Abstract
Phage therapy, long overshadowed by antibiotics in Western medicine, has a well-established history in some Eastern European countries and is now being revitalized as a promising strategy against antimicrobial resistance (AMR). This resurgence of phage therapy is driven by the urgent need for innovative countermeasures to AMR, which will cause an estimated 10 million deaths annually by 2050. However, the emergence of phage-resistant variants presents challenges similar to AMR, thus necessitating a deeper understanding of phage resistance mechanisms and control strategies. The highest priority must be to prevent the emergence of phage resistance. Although phage cocktails targeting multiple receptors have demonstrated a certain level of phage resistance suppression, they cannot completely suppress resistance in clinical settings. This highlights the need for strategies beyond simple resistance suppression. Notably, recent studies examining fitness trade-offs associated with phage resistance have opened new avenues in phage therapy that offer the potential of restoring antibiotic susceptibility and attenuating pathogen virulence despite phage resistance. Thus, controlling phage resistance may rely on both its suppression and strategic redirection. This review summarizes key concepts in the control of phage resistance and explores evolutionary engineering as a means of optimizing phage therapy, with a particular focus on Pseudomonas infections. Harnessing evolutionary dynamics by intentionally breaking the spontaneous evolutionary trajectories of target bacterial pathogens could potentially reshape bacterial adaptation by acquisition of phage resistance, unlocking potential in the application of phage therapy.
1. Bacteriophages, Phage Therapy, and Phage-Resistant Variants
1.1. Bacteriophages
Bacteriophages (phages) are prokaryotic viruses that specifically infect and lyse bacteria. Phages are the most abundant biological entities on Earth, with an estimated total exceeding 1031 particles [1]. Ubiquitously distributed in a variety of environments, phages play a significant role in shaping bacterial community dynamics [2,3]. The basic structure of phages consists of a capsid that encloses genetic material and an associated tail. The tail is particularly crucial for host recognition, as it binds specifically to bacterial surface receptors and facilitates genome injection [4]. Phage genomes exhibit considerable diversity, comprising double-stranded DNA (dsDNA), single-stranded DNA, and RNA. dsDNA-tailed phages have traditionally been classified into three major families based on tail morphology: Myoviridae (contractile tail), Siphoviridae (long, non-contractile tail), and Podoviridae (short tail). Notably, the majority of phages used for therapeutic applications belong to these morphotypes [5]. However, in 2022, the International Committee on Taxonomy of Viruses abolished the morphological classification scheme for phages and adopted a new system based on genomic information [6].
1.2. Phage Therapy
Phages were first discovered by Félix d’Hérelle and Frederick Twort in the early 20th century and initially studied as potential antibacterial agents [7,8]. However, their application declined in Western countries following the discovery of penicillin and subsequent widespread adoption of antibiotics. Nevertheless, interest in phage therapy has recently resurged due to the increasing prevalence of bacteria exhibiting antimicrobial resistance (AMR), which will cause an estimated 10 million deaths annually by 2050 [5,9,10]. Additionally, phage therapy offers a targeted treatment approach that selectively eliminates pathogenic bacteria while minimizing the dysbiosis associated with conventional antibiotics because the host range of phages is specific and limited to a single bacterial genus or species [11,12].
The life cycle of phages exhibits two primary stages: the lytic cycle, in which the host cell is lysed upon phage replication, and the lysogenic cycle, in which the genome of temperate phages is integrated into the host genome to establish a symbiotic relationship. Importantly, integration of the genome of temperate phages can alter bacterial gene expression and facilitate the horizontal transfer of virulence or resistance genes, rendering these phages generally unsuitable for therapeutic applications in their native (wild-type) form. By contrast, strictly lytic (virulent) phages, which efficiently lyse bacteria without integration of the genome, are therefore considered the primary candidates for therapeutic applications [5,13]. However, recent advances in synthetic biology have enabled the engineering of temperate phages to eliminate their lysogenic potential, thereby converting them into synthetic virulent phages suitable for therapeutic applications [14].
1.3. Phage-Resistant Variants
A major obstacle to the successful application of phage therapy is the emergence of phage-resistant variants [15,16,17]. These resistant phenotypes commonly arise during phage treatment through alterations, deletions, or modifications of bacterial surface structures—such as outer membrane proteins (omps), lipopolysaccharides (LPS), pili, flagella, and transporters—that serve as phage receptors [18,19,20,21,22], thereby preventing successful phage adsorption. In addition to receptor-based resistance, bacterial sensitivity to phage infection is also shaped by intracellular defense mechanisms. These include the degradation of invading phage genomes via CRISPR-Cas and restriction–modification systems [23], as well as interference with the phage life cycle through the modulation of host cellular processes [24,25]. Collectively, these anti-phage defense systems play a critical role in determining the baseline susceptibility of bacteria to phage infection. Therefore, to effectively manage the emergence of resistance during phage therapy, it is essential to deepen our understanding of the molecular mechanisms underlying resistance, particularly those involving receptor modifications.
In this review, resistance specifically refers to the acquisition of traits that enable a pathogenic bacterium to withstand therapeutic concentrations of a phage (or antibiotic), as distinct from intrinsic insusceptibility.
2. Pseudomonas Infections Associated with AMR
2.1. Pseudomonas aeruginosa
Pseudomonas aeruginosa is a rod-shaped, Gram-negative, γ-proteobacterium of the family Pseudomonadaceae [26]. The bacterium is a facultative aerobe capable of both aerobic and anaerobic respiration and renowned for exceptional metabolic versatility, evidenced by its ability to utilize a wide range of organic compounds. Due to its adaptability, P. aeruginosa can persist in diverse environments, including soil, water, vegetation, and human-associated niches such as the skin and mucosal surfaces [27,28]. P. aeruginosa is frequently isolated from hospital reservoirs such as sinks, humidifiers, and respiratory equipment, contributing to its role as a major cause of nosocomial infections. Clinically, P. aeruginosa is a prototypical opportunistic pathogen and leading cause of both acute and chronic infections in individuals with underlying conditions such as cystic fibrosis, burn injuries, cancer, chronic obstructive pulmonary disease, and ventilator-associated infections [28,29].
The virulence of P. aeruginosa is mediated by a complex array of factors that facilitate tissue invasion, immune evasion, and biofilm formation, which often results in persistent infections with high morbidity and mortality rates, reaching 40% in some populations [26,28]. The high level of intrinsic resistance to many classes of antibiotics and the bacterium’s extraordinary capacity to acquire new resistance mechanisms represent critical concerns in the management of P. aeruginosa infections [30]. As such, P. aeruginosa is one of the most challenging pathogens affecting clinical practice, making it a priority target for alternative therapeutic strategies, including phage therapy [5,13,31].
2.2. AMR in Pseudomonas Infections
The high level of intrinsic insusceptibility of P. aeruginosa limits the efficacy of multiple antibiotics, including carbapenems and third-generation cephalosporins. This insusceptibility is primarily attributed to factors such as a low-permeability outer membrane, constitutively expressed efflux pumps, and antibiotic-modifying enzymes [29,30]. Additionally, acquired resistance can arise through chromosomal mutations, which can lead to the overexpression of these intrinsic efflux pumps, or the acquisition of mobile genetic elements, such as plasmids and integrons [29,32]. Acquired resistance produces a variety of phenotypes, such as multidrug-resistant (MDR), extensively drug-resistant (XDR), or pan-drug-resistant (PDR) [29,33]. Given its advanced resistance mechanisms, the World Health Organization classifies P. aeruginosa as a high-priority organism on the “priority pathogens list”. Furthermore, P. aeruginosa is included in the “ESKAPE pathogens” group along with Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, and Enterobacter spp., pathogens considered high-priority targets in the development of new antibiotics [34].
