieva hold a patent related to the impact of single nucleotide polymorphisms on measles vaccine immunity. Kennedy has received funding from Merck Research Laboratories to study waning immunity to mumps vaccine. Kennedy also offers consultative advice on vaccine development to Merck & Co., Inc. and Sanofi Pasteur. These activities have been reviewed by the Mayo Clinic Conflict of Interest Review Board and are conducted in compliance with Mayo Clinic Conflict of Interest policies. All other authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| CRS | Congenital rubella syndrome |
| ELISA | Enzyme-linked immunosorbent assay |
| ICC | Intra-class correlation coefficient |
| IgG | Immunoglobulin G |
| lncRNA | Long non-coding RNA |
| mc | Model coefficient |
| mRNA | Messenger ribonucleic acid |
| NGS | Next-generation sequencing |
| PBMCs | Peripheral blood mononuclear cells |
| Q1 | First quartile (25th percentile) |
| Q3 | Third quartile (75th percentile) |
| RNA | Ribonucleic acid |
| RV | Rubella virus |
| SFUs | Spot forming units |
| TLR | Toll-like receptor |
Appendix A
Demographics and Immune Response Characterization of the Study Subjects

Table A1.
Demographic and screening of the study subjects.
Table A1.
Demographic and screening of the study subjects.
| Low (n = 53) | High (n = 45) | Total (n = 98) | |
|---|---|---|---|
| Sex | |||
| Female | 53 (100.0%) | 45 (100.0%) | 98 (100.0%) |
| Race | |||
| Asian | 0 (0.0%) | 1 (2.2%) | 1 (1.0%) |
| White | 53 (100.0%) | 44 (97.8%) | 97 (99.0%) |
| Ethnicity | |||
| Non-Hispanic nor Latino | 51 (96.2%) | 44 (97.8%) | 95 (96.9%) |
| Hispanic or Latino | 2 (3.8%) | 1 (2.2%) | 3 (3.1%) |
| Age at enrollment/3rd MMR vaccine dose (years) | |||
| Median | 35.7 | 33.9 | 35.2 |
| Q1, Q3 | 31.4, 40.3 | 30.4, 40.9 | 30.7, 40.4 |
| Range | 21.5–44.9 | 22.6–45.1 | 21.5–45.1 |
| Age at 1st rubella vaccination (months) | |||
| Median | 15.8 | 15.4 | 15.7 |
| Q1, Q3 | 15.1, 18.0 | 14.9, 16.0 | 15.0, 17.0 |
| Range | 11.9–352.9 | 4.6–312.3 | 4.6–352.9 |
| Age at 2nd rubella vaccination (years) | |||
| Median | 12.2 | 12.4 | 12.2 |
| Q1, Q3 | 9.8, 16.6 | 11.0, 17.2 | 10.1, 17.0 |
| Range | 4.3–35.1 | 1.5–28.3 | 1.5–35.1 |
| Time from 2nd rubella vaccination to enrollment/3rd MMR vaccine dose (years) | |||
| Median | 23.2 | 22.7 | 23.2 |
| Q1, Q3 | 19.9, 25.1 | 18.0, 26.3 | 18.7, 25.5 |
| Range | 1.9–39.0 | 3.9–40.6 | 1.9–40.6 |
| Prior rubella ELISA Ab titers 1 | |||
| Median | 0.2 | 1.3 | 0.3 |
| Q1, Q3 | 0.1, 0.3 | 1.1, 1.6 | 0.2, 1.2 |
| Range | 0.1–0.3 | 0.8–3.4 | 0.1–3.4 |
Bold text indicates the heading for each demographic or clinical variable. 1 Prior rubella antibody titers were measured by ELISA and are expressed as optical density units.

Table A2.
Immune response characterization of the study subjects.
Table A2.