The global prevalence of antimicrobial-resistant P. aeruginosa strains continues to increase. According to a 2021 European Centre for Disease Prevention and Control report, 18.7% of P. aeruginosa isolates examined were carbapenem-resistant, and 13.4% of isolates were resistant to three or more classes of antibiotics [35]. Moreover, reports of carbapenem-resistant P. aeruginosa in the veterinary and agricultural sectors are increasing [36], indicating that the incidence of AMR is also increasing in fields other than human medicine.
The complexity of treating bacterial infections is shaped by multiple clinical and pharmacological factors, not solely by the pathogen’s AMR profile. Patient-specific characteristics, such as allergies or comorbidities, can constrain prescription decisions. Likewise, the ability of an antimicrobial to diffuse to the site of infection at an effective concentration is a critical determinant of therapeutic success. Indeed, many infections are considered “difficult-to-treat” even when caused by strains fully susceptible to tested antibiotics. It is within this multifaceted clinical landscape that the emergence of difficult-to-treat resistant (DTR) P. aeruginosa strains in recent years has added another significant layer of complexity to treatment strategies. These strains are resistant to nearly all commonly used antibiotics, including carbapenems, cephalosporins, and quinolones, although they remain susceptible to colistin and novel beta-lactam–beta-lactamase inhibitor combinations such as ceftolozane-tazobactam and ceftazidime-avibactam [37,38]. The Infectious Diseases Society of America introduced the concept of DTR P. aeruginosa to highlight strains that present with a complex resistance profile with limited treatment options, distinguishing these strains from MDR and XDR strains. DTR P. aeruginosa is primarily isolated in hospital settings, particularly in intensive care units, where excessive antibiotic use and prolonged hospital stays contribute to the selection of DTR strains. The spread of DTR P. aeruginosa strains emphasizes the critical need for tailored treatment approaches and stringent antimicrobial stewardship practices to prevent further dissemination and mitigate the impact of resistant organisms on patient outcomes [39].
3. Prevention of Phage Resistance Using Phage Cocktails
In the context of the global spread of antimicrobial-resistant P. aeruginosa, increasing international interest has focused on the clinical application of phages [5,13,29]. However, pathogenic bacteria can readily acquire phage resistance, notably through receptor alterations [15,17]. Indeed, in vitro experiments examining co-culture of various phages and target P. aeruginosa strains demonstrated that bacterial regrowth can occur within a few hours after phage inoculation [40,41], suggesting that phage resistance emerges rapidly, potentially through genomic mutations or, in certain bacterial species, via epigenetic mechanisms in receptor-encoding genes [20,22,42,43]. The phenomenon of phage resistance bears striking resemblance to the historical emergence of antibiotic-resistant bacteria, indicating that development of proactive strategies aimed at anticipating and mitigating phage resistance will be essential for sustainable phage therapy. In the absence of such strategies, phage therapy may follow the same trajectory as antibiotic therapy; that is, initial effectiveness followed by loss of efficacy due to the emergence of resistant bacteria. The control of phage resistance thus represents one of the most critical obstacles to the widespread adoption of phage therapy.
Among possible strategies to overcome phage resistance, the most fundamental and essential involves the early prevention or suppression of resistance emergence. A commonly adopted approach to prevent or suppress phage resistance involves the use of phage cocktails—combinations of multiple phages that target distinct bacterial surface receptors—to reduce the likelihood of resistance development [5,13,15]. This strategy is also applicable to P. aeruginosa, and a prevailing principle for effective cocktail design is that each component phage should exploit a different bacterial receptor. However, this principle warrants careful consideration as the definition of a “different” receptor is complicated. For instance, even within a single receptor class like LPS, individual phages may recognize distinct moieties, such as the O-antigen versus the core oligosaccharides, suggesting resistance caused by O-antigen truncation to one may not confer resistance to the other. Furthermore, resistance often arises not from the complete deletion of a receptor but from other mechanisms, such as modified receptor structures or reduced expression levels. These alterations might prevent recognition by one phage while still permitting binding by another that targets the same protein class. Consequently, the acquisition of resistance to a single phage does not necessarily lead to cross-resistance against other phages targeting the same nominal receptor. Indeed, some studies suggest that simultaneous predation by multiple phages can lead to the selection of mutants that circumvent this design logic [44]. Nevertheless, these complexities do not invalidate the cocktail strategy. Instead, they underscore the need for a more sophisticated understanding of phage-host interactions to guide cocktail formulation. This deeper insight could enable the design of cocktails that more potently suppress resistance or even “steer” bacterial evolution toward a clinically manageable state, highlighting a rich area for future investigation. Accordingly, identifying the receptor genes utilized by P. aeruginosa phages serves as the rational starting point for such design. Previous studies have shown that phages targeting P. aeruginosa exploit multiple receptors, including cell surface LPS—particularly the O-antigen and core oligosaccharides [20,45,46], outer membrane transporters [47], and type IV pili [48,49]. These findings strongly suggest that combining phages that target different classes of receptors, such as LPS and outer membrane proteins, could substantially suppress the emergence of phage resistance in P. aeruginosa. Empirical evidence suggests this hypothesis applies to other Gram-negative bacteria as well. For example, the emergence of phage resistance was significantly suppressed in vitro using a combination of phages that target flagella and LPS [50] in Cronobacter and phages targeting the O-antigen, BtuB, and OmpC in Salmonella [51]. Notably, whereas in vitro co-culture with individual phages (ΦCR8 and ΦS13) allowed the emergence of resistant Cronobacter mutants within 5 h, their combination as a cocktail delayed bacterial regrowth for over 11 h [50]. Moreover, in silico modeling studies have demonstrated that cocktails composed of Pseudomonas-targeting phages that recognize distinct receptors reduce the probability of simultaneous acquisition of resistance to each phage [52]. A comprehensive study further revealed that resistance of P. aeruginosa to 27 different phages exhibited a modular network pattern [53]. In this network structure, phages within the same module tended to exhibit similar resistance profiles, whereas cross-resistance was less likely between modules. These modules corresponded to genetic mutations in distinct receptor-associated genes, such as those related to LPS or type IV pili, indicating that the evolution of phage resistance is strongly associated with phage receptor specificity. Paradoxically, this insight hints at a strategic advantage for phage therapy: phage combinations that target distinct modules (i.e., different classes of receptors) would likely be the most effective for minimizing the risk of resistance development. In support of this hypothesis, recent studies have suggested that combinations of genetically diverse P. aeruginosa-targeting phages demonstrate enhanced cocktail effects [54], thus further supporting the receptor-based cocktail design principle. Nevertheless, a recent study using Pseudomonas alcaligenes, a close relative of P. aeruginosa, reported that sequential administration of individual phages was more effective at preventing the emergence of resistance than application of a multi-phage cocktail [55]. This finding suggests that the optimal dosing protocol—simultaneous versus sequential administration—remains an open question that warrants further investigation and standardization. There is clearly room for refinement in phage therapy strategies aimed at suppressing the emergence of resistant bacteria.
4. Phage Resistance in Clinical Pseudomonas Infections
The administration of phage cocktails as a strategy to mitigate the challenge of phage resistance has been extensively explored (Table 1). Indeed, five cases of P. aeruginosa infection treated at the University of California San Diego between 2018 and 2020 involved the use of phage cocktails for diverse conditions, including post-transplant pneumonia, cystic fibrosis, and ventricular assist device (VAD) infections (Table 1, Cases 4–6) [56,57]. While these therapies, predominantly administered with antibiotics, led to successful outcomes like bacteremia resolution, recurrence was observed in VAD-related infections, potentially due to poor phage penetration into biofilms. The efficacy of combined phage and antibiotic therapy was also demonstrated in other cases of P. aeruginosa infection, including those involving ventilator-associated pneumonia (VAP), prosthetic knee infection and Kartagener syndrome (Table 1, Cases 3, 9, 14, and 25) [58,59,60,61]. In addition, case reports of phage cocktail therapy for P. aeruginosa infections continue to accumulate. A notable example is a large Belgian-led cohort study, where P. aeruginosa was the most frequently treated pathogen (49 of 100 cases), utilizing various phage combinations or predefined cocktails (Table 1, Cases 15–23) [62,63,64,65,66]. Numerous other reports highlight successful outcomes across different clinical contexts. These include the local administration of a single phage with ceftazidime for a chronic infection (Table 1, Case 1) [67], and the use of cocktails to treat a refractory infection with a bronchopleural fistula (Table 1, Case 10) [68], MDR P. aeruginosa infection (Table 1, Case 11) [69], and a pediatric case (Table 1, Case 2) [70]. Further successes have been documented in treating a recurrent left ventricular assist device (LVAD)-related infection, where adjunctive surgery was also performed, and a postoperative infection following a Bentall procedure (Table 1, Cases 13 and 26) [71,72]. Conversely, cases in which phage therapy has exhibited suboptimal or limited efficacy have also been reported. In one case of a vascular graft infection, bacteremia recurred despite combined therapy; isolates from the recurrence showed both phage resistance and enhanced biofilm formation (Table 1, Case 12) [73]. Similarly, the emergence of phage-resistant variants was implicated in the treatment failure of a chronic LVAD infection (Table 1, Case 24) [57,74]. Analysis of cases from the Belgian cohort revealed that single nucleotide polymorphisms or deletions in phage receptor genes were the presumed cause of resistance, preventing pathogen eradication (Table 1, Cases 15–17) [62]. Interestingly, however, two of these patients showed clinical improvement despite the failure to eradicate the pathogen, suggesting benefits beyond bacterial clearance (Table 1, Cases 16 and 17).

Table 1.
Clinical cases of phage therapy for Pseudomonas aeruginosa infections: therapeutic protocols and outcomes. NA; not applicable.
Common factors that contribute to treatment failure in challenging cases such as those described above include suboptimal delivery of phages to the infection site, rapid emergence of phage resistance, and associated bacterial phenotypic shifts, particularly with regard to increased ability to form biofilms. Notably, the acquisition of phage resistance appears to critically affect the chances of therapeutic success in challenging cases. Furthermore, the diminishment of therapeutic efficacy due to the emergence of phage resistance, even with the use of phage cocktails, is not confined to P. aeruginosa but also observed in phage therapy for the treatment of other bacterial pathogens [16,75,76,77]. The potential emergence of phage resistance thus underscores the necessity of developing strategies that not only fundamentally curtail the development of phage resistance but also effectively manage phage-resistant bacterial populations. Moreover, the PhagoBurn clinical trial serves as a critical reminder that beyond these biological and clinical hurdles, ensuring the therapeutic efficacy of phage cocktails also depends heavily on pharmaceutical factors such as their optimal design and long-term stability [78].
5. Fitness Trade-Offs with Phage Resistance
The Russian-American geneticist Theodosius Dobzhansky (1900–1975) famously stated, “Nothing in biology makes sense except in the light of evolution”. This evolutionary perspective is profoundly relevant to infection control strategies involving phage therapy. Specifically, this perspective underscores the principle that any approach to managing infections, particularly phage therapy, that disregards evolutionary dynamics is unlikely to achieve sustained and effective therapeutic outcomes. The “fitness” of bacteria (i.e., their capacity to adapt to a given environment) is continually enhanced through the accumulation of diverse phenotypic variations and adaptations essential for survival. The interactions between phages and bacteria represent an integral component of this adaptive process, in which both entities are locked in a perpetual “evolutionary arms race” [79,80,81]. The acquisition of specific traits frequently incurs biological costs that in turn lead to “evolutionary constraints” or “fitness costs”, with the enhancement of one function potentially coming at the expense of another. Such trade-offs are a universally observed phenomenon in biological evolution [82,83]. Notably, recent research revealed that so-called “fitness trade-offs” associated with the acquisition of phage resistance—the ability to evade phage infection—can paradoxically lead to an attenuation of bacterial virulence or reductions in antibiotic resistance levels in antimicrobial resistant bacteria [15,17,83,84]. These fitness trade-offs represent new avenues for phage therapy. Consequently, phage therapy should not be conceptualized solely as a “bactericidal tool” for eliminating pathogenic bacteria but also as a potent “selective pressure” capable of affecting the trajectory of bacterial evolution. Therapeutic strategies employing phages must therefore consider not only the direct lytic activity of the therapy but also the broader evolutionary implications, including potential decreases in pathogenicity and antibiotic resistance stemming from the development of phage resistance. In essence, the infection site serves as a dynamic stage upon which phage–pathogen interactions unfold on an evolutionary timescale. A holistic viewpoint that encompasses this complex interplay is paramount to devising and implementing sustainable infection control strategies.
5.1. Fitness Trade-Offs Between Phage Resistance and Bacterial Virulence: Attenuating Virulence
The pathogenicity of P. aeruginosa is determined by an intricate pathway involving adhesion to host cells, tissue invasion, and immune evasion. The virulence of P. aeruginosa is often mediated by bacterial transmembrane proteins, cellular appendages such as flagella and pili, and polysaccharides such as LPS [29]. Increasing evidence suggests that these virulence factors serve as receptors for phage infection [20,45,46,49]. Consequently, the development of resistance to these phages can lead to an “evolutionary trade-off” characterized by attenuated virulence, and numerous instances of this phenomenon have been reported (Figure 1 and Table 2).
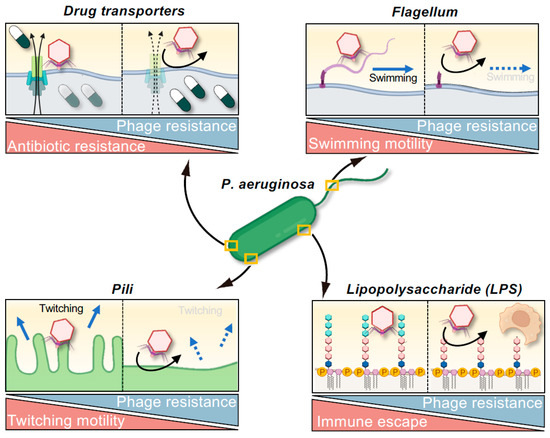
Figure 1.
Evolutionary trade-offs associated with phage resistance in Pseudomonas aeruginosa. The acquisition of phage resistance can compromise bacterial traits such as antibiotic resistance, motility, and immune evasion. Resistance mechanisms that affect pili or flagella often impair bacterial twitching or swimming motility. Modifications to outer membrane structures, including LPS and drug transporters, can reduce the capacity for immune evasion or increase antibiotic susceptibility. These trade-offs offer therapeutic opportunities by attenuating virulence and reversing AMR, even when phage-resistant variants emerge.