Immune response characterization of the study subjects.
| Low (n = 53) | High (n = 45) | Total (n = 98) | |
|---|---|---|---|
| Day 28–Day 0 Karber NT50 1 | |||
| N-Miss | 1 | 3 | 4 |
| Median | 153.9 | 122.8 | 136.8 |
| Q1, Q3 | 75.1, 269.3 | 68.9, 216.5 | 70.7, 246.1 |
| Day 28–Day 0 rubella IgG titer (IU/mL) | |||
| N-Miss | 1 | 3 | 4 |
| Median | 95.1 | 49.3 | 66.0 |
| Q1, Q3 | 46.8, 176.7 | 24.7, 84.2 | 36.3, 123.9 |
| Day 28-Day 0 avidity index (%) | |||
| N-Miss | 4 | 3 | 7 |
| Median | 11.2 | 5.6 | 7.9 |
| Q1, Q3 | 6.2, 16.4 | 2.6, 8.1 | 4.2, 13.6 |
| Day 28 memory B cell ELISpot (SFUs/200,000 cells/PBMCs) 2 | |||
| N-Miss | 10 | 12 | 22 |
| Median | 27.5 | 40.0 | 29.8 |
| Q1, Q3 | 13.0, 43.8 | 18.5, 66.5 | 16.0, 50.6 |
Bold text indicates the heading for each demographic or clinical variable. 1 The neutralization titer represented the highest dilution at which the input virus signal was reduced by at least 50% within the dilution series (NT50). 2 ELISpot results are expressed in spot forming units (SFUs) per 200,000 peripheral blood mononuclear cells (PBMCs).
References
- Plotkin, S.A.; Orenstein, W.; Offit, P.A. Vaccines, 6th ed; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Vynnycky, E.; Adams, E.J.; Cutts, F.T.; Reef, S.E.; Navar, A.M.; Simons, E.; Yoshida, L.-M.; Brown, D.W.J.; Jackson, C.; Strebel, P.M.; et al. Using Seroprevalence and Immunisation Coverage Data to Estimate the Global Burden of Congenital Rubella Syndrome, 1996–2010: A Systematic Review. PLoS ONE 2016, 11, e0149160. [Google Scholar] [CrossRef]
- Vynnycky, E.; Knapp, J.K.; Papadopoulos, T.; Cutts, F.T.; Hachiya, M.; Miyano, S.; Reef, S.E. Estimates of the global burden of Congenital Rubella Syndrome, 1996–2019. Int. J. Infect. Dis. 2023, 137, 49–156. [Google Scholar] [CrossRef]
- Otani, N.; Shima, M.; Ueda, T.; Nakajima, K.; Takesue, Y.; Yamamoto, T.; Okuno, T. Changes in the Epidemiology of Rubella: The Influence of Vaccine-Introducing Methods and COVID-19. Vaccines 2023, 11, 1358. [Google Scholar] [CrossRef]
- Rey-Benito, G.; Pastor, D.; Whittembury, A.; Durón, R.; Pacis-Tirso, C.; Bravo-Alcántara, P.; Ortiz, C.; Andrus, J. Sustaining the Elimination of Measles, Rubella and Congenital Rubella Syndrome in the Americas, 2019–2023: From Challenges to Opportunities. Vaccines 2024, 12, 690. [Google Scholar] [CrossRef]
- Popova, G.; Retallack, H.; Kim, C.N.; Wang, A.; Shin, D.; DeRisi, J.L.; Nowakowski, T. Rubella virus tropism and single-cell responses in human primary tissue and microglia-containing organoids. eLife 2023, 12, RP87696. [Google Scholar] [CrossRef]
- Perelygina, L.; Faisthalab, R.; Abernathy, E.; Chen, M.-H.; Hao, L.; Bercovitch, L.; Bayer, D.K.; Noroski, L.M.; Lam, M.T.; Cicalese, M.P.; et al. Rubella Virus Infected Macrophages and Neutrophils Define Patterns of Granulomatous Inflammation in Inborn and Acquired Errors of Immunity. Front. Immunol. 2021, 12, 796065. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.N.; Haralambieva, I.H.; Grill, D.E.; Ovsyannikova, I.G.; Kennedy, R.B.; Poland, G.A. Seroprevalence and durability of rubella virus antibodies in a highly immunized population. Vaccine 2019, 37, 3876–3882. [Google Scholar] [CrossRef]
- Crooke, S.N.; Riggenbach, M.M.; Ovsyannikova, I.G.; Warner, N.D.; Chen, M.-H.; Hao, L.; Icenogle, J.P.; Poland, G.A.; Kennedy, R.B. Durability of humoral immune responses to rubella following MMR vaccination. Vaccine 2020, 38, 8185–8193. [Google Scholar] [CrossRef] [PubMed]
- Haralambieva, I.H.; Ovsyannikova, I.G.; Kennedy, R.B.; Goergen, K.M.; Grill, D.E.; Chen, M.-H.; Hao, L.; Icenogle, J.; Poland, G.A. Rubella virus-specific humoral immune responses and their interrelationships before and after a third dose of measles-mumps-rubella vaccine in women of childbearing age. Vaccine 2020, 38, 1249–1257. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention. Serology Testing for Rubella. Rubella (German Measles, Three-Day Measles) 2024. 10 June 2024. Available online: https://www.cdc.gov/rubella/php/laboratories/serology-testing.html (accessed on 13 June 2025).