Type IV pili are key virulence factors of P. aeruginosa, and they have been implicated as mediating twitching motility and adhesion to host cells. Resistance to type IV pili-targeting phages can result from mutations in genes such as pilB or pilT, which leads to diminished twitching motility [49,85,86,87,88,89]. Similarly, strains in which flagella-encoding genes such as flgC or motABCD have been knocked out exhibit a loss of swimming motility [90,91]; this change can impact interactions with host immunity, such as the evasion of released neutrophil extracellular traps [90]. Furthermore, biofilm formation is crucial for the establishment of chronic infections and protection against environmental stress, and the acquisition of resistance to phages that target pili, flagella, or LPS is consistently associated with a diminished capacity for biofilm formation [46,85,89,91,92]. This diminished capacity to form biofilm is often attributed to the loss of motility or changes in surface polysaccharide structures necessary for biofilm formation. Moreover, P. aeruginosa strains that have developed resistance to LPS-targeting phages through mutations in the waaL gene (which encodes O-antigen ligase) are more susceptible to phagocytosis by bone marrow-derived dendritic cells [91]. This exemplifies a concept known as “immunophage synergy”. Indeed, Roach et al. demonstrated that although treatment with phages alone is sometimes ineffective, successful infection control can be achieved through synergism with host immunity, particularly with regard to neutrophil activation [93], which has clinically significant implications.
These observations reveal a strategic advantage for phage therapy: when phages target virulence structures, the evolution of phage resistance can inherently lead to attenuated pathogenicity. From a clinical standpoint, the prospect of resistant bacterial strains exhibiting debilitated infectivity offers a promising paradigm for future therapeutic design, often referred to as an “evolutionary attenuation strategy” or leveraging “evolutionary trade-offs” in phage therapy. This principle may be reflected in Cases 16 and 17 summarized in Table 1, in which clinical improvement was noted despite the failure to eradicate the target P. aeruginosa strain and the suspected emergence of phage-resistant variants [62], suggesting that the resistant bacteria may have become less virulent.
5.2. Fitness Trade-Offs Between Phage Resistance and Antibiotic Sensitivity: Reversing AMR
Drug efflux pumps are prominent components of resistance mechanisms in MDR bacteria that reduce the antibiotic susceptibility of a bacterium by actively exporting antimicrobial agents from the cell [29]. In P. aeruginosa, the MexAB-OprM and MexXY-OprM efflux systems are particularly significant in this regard, contributing to resistance against a broad spectrum of antibiotics [94,95]. Intriguingly, these efflux pumps can also serve as receptors for certain phages. Consequently, when phages target these structures, bacteria may develop phage resistance through mutations in the genes encoding the pump components or downregulation of their expression. This can result in impaired drug efflux and restoration of antibiotic susceptibility, illustrating compensatory changes associated with adaptive evolution (Figure 1 and Table 2). A prime example of this phenomenon is phage OMKO1, which infects P. aeruginosa via OprM, an outer membrane protein common to both the MexAB-OprM and MexXY-OprM efflux systems [47]. Strains of P. aeruginosa resistant to ΦOMKO1 reportedly exhibit up to 50-fold increased susceptibility to various antibiotics, including ciprofloxacin, tetracycline, ceftazidime, and erythromycin. Such an increase in antibiotic susceptibility has been corroborated using in vitro studies and Galleria mellonella infection models [47,96]. Clinically, the case of a patient with a vascular graft infection treated with a combination of phage OMKO1 and ceftazidime (Table 1, Case 1) suggested that the therapeutic efficacy of ceftazidime was in part attributable to antibiotic re-sensitization due to phage resistance, in addition to direct phage-mediated bacterial lysis [67].
A recently identified phage, PIAS, was found to target the P. aeruginosa transporter protein MexY, which is a core component of the MexXY-OprM drug efflux system implicated in the efflux of aminoglycosides and fluoroquinolones [97]. Resistance to ΦPIAS often involves mutations or deletions in the mexY gene, leading to marked bactericidal effects when phage therapy is combined with administration of antibiotics such as fosfomycin, gentamicin, tetracycline, and ceftazidime. Sensitization of P. aeruginosa to antibiotics can also occur even when phages do not directly target efflux pumps, as large-scale deletions of genomic regions adjacent to efflux pump genes can indirectly compromise efflux system functionality [98]. In this case, resistance of P. aeruginosa isolate Pa12 to phage ΦS12-3 was found to be associated with a large chromosomal deletion encompassing the galU gene (designated BigD: bacteriophage-induced galU deficiency), which resulted in concomitant loss of the MexXY efflux pump and consequently increased susceptibility to fluoroquinolones such as levofloxacin, enrofloxacin, and orbifloxacin. Thus, therapies that promote phage-induced deletions in chromosome could potentially restore the clinical utility of previously ineffective antibiotics [98,99,100,101].
Paradoxically, evolution guided by phage infection can confer the therapeutic benefit of antibiotic re-sensitization. Increased attention is focusing on this “evolutionarily steered therapeutic strategy” as an innovative approach to combat MDR bacteria. Additionally, if mutations exist within the quinolone resistance-determining region (QRDR), the resulting loss of efflux pump function and subsequent intracellular drug accumulation may not restore quinolone susceptibility [99]. Nevertheless, recent case reports have documented instances in which phage resistance has led to the reversion of QRDR mutations to susceptible genotypes (Table 1, Case 17) [62], thereby suggesting avenues for further exploration. Consequently, continued characterization of phages and the development of evolutionarily informed therapeutic strategies are highly anticipated. In addition, a series of personalized inhaled phage therapies strongly supports phage-driven trade-offs in clinical cases, which might affect clinical endpoints [102].

Table 2.
Expected trade-offs associated with phage resistance in Pseudomonas aeruginosa: virulence attenuation and antibiotic resensitization.
Table 2.
Expected trade-offs associated with phage resistance in Pseudomonas aeruginosa: virulence attenuation and antibiotic resensitization.
| Expected Trade-Offs in Phage Resistant P. aeruginosa | Reported or Potential Phages | Associated Genes | Remarks | References | |
|---|---|---|---|---|---|
| Attenuating virulence | Twitching motility | Pili-targeting Pseudomonas phages | pilA, pilT, pilB, pilZ, pilO, pilN, pilY1, pilX, pilM, pilR | Motility reduction accompanying phage resistance. | [49,85,86,87,88,89] |
| Swimming motility | Flagella-recognizing Pseudomonas phages | flgC, motABCD | Reduced motility, potentially aiding evasion of neutrophil NETs. | [90,91] | |
| Biofilm formation | LPS-recognizing Pseudomonas phages | wzy | Phage resistance linked to decreased biofilm formation. | [46] | |
| Flagella-recognizing Pseudomonas phages | motABCD | Flagellar inactivation leads to reduced biofilm formation. | [91] | ||
| Pili-recognizing Pseudomonas phages | pilT, pilB, pilO, pilN, pilY1, pilX, pilM, pilR | Reduced biofilm production accompanying phage resistance. | [85,89,92] | ||
| Phagocytosis | LPS-recognizing Pseudomonas phages | waaL | Truncated LPS enhances phagocytosis by mouse BMDCs. | [91] | |
| Reversing AMR | Ciprofloxacin sensitivity | OprM-recognizing Pseudomonas phages | oprM | Resistance to ΦOMKO1 leads to efflux pump loss and antibiotic sensitization. | [47] |
| Tetracycline sensitivity | |||||
| Ceftazidime sensitivity | |||||
| Erythromycin sensitivity | |||||
| Levofloxacin sensitivity | LPS-recognizing Pseudomonas phages | Bacteriophage-induced galU deficiency (BigD) regions, including mexX and mexY. | Phage resistance via chromosomal deletion enhances susceptibility to quinolones and other antibiotics but may concurrently promote biofilm formation. | [85,98,99,100] | |
| Orbifloxacin sensitivity | |||||
| Enrofloxacin sensitivity | |||||
| Colistin sensitivity | [101] | ||||
| Tetracycline sensitivity | MexY-recognizing Pseudomonas phages | mexY | mexY mutations or Brmts induced by MexY-targeting phages confer antibiotic susceptibility. | [97] | |
| Fosfomycin sensitivity | |||||
| Ceftazidime sensitivity | |||||
| Gentamicin sensitivity | |||||
| Quinolone sensitivity | Unknown (not identified in detail) | Quinolone resistance determining region (QRDR), H87D conversion. | Phage-resistant P. aeruginosa clinical isolates exhibit QRDR mutation conversion after phage therapy. | [62] | |
5.3. Concerns Regarding Negative Trade-Ups and Unintended Consequences
While a fitness trade-off strategy can be helpful, it also carries potential risks. Previous studies have suggested that if we only look at the beneficial parts of evolution, we might miss possible weaknesses. For instance, large chromosomal deletions caused by phage infection can lead to undesirable phenotypic changes, such as enhanced biofilm formation in P. aeruginosa [85]. This emphasizes the critical need for attention, as some phage resistance may drive a negative “trade-up” that increases bacterial resistance to one or more antibiotics, instead of the desired trade-off [17,98]. A notable example involves the coliphages T6 and U115, which target the Tsx porin. This porin is also the entry point for the antibiotic albicidin. Consequently, bacteria that evolved resistance to these phages by mutating tsx also exhibited cross-resistance to albicidin, which is a clearly unfavorable “trade-up” in clinical settings [103]. It has also been reported that, while a majority of mutants resistant to the TolC-targeting coliphage U136B became more susceptible to antibiotics, as TolC functions as a drug efflux transporter, a few variants were observed to become more drug-resistant, likely due to synergistic pleiotropy [104]. These highlight that accurately predicting phage-driven evolutionary trajectories remains a significant challenge. Therefore, to refine our predictions for phage therapies and to minimize unintended effects, a deeper understanding of these intricate interactions is essential.