- Lambert, N.; Strebel, W.; Orenstein, J.; Icenogle; Poland, G.A. Rubella. Lancet 2015, 385, 2297–2307. [Google Scholar] [CrossRef] [PubMed]
- Lambert, N.D.; Haralambieva, I.H.; Kennedy, R.B.; Ovsyannikova, I.G.; Pankratz, V.S.; Poland, G.A. Polymorphisms in HLA-DPB1 are associated with differences in rubella virus-specific humoral immunity after vaccination. J. Infect. Dis. 2015, 211, 898–905. [Google Scholar] [CrossRef]
- Haralambieva, I.H.; Lambert, N.D.; Ovsyannikova, I.G.; Kennedy, R.B.; Larrabee, B.R.; Pankratz, V.S.; Poland, G.A.; Kimman, T. Associations between single nucleotide polymorphisms in cellular viral receptors and attachment factor-related genes and humoral immunity to rubella vaccination. PLoS ONE 2014, 9, e99997. [Google Scholar] [CrossRef]
- Kennedy, R.B.; Ovsyannikova, I.G.; Haralambieva, I.H.; Lambert, N.D.; Pankratz, V.S.; Poland, G.A. Genetic polymorphisms associated with rubella virus-specific cellular immunity following MMR vaccination. Hum. Genet. 2014, 133, 1407–1417. [Google Scholar] [CrossRef]
- Kennedy, R.B.; IOvsyannikova, G.; Haralambieva, I.H.; Lambert, N.D.; Pankratz, V.S.; Poland, G.A. Genome-wide SNP associations with rubella-specific cytokine responses in measles-mumps-rubella vaccine recipients. Immunogenetics 2014, 66, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Hansen, K.D.; Irizarry, R.A.; Wu, Z. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics 2012, 13, 204–216. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Xu, S.; Hu, E.; Cai, Y.; Xie, Z.; Luo, X.; Zhan, L.; Tang, W.; Wang, Q.; Liu, B.; Wang, R.; et al. Using clusterProfiler to characterize multiomics data. Nat. Protoc. 2024, 19, 3292–3320. [Google Scholar] [CrossRef]
- Fiebelkorn, A.P.; Coleman, L.A.; Belongia, E.A.; Freeman, S.K.; York, D.; Bi, D.; Zhang, C.; Ngo, L.; Rubin, S. Mumps antibody response in young adults after a third dose of measles-mumps-rubella vaccine. Open Forum Infect. Dis. 2014, 1, ofu094. [Google Scholar] [CrossRef]
- Kaaijk, P.; Wijmenga-Monsuur, A.J.; Hulscher, H.I.T.; Kerkhof, J.; Smits, G.; Nicolaie, M.A.; van Houten, M.A.; van Binnendijk, R.S. Antibody Levels at 3-Years Follow-Up of a Third Dose of Measles-Mumps-Rubella Vaccine in Young Adults. Vaccines 2022, 10, 132. [Google Scholar] [CrossRef]
- Fiebelkorn, A.P.; Coleman, L.A.; Belongia, E.A.; Freeman, S.K.; York, D.; Bi, D.; Kulkarni, A.; Audet, S.; Mercader, S.; McGrew, M.; et al. Measles Virus Neutralizing Antibody Response, Cell-Mediated Immunity, and Immunoglobulin G Antibody Avidity Before and After Receipt of a Third Dose of Measles, Mumps, and Rubella Vaccine in Young Adults. J. Infect. Dis. 2016, 213, 1115–1123. [Google Scholar] [CrossRef]
- Cutolo, M.; Campitiello, R.; Gotelli, E.; Soldano, S. The Role of M1/M2 Macrophage Polarization in Rheumatoid Arthritis Synovitis. Front. Immunol. 2022, 13, 867260. [Google Scholar] [CrossRef]
- Ledesma-Colunga, M.G.; Baschant, U.; Weidner, H.; Alves, T.C.; Mirtschink, P.; Hofbauer, L.C.; Rauner, M. Transferrin receptor 2 deficiency promotes macrophage polarization and inflammatory arthritis. Redox. Biol. 2023, 60, 102616. [Google Scholar] [CrossRef] [PubMed]