6. Core Molecular Machinery Involved in the Acquisition of Bacterial Phage Resistance
The concept of spontaneous symmetry breaking, originally proposed in the context of theoretical physics, refers to the spontaneous transition of a system that is symmetric in its overall structure to an asymmetric state without any explicit external bias [105,106]. A commonly used analogy involves a round table with four glasses placed equidistantly. Although each glass is equally accessible, once the first individual reaches for the glass on their left, other individuals are likely to follow suit, resulting in a collective preference for one side. Despite the absence of any predetermined bias, symmetry is thus “spontaneously” broken. This concept offers a compelling analogy for understanding the evolutionary responses of pathogenic bacteria to phage predation.
In principle, a wide variety of genetic routes could enable bacteria to acquire phage resistance—including changes in the structure of LPS or membrane transporters, or dysregulation of transcriptional regulators [18,42,47]. Even within a single resistance pathway, such as LPS modification, multiple mutations—differing in the genes involved or specific nucleotide changes—can give rise to functionally similar outcomes [20,22,98]. In practice, however, the emergence of a particular mutation often leads to its rapid dominance within the population, resulting in a highly skewed distribution of resistance patterns. This emergent asymmetry—despite a theoretically “symmetric” landscape of mutational possibilities—can be viewed as a form of spontaneous breaking of the symmetry of evolutionary selection. In this sense, the concept of spontaneous evolutionary symmetry breaking in pathogenic bacteria could serve as a framework for understanding how selective pressure channels the adaptive trajectories of bacterial populations. Importantly, such evolutionary shifts do not always result in beneficial outcomes from a clinical standpoint, such as trade-offs that increase antibiotic susceptibility or reduce virulence (Table 2). Indeed, spontaneous evolutionary symmetry breaking can occasionally yield bacterial variants exhibiting enhanced antibiotic resistance and adaptability, thereby complicating infection control efforts [85,98,104]. To address this challenge through strategic design of phages or phage cocktails—or by externally manipulating key stages of the phage infection process—it may be possible to intentionally bias the evolutionary trajectory of a bacterial population towards desired clinical outcomes. This approach represents a form of evolutionary engineering that could potentially reshape bacterial adaptation by acquisition of phage resistance. In the following sections, we illustrate this concept by examining the molecular mechanisms of mutation acquisition and DNA damage repair in relation to the acquisition of phage resistance. Based on these findings, we propose a working model that outlines how the molecular machinery of DNA repair could be deeply involved in the evolutionary trade-offs associated with acquiring phage resistance.
6.1. Phage Resistance via the DNA Damage Response and Chromosomal Rearrangement
In P. aeruginosa, phage resistance can be acquired through large-scale chromosomal deletions [85,97,98,100,101,107,108], mediated by the DNA mismatch repair system, subsequent double-strand breaks (DSBs), and their associated repair mechanisms (Figure 2). Two key mismatch repair components, MutS and MutL, preserve genomic integrity by detecting and correcting misincorporated nucleotide pairs during DNA replication. The recognition of mismatches by MutS and the formation of a MutS-MutL complex initiate localized nucleotide excision, which can go on to form DSBs [109,110,111]. DSBs are repaired through two major pathways: homologous recombination (HR), primarily mediated by RecA and RecBCD, and non-homologous end joining (NHEJ), mediated by Ku and LigD [112,113,114,115]. In the HR pathway, RecBCD unwinds DNA from the DSB ends, and upon recognition of the Chi sequence (5’-GCTGGTGG-3’ in Escherichia coli) [116], its mode of activity switches to facilitate RecA loading and homologous strand exchange. This mechanism enables high-fidelity repair and restoration of the original sequence. When HR predominates, the emergence and selection of deletion mutants are relatively suppressed, even under phage pressure. Conversely, the NHEJ pathway directly ligates non-homologous DNA ends through the activity of Ku and LigD, which often results in deletions or insertions. This error-prone repair mechanism frequently leads to the loss of extensive chromosomal regions, including those encoding phage receptors, thereby conferring phage resistance. Under selective pressure from phage infection, mutants harboring such large-scale deletions are likely to be positively selected. Indeed, Shen et al. reported that overexpression of mutL in P. aeruginosa PAO1 dramatically increases the frequency of mutants with large chromosomal deletions. Similarly, overexpression of ku or ligD increases the likelihood of such deletions. Moreover, deletion of recA—thus disabling the HR pathway—also markedly upregulates the selection of large-scale deletion mutants [107]. Collectively, these findings suggest that the modulation of the dynamics of DNA repair plays a crucial role in shaping bacterial evolutionary responses to phage predation, particularly in promoting resistance via genome restructuring.
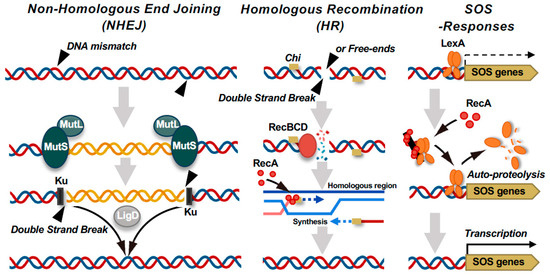
Figure 2.
DNA double-strand break (DSB) repair pathways and SOS response activation in bacteria linked to phage resistance. Bacteria utilize multiple pathways to repair DSBs. The non-homologous end joining (NHEJ) pathway involves Ku and LigD, which directly ligate DNA ends, whereas MutS–MutL appears to suppress aberrant recombination. By contrast, homologous recombination (HR), which is initiated by the RecBCD complex, processes DNA ends and facilitates RecA-mediated strand invasion. Notably, RecBCD also functions as a defense factor by recognizing and degrading foreign linear DNA, such as DNA of injected phage genomes. RecA activation leads to LexA autocleavage, triggering the SOS response and inducing genes involved in DNA repair and damage tolerance.