- Radoshitzky, S.R.; Abraham, J.; Spiropoulou, C.F.; Kuhn, J.H.; Nguyen, D.; Li, W.; Nagel, J.; Schmidt, P.J.; Nunberg, J.H.; Andrews, N.C.; et al. Transferrin receptor 1 is a cellular receptor for New World haemorrhagic fever arenaviruses. Nature 2007, 446, 92–96. [Google Scholar] [CrossRef]
- Mazel-Sanchez, B.; Niu, C.; Williams, N.; Bachmann, M.; Choltus, H.; Silva, F.; Serre-Beinier, V.; Karenovics, W.; Iwaszkiewicz, J.; Zoete, V.; et al. Influenza A virus exploits transferrin receptor recycling to enter host cells. Proc. Natl. Acad. Sci. USA 2023, 120, e2214936120. [Google Scholar] [CrossRef]
- Wang, X.; Wen, Z.; Cao, H.; Luo, J.; Shuai, L.; Wang, C.; Ge, J.; Wang, X.; Bu, Z.; Wang, J.; et al. Transferrin Receptor Protein 1 Is an Entry Factor for Rabies Virus. J. Virol. 2023, 97, e0161222. [Google Scholar] [CrossRef]
- Martin, D.N.; Uprichard, S.L. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc. Natl. Acad. Sci. USA 2013, 110, 10777–11782. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Qian, C.; Wang, X.; Qian, Z.M. Transferrin receptors. Exp. Mol. Med. 2025, 57, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Claireaux, M.; Robinot, R.; Kervevan, J.; Patgaonkar, M.; Staropoli, I.; Brelot, A.; Nouël, A.; Gellenoncourt, S.; Tang, X.; Héry, M.; et al. Low CCR5 expression protects HIV-specific CD4+ T cells of elite controllers from viral entry. Nat. Commun. 2022, 13, 521. [Google Scholar] [CrossRef]
- Munoz-Alia, M.A.; Nace, R.A.; Zhang, L.; Russell, S.J. Serotypic evolution of measles virus is constrained by multiple co-dominant B cell epitopes on its surface glycoproteins. Cell Rep. Med. 2021, 2, 100225. [Google Scholar] [CrossRef] [PubMed]
- Ho-Terry, L.; Terry, G.M.; Cohen, A.; Londesborough, P. Immunological characterisation of the rubella E 1 glycoprotein. Brief report. Arch. Virol. 1986, 90, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Chaye, H.; Ou, D.; Chong, P.; Gillam, S. Human T- and B-cell epitopes of E1 glycoprotein of rubella virus. J. Clin. Immunol. 1993, 13, 93–100. [Google Scholar] [CrossRef]
- Dube, M.; Etienne, L.; Fels, M.; Kielian, M. Calcium-Dependent Rubella Virus Fusion Occurs in Early Endosomes. J. Virol. 2016, 90, 6303–6313. [Google Scholar] [CrossRef]
- DuBois, R.M.; Vaney, M.-C.; Tortorici, M.A.; Al Kurdi, R.; Barba-Spaeth, G.; Krey, T.; Rey, F.A. Functional and evolutionary insight from the crystal structure of rubella virus protein E1. Nature 2013, 493, 552–556. [Google Scholar] [CrossRef]
- Saffari, A.; Arno, M.; Nasser, E.; Ronald, A.; Wong, C.C.Y.; Schalkwyk, L.C.; Mill, J.; Dudbridge, F.; Meaburn, E.L. RNA sequencing of identical twins discordant for autism reveals blood-based signatures implicating immune and transcriptional dysregulation. Mol. Autism. 2019, 10, 38. [Google Scholar] [CrossRef]
- Pishesha, N.; Harmand, T.J.; Ploegh, H.L. A guide to antigen processing and presentation. Nat. Rev. Immunol. 2022, 22, 751–764. [Google Scholar] [CrossRef]
- Paolini, R.; Bernardini, G.; Molfetta, R.; Santoni, A. NK cells and interferons. Cytokine Growth Factor Rev. 2015, 26, 113–120. [Google Scholar] [CrossRef]