6.2. RecA-Mediated Mutagenesis: Hypermutable State and the SOS Response
Loss-of-function mutations in MutL or MutS compromise the repair of replication errors, thus driving bacteria into a hypermutable state. In this hypermutable state, point mutation accumulation accelerates markedly, leading to the emergence of diverse resistance mutations [117,118]. Notably, when mutS harbors mobile genetic elements, external stressors such as phage infection can trigger transposition events that disrupt mutS function, resulting in a rapid and extensive increase in point mutations [119]. This mutation-inducing process is tightly regulated by the bacterial SOS response, which is triggered when RecA binds to damaged DNA, resulting in autocleavage of the LexA repressor and induction of a suite of DNA repair and mutagenic genes (Figure 2) [120,121,122,123]. The induced genes include those encoding error-prone DNA polymerases such as Pol IV (dinB) and Pol V (umuDC), which introduce mutations [124,125]. Classical studies have elucidated the multifaceted roles of RecA in this context. Ennis et al. demonstrated that RecA tightly regulates the mutation frequency of UV-damaged phage λ [126]. They found that even when LexA was non-functional, no mutagenesis occurred in the absence of activated RecA. Conversely, introduction of a constitutively active RecA mutant sustained mutagenesis even in a LexA-deficient background. These findings indicate that RecA not only initiates the SOS response via LexA cleavage but also directly contributes to mutagenesis as a secondary function. Importantly, RecA exhibits extraordinarily high fidelity in recognizing homologous sequences. In vitro studies have shown that RecA can detect even a single base mismatch during the early stages of HR [127]. Mismatches near the 3’-end significantly suppress recombination, suggesting that RecA prevents erroneous DNA repair by blocking strand exchange when the sequence homology is low. RecA thus functions as a “kinetic proofreader” that actively excludes sequences of imperfect homology. Recent structural studies revealed that RecA assesses sequence homology with remarkable precision, even during the homology search phase, without forcibly unwinding dsDNA [128]. Instead, RecA only minimally disrupts the DNA duplex to exert no internal stress while probing for homology within single-stranded DNA. This mechanism likely plays a key role in maintaining genome stability during recombination and DNA repair processes.
In summary, RecA functions not only as the canonical trigger of the SOS response via LexA cleavage but also plays a central role in mutagenesis and the maintenance of genome stability. By coupling precise homology recognition with mutagenic potential, RecA balances two seemingly contradictory activities—introduction of mutations for adaptation versus ensuring genomic stability for survival—and thus serves as an evolutionary hub that finely tunes the bacterial response to environmental challenges.
6.3. The Dual Nature of the RecBCD Complex: DNA Repair and Phage Interference
The Rec system, particularly the RecBCD complex, serves not only as a primary DNA DSB repair pathway but also plays a key role in the phage infection defense mechanism. In E. coli, RecBCD exhibits two distinct modes of action: a destructive mode, in which the complex acts as a potent nuclease that degrades free DNA ends into single-stranded fragments, and a recombinogenic mode, in which encountering a chi site triggers the loading of RecA and initiation of HR repair (Figure 2) [129]. The destructive mode provides multifaceted protection against phage invasion by degrading phage genomes, supplying protospacer sequences to CRISPR libraries, and suppressing rolling-circle replication [130,131,132]. The recombinogenic mode, by contrast, is activated upon recognition of the chi sequence, which is overrepresented in the host genome but rare in phage DNA. This difference enables RecBCD to distinguish “self” from “non-self”, thus integrating repair and immune functions in a context-dependent manner. Recent studies have revealed that certain phages encode proteins collectively referred to as “anti-RecBCD” factors, which interfere with the activity of the RecBCD complex to enable phages to evade bacterial defenses [133]. However, the broader implications of such phage countermeasures—particularly their impact on RecBCD-mediated functions such as mutagenesis and chromosomal rearrangement—remain largely unexplored. As discussed in this chapter, DNA repair mechanisms, including HR and NHEJ, underpin the molecular evolution through which bacteria acquire resistance to phage infection. Targeting these pathways to deliberately disrupt bacterial spontaneous evolutionary selection could provide a novel conceptual framework for studying the evolution of pathogenic bacteria towards desired clinical outcomes.
6.4. The Evolutionary Arms Race and Cooperation Between E. coli and Escherichia-Targeting Phages
While the main focus of this review is on P. aeruginosa, the extensively studied evolutionary arms race between the model organism E. coli and its phages offers invaluable insights. This relationship serves as a rich source of examples for finely tuned molecular adaptation strategies and provides a crucial framework for understanding the principles of the DNA repair-based working model discussed earlier. Therefore, the interactions between E. coli and its phages offer valuable lessons for advancing evolutionary engineering in synthetic biology and for developing intervention strategies applicable to other pathogens like P. aeruginosa. The efficiency of T7 phage DNA replication is enhanced through utilization of E. coli thioredoxin as an accessory factor to compensate for the low processivity of the phage’s own DNA polymerase [134]. To outcompete the host cell’s RNA polymerase and ensure successful transcription of phage genes, T7 has evolved proteins such as Gp2 and Gp5.7, which sequentially inhibit the host’s transcriptional machinery [135,136]. Furthermore, coordinated interactions between multifunctional proteins—such as T7 RNA polymerase and lysozyme—allow for temporal and spatial regulation of key steps in the phage life cycle, including transcription, replication, and DNA packaging [137]. These molecular interactions and network-level control strategies, which have been forged through co-evolution, can be viewed as products of evolutionary optimization in nature. Recent technological advances have increased the feasibility of inserting new genes into phage genomes or knocking out existing genes [138,139,140]. From an evolutionary perspective, such manipulations effectively “skip” or “rewind” the natural evolutionary processes of phages. By elucidating and then mimicking, and reconstructing these mechanisms, we can uncover essential design principles that will enable the future development of evolution-informed phage therapies. Ultimately, this approach promises to provide powerful insights into how evolution itself can be harnessed and deliberately manipulated for infection control.
7. Concluding Remarks
One of the most critical challenges limiting the use of phage therapy to treat antimicrobial-resistant bacteria is the emergence of phage resistance. Although it is essential to first suppress the development of phage resistance, even when resistance does arise, strategies are available to actively exploit evolutionary trade-offs. Specifically, the fitness costs associated with phage resistance can be harnessed to deliberately direct bacterial evolution toward reduced virulence and restored antibiotic susceptibility. Such evolutionary interventions not only contribute to restoring the efficacy of conventional antibiotics but also enhance the sustainability of phage therapy by exploiting fitness costs, thereby providing a foundation for preserving effective treatment options into the future. It is important to acknowledge, however, that the practical application of this framework will probably face challenges: beneficial evolutionary trade-offs may not be universally achievable for all phage-bacteria pairings, and furthermore, the reliable induction of specific resistance mutations that consistently lead to such advantageous trade-offs may not always be feasible. On the other hand, a greater understanding of the evolutionary mechanisms that direct the interplay between pathogenic bacteria and phages would offer more than just a means of overcoming resistance; such an understanding would also pave the way for the development of innovative approaches that enhance the overall impact of phage-based treatments. Ultimately, intentionally breaking the spontaneous evolutionary trajectories of bacterial pathogens to redirect their adaptation may unlock unforeseen frontiers in the design of phage therapies against AMR.
Author Contributions
Conceptualization, J.F., D.Y. and K.N.; validation, D.Y., H.Y., N.K., M.S., H.K., K.N. and H.I.; writing—original draft preparation, J.F.; writing—review and editing, D.Y., H.Y., N.K., M.S., H.K., K.N. and H.I.; visualization, J.F.; supervision, J.F. and H.I.; project administration, J.F.; funding acquisition, J.F. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the MEXT/JSPS KAKENHI, grant number JP23K23791 (to JF).
Acknowledgments
All illustrations were created using BioRender.com (https://www.biorender.com/).
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships.
References
- Mushegian, A.R. Are there 10(31) virus particles on earth, or more, or fewer? J. Bacteriol. 2020, 202, e00052-20. [Google Scholar] [CrossRef] [PubMed]
- Parikka, K.J.; Le Romancer, M.; Wauters, N.; Jacquet, S. Deciphering the virus-to-prokaryote ratio (vpr): Insights into virus-host relationships in a variety of ecosystems. Biol. Rev. Camb. Philos. Soc. 2017, 92, 1081–1100. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A. Marine viruses—Major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef]
- Nobrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E.; Brouns, S.J.J. Targeting mechanisms of tailed bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage therapy: From biological mechanisms to future directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ictv bacterial viruses subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef]
- Twort, F.W. Further Investigations on the nature of ultra-microscopic viruses and their cultivation. J. Hyg. 1936, 36, 204–235. [Google Scholar] [CrossRef]
- d’Hérelle, F. Sur un microbe invisible antagoniste des bacilles dysenteriques. C. R. Acad. Sci. 1917, 165, 373–375. (In French) [Google Scholar]
- Reardon, S. Phage therapy gets revitalized. Nature 2014, 510, 15–16. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. 2015, 1, 1–20. Available online: https://amr-review.org (accessed on 23 June 2025).
- Fujiki, J.; Schnabl, B. Phage therapy: Targeting intestinal bacterial microbiota for the treatment of liver diseases. JHEP Rep. 2023, 5, 100909. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Young, R.; Schnabl, B. Bacteriophages and their potential for treatment of gastrointestinal diseases. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage therapy: A renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Fujiki, J.; Nakamura, K.; Nakamura, T.; Iwano, H. Fitness trade-offs between phage and antibiotic sensitivity in phage-resistant variants: Molecular action and insights into clinical applications for phage therapy. Int. J. Mol. Sci. 2023, 24, 15628. [Google Scholar] [CrossRef]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017, 61, e00954-17. [Google Scholar] [CrossRef] [PubMed]
- Oromi-Bosch, A.; Antani, J.D.; Turner, P.E. Developing phage therapy that overcomes the evolution of bacterial resistance. Annu. Rev. Virol. 2023, 10, 503–524. [Google Scholar] [CrossRef]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef]
- Esteves, N.C.; Scharf, B.E. Flagellotropic Bacteriophages: Opportunities and challenges for antimicrobial applications. Int. J. Mol. Sci. 2022, 23, 7084. [Google Scholar] [CrossRef]
- Uchiyama, J.; Suzuki, M.; Nishifuji, K.; Kato, S.I.; Miyata, R.; Nasukawa, T.; Yamaguchi, K.; Takemura-Uchiyama, I.; Ujihara, T.; Shimakura, H.; et al. Analyses of short-term antagonistic evolution of Pseudomonas aeruginosa strain pao1 and phage kpp22 (myoviridae family, pb1-like virus genus). Appl. Environ. Microbiol. 2016, 82, 4482–4491. [Google Scholar] [CrossRef]
- Esteves, N.C.; Porwollik, S.; McClelland, M.; Scharf, B.E. The multi-drug efflux system AcrABZ-TolC is essential for infection of Salmonella typhimurium by the flagellum-dependent bacteriophage chi. J. Virol. 2021, 95, e00394-21. [Google Scholar] [CrossRef]
- Forti, F.; Bertoli, C.; Cafora, M.; Gilardi, S.; Pistocchi, A.; Briani, F. Identification and impact on Pseudomonas aeruginosa virulence of mutations conferring resistance to a phage cocktail for phage therapy. Microbiol. Spectr. 2023, 11, e0147723. [Google Scholar] [CrossRef] [PubMed]
- Ofir, G.; Sorek, R. Contemporary phage biology: From classic models to new insights. Cell 2018, 172, 1260–1270. [Google Scholar] [CrossRef]
- Yuping, L.; Guan, L.; Becher, I.; Makarova, K.S.; Cao, X.; Hareendranath, S.; Guan, J.; Stein, F.; Yang, S.; Boergel, A.; et al. Jumbo phage killer immune system targets early infection of nucleus-forming phages. Cell 2025, 188, 2127–2140.e21. [Google Scholar] [CrossRef]
- Wang, M.; Ji, Q.; Liu, P.; Liu, Y. Nad(+) depletion and defense in bacteria. Trends Microbiol. 2023, 31, 435–438. [Google Scholar] [CrossRef]
- Gellatly, S.L.; Hancock, R.E. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef]
- Yoon, S.S.; Hennigan, R.F.; Hilliard, G.M.; Ochsner, U.A.; Parvatiyar, K.; Kamani, M.C.; Allen, H.L.; DeKievit, T.R.; Gardner, P.R.; Schwab, U.; et al. Pseudomonas aeruginosa anaerobic respiration in biofilms: Relationships to cystic fibrosis pathogenesis. Dev. Cell 2002, 3, 593–603. [Google Scholar] [CrossRef]
- Wood, S.J.; Kuzel, T.M.; Shafikhani, S.H. Pseudomonas aeruginosa: Infections, animal modeling, and therapeutics. Cells 2023, 12, 199. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Xiao, W.; Zhou, C.; Pu, Q.; Deng, X.; Lan, L.; Liang, H.; Song, X.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Raudonis, R.; Glick, B.R.; Lin, T.J.; Cheng, Z. Antibiotic resistance in Pseudomonas aeruginosa: Mechanisms and alternative therapeutic strategies. Biotechnol. Adv. 2019, 37, 177–192. [Google Scholar] [CrossRef]
- Subedi, D.; Vijay, A.K.; Willcox, M. Overview of mechanisms of antibiotic resistance in Pseudomonas aeruginosa: An ocular perspective. Clin. Exp. Optom. 2018, 101, 162–171. [Google Scholar] [CrossRef]
- Livermore, D.M. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: Our worst nightmare? Clin. Infect. Dis. 2002, 34, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Karageorgopoulos, D.E. Pandrug resistance (pdr), extensive drug resistance (xdr), and multidrug resistance (mdr) among gram-negative bacilli: Need for international harmonization in terminology. Clin. Infect. Dis. 2008, 46, 1121–1122. [Google Scholar] [CrossRef]
- Miller, W.R.; Arias, C.A. Eskape pathogens: Antimicrobial resistance, epidemiology, clinical impact and therapeutics. Nat. Rev. Microbiol. 2024, 22, 598–616. [Google Scholar] [CrossRef]
- Elfadadny, A.; Ragab, R.F.; AlHarbi, M.; Badshah, F.; Ibanez-Arancibia, E.; Farag, A.; Hendawy, A.O.; De Los Rios-Escalante, P.R.; Aboubakr, M.; Zakai, S.A.; et al. Antimicrobial resistance of Pseudomonas aeruginosa: Navigating clinical impacts, current resistance trends, and innovations in breaking therapies. Front. Microbiol. 2024, 15, 1374466. [Google Scholar] [CrossRef]