- Mackay, C.R.; Imhof, B.A. Cell adhesion in the immune system. Immunol. Today 1993, 14, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Lamm, M.E.; Nedrud, J.G.; Kaetzel, C.S.; Mazanec, M.B. IgA and mucosal defense. APMIS 1995, 103, 241–246. [Google Scholar] [CrossRef]
- Zilliox, M.J.; Parmigiani, G.; Griffin, D.E. Gene expression patterns in dendritic cells infected with measles virus compared with other pathogens. Proc. Natl. Acad. Sci. USA 2006, 103, 3363–3368. [Google Scholar] [CrossRef] [PubMed]
- Haralambieva, I.H.; Zimmermann, M.T.; Ovsyannikova, I.G.; Grill, D.E.; Oberg, A.L.; Kennedy, R.B.; Poland, G.A.; Tregoning, J.S. Whole Transcriptome Profiling Identifies CD93 and Other Plasma Cell Survival Factor Genes Associated with Measles-Specific Antibody Response after Vaccination. PLoS ONE 2016, 11, e0160970. [Google Scholar] [CrossRef]
- Haralambieva, I.H.; Chen, J.; Quach, H.Q.; Ratishvili, T.; Warner, N.D.; Ovsyannikova, I.G.; Poland, G.A.; Kennedy, R.B. Early B cell transcriptomic markers of measles-specific humoral immunity following a 3(rd) dose of MMR vaccine. Front. Immunol. 2024, 15, 1358477. [Google Scholar] [CrossRef]
- Haralambieva, I.H.; Ovsyannikova, I.G.; Kennedy, R.B.; Larrabee, B.R.; Zimmermann, M.T.; Grill, D.E.; Schaid, D.J.; Poland, G.A. Genome-wide associations of CD46 and IFI44L genetic variants with neutralizing antibody response to measles vaccine. Hum. Genet. 2017, 136, 421–435. [Google Scholar] [CrossRef]
- Tian, C.; Hromatka, B.S.; Kiefer, A.K.; Eriksson, N.; Noble, S.M.; Tung, J.Y.; Hinds, D.A. Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat. Commun. 2017, 8, 599. [Google Scholar] [CrossRef]
- Coombes, B.J.; Ovsyannikova, I.G.; Schaid, D.J.; Warner, N.D.; Poland, G.A.; Kennedy, R.B. Polygenic prediction of cellular immune responses to mumps vaccine. Genes Immun. 2025, 26, 413–417. [Google Scholar] [CrossRef]
- Ovsyannikova, I.G.; Haralambieva, I.H.; Schaid, D.J.; Warner, N.D.; Poland, G.A.; Kennedy, R.B. Genome-wide determinants of cellular immune responses to mumps vaccine. Vaccine 2023, 41, 6579–6588. [Google Scholar] [CrossRef] [PubMed]
- Palm, A.E.; Henry, C. Remembrance of Things Past: Long-Term B Cell Memory After Infection and Vaccination. Front. Immunol. 2019, 10, 1787. [Google Scholar] [CrossRef]
- De La Cruz-Rivera, P.C.; Kanchwala, M.; Liang, H.; Kumar, A.; Wang, L.F.; Xing, C.; Schoggins, W.J. The IFN Response in Bats Displays Distinctive IFN-Stimulated Gene Expression Kinetics with Atypical RNASEL Induction. J. Immunol. 2018, 200, 209–217. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, S.M.; Rafnar, T.; Langdon, J.; Lichtenstein, L.M. Molecular identification of an IgE-dependent histamine-releasing factor. Science 1995, 269, 688–690. [Google Scholar] [CrossRef]
- Kang, H.S.; Lee, M.J.; Song, H.; Han, S.H.; Kim, Y.M.; Im, J.Y.; Choi, I. Molecular identification of IgE-dependent histamine-releasing factor as a B cell growth factor. J. Immunol. 2001, 166, 6545–6554. [Google Scholar] [CrossRef] [PubMed]
- Atianand, M.K.; Fitzgerald, K.A. Fitzgerald. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol. Med. 2014, 20, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Heward, J.A.; Lindsay, M.A. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014, 35, 408–419. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).