- Elfadadny, A.; Uchiyama, J.; Goto, K.; Imanishi, I.; Ragab, R.F.; Nageeb, W.M.; Iyori, K.; Toyoda, Y.; Tsukui, T.; Ide, K.; et al. Antimicrobial resistance and genotyping of Pseudomonas aeruginosa isolated from the ear canals of dogs in Japan. Front. Vet. Sci. 2023, 10, 1074127. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious diseases society of America 2023 guidance on the treatment of antimicrobial resistant gram-negative infections. Clin. Infect. Dis. 2023, 1, 1–53. [Google Scholar] [CrossRef]
- Eid, R.; Dabar, G.; Hanna, L.R.; Saliba, G.; Riachy, M.; Choucair, J.; Saliba, R. Comparison of antimicrobial resistance in Pseudomonas aeruginosa from intensive care and non-intensive care units and its impact on treatment decisions. Sci. Rep. 2025, 15, 11288. [Google Scholar] [CrossRef]
- Furusawa, T.; Iwano, H.; Hiyashimizu, Y.; Matsubara, K.; Higuchi, H.; Nagahata, H.; Niwa, H.; Katayama, Y.; Kinoshita, Y.; Hagiwara, K.; et al. Phage therapy is effective in a mouse model of bacterial equine keratitis. Appl. Environ. Microbiol. 2016, 82, 5332–5339. [Google Scholar] [CrossRef]
- Fujiki, J.; Furusawa, T.; Munby, M.; Kawaguchi, C.; Matsuda, Y.; Shiokura, Y.; Nakamura, K.; Nakamura, T.; Sasaki, M.; Usui, M.; et al. Susceptibility of Pseudomonas aeruginosa veterinary isolates to pbunavirus pb1-like phages. Microbiol. Immunol. 2020, 64, 778–782. [Google Scholar] [CrossRef]
- Cota, I.; Sanchez-Romero, M.A.; Hernandez, S.B.; Pucciarelli, M.G.; Garcia-Del Portillo, F.; Casadesus, J. Epigenetic control of Salmonella enterica o-antigen chain length: A tradeoff between virulence and bacteriophage resistance. PLoS Genet. 2015, 11, e1005667. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Fernandez, R.; Olivenza, D.R.; Weyer, E.; Singh, A.; Casadesus, J.; Sanchez-Romero, M.A. Evolution of a bistable genetic system in fluctuating and nonfluctuating environments. Proc. Natl. Acad. Sci. USA 2024, 121, e2322371121. [Google Scholar] [CrossRef]
- Hall, A.R.; De Vos, D.; Friman, V.P.; Pirnay, J.P.; Buckling, A. Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa In Vitro and in wax moth larvae. Appl. Environ. Microbiol. 2012, 78, 5646–5652. [Google Scholar] [CrossRef]
- Yang, Y.; Shen, W.; Zhong, Q.; Chen, Q.; He, X.; Baker, J.L.; Xiong, K.; Jin, X.; Wang, J.; Hu, F.; et al. Development of a bacteriophage cocktail to constrain the emergence of phage-resistant Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 327. [Google Scholar] [CrossRef]
- Li, G.; Shen, M.; Yang, Y.; Le, S.; Li, M.; Wang, J.; Zhao, Y.; Tan, Y.; Hu, F.; Lu, S. Adaptation of Pseudomonas aeruginosa to phage pap1 predation via o-antigen polymerase mutation. Front. Microbiol. 2018, 9, 1170. [Google Scholar] [CrossRef]
- Chan, B.K.; Sistrom, M.; Wertz, J.E.; Kortright, K.E.; Narayan, D.; Turner, P.E. Phage selection restores antibiotic sensitivity in mdr Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 26717. [Google Scholar] [CrossRef]
- Markwitz, P.; Lood, C.; Olszak, T.; van Noort, V.; Lavigne, R.; Drulis-Kawa, Z. Genome-driven elucidation of phage-host interplay and impact of phage resistance evolution on bacterial fitness. ISME J. 2022, 16, 533–542. [Google Scholar] [CrossRef]
- Martins, L.F.; Dos Santos Junior, A.P.; Nicastro, G.G.; Scheunemann, G.; Angeli, C.B.; Rossi, F.P.N.; Quaggio, R.B.; Palmisano, G.; Sgro, G.G.; Ishida, K.; et al. Phages zc01 and zc03 require type-iv pilus for Pseudomonas aeruginosa infection and have a potential for therapeutic applications. Microbiol. Spectr. 2024, 12, e0152724. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Son, B.; Kim, Y.; Kim, H.; Nam, G.; Shin, H.; Ryu, S. Targeted dual-receptor phage cocktail against Cronobacter sakazakii: Insights into phage-host interactions and resistance mechanisms. Front. Microbiol. 2024, 15, 1468686. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Soto, C.E.; McClelland, M.; Kropinski, A.M.; Lin, J.T.; Khursigara, C.M.; Anany, H. Multireceptor phage cocktail against Salmonella enterica to circumvent phage resistance. Microlife 2024, 5, uqae003. [Google Scholar] [CrossRef]
- Marchi, J.; Minh, C.N.N.; Debarbieux, L.; Weitz, J.S. Multi-strain phage induced clearance of bacterial infections. PLoS Comput. Biol. 2025, 21, e1012793. [Google Scholar] [CrossRef]
- Wright, R.C.T.; Friman, V.P.; Smith, M.C.M.; Brockhurst, M.A. Cross-resistance is modular in bacteria-phage interactions. PLoS Biol. 2018, 16, e2006057. [Google Scholar] [CrossRef]
- Naknaen, A.; Samernate, T.; Wannasrichan, W.; Surachat, K.; Nonejuie, P.; Chaikeeratisak, V. Combination of genetically diverse Pseudomonas phages enhances the cocktail efficiency against bacteria. Sci. Rep. 2023, 13, 8921. [Google Scholar] [CrossRef]
- Ulrich, L.; Steiner, L.X.; Giez, C.; Lachnit, T. Optimizing bacteriophage treatment of resistant Pseudomonas. mSphere 2024, 9, e0070723. [Google Scholar] [CrossRef] [PubMed]
- Aslam, S.; Lampley, E.; Wooten, D.; Karris, M.; Benson, C.; Strathdee, S.; Schooley, R.T. Lessons learned from the first 10 consecutive cases of intravenous bacteriophage therapy to treat multidrug-resistant bacterial infections at a single center in the United States. Open Forum Infect. Dis. 2020, 7, ofaa389. [Google Scholar] [CrossRef]
- Aslam, S.; Roach, D.; Nikolich, M.P.; Biswas, B.; Schooley, R.T.; Lilly-Bishop, K.A.; Rice, G.K.; Cer, R.Z.; Hamilton, T.; Henry, M.; et al. Pseudomonas aeruginosa ventricular assist device infections: Findings from ineffective phage therapies in five cases. Antimicrob. Agents Chemother. 2024, 68, e0172823. [Google Scholar] [CrossRef]
- Maddocks, S.; Fabijan, A.P.; Ho, J.; Lin, R.C.Y.; Ben Zakour, N.L.; Dugan, C.; Kliman, I.; Branston, S.; Morales, S.; Iredell, J.R. Bacteriophage therapy of ventilator-associated pneumonia and empyema caused by Pseudomonas aeruginosa. Am. J. Respir. Crit. Care Med. 2019, 200, 1179–1181. [Google Scholar] [CrossRef] [PubMed]
- Ferry, T.; Kolenda, C.; Batailler, C.; Gaillard, R.; Gustave, C.A.; Lustig, S.; Fevre, C.; Petitjean, C.; Leboucher, G.; Laurent, F.; et al. Case report: Arthroscopic “debridement antibiotics and implant retention” with local injection of personalized phage therapy to salvage a relapsing Pseudomonas aeruginosa prosthetic knee infection. Front. Med. 2021, 8, 569159. [Google Scholar] [CrossRef]
- Kohler, T.; Luscher, A.; Falconnet, L.; Resch, G.; McBride, R.; Mai, Q.A.; Simonin, J.L.; Chanson, M.; Maco, B.; Galiotto, R.; et al. Personalized aerosolised bacteriophage treatment of a chronic lung infection due to multidrug-resistant Pseudomonas aeruginosa. Nat. Commun. 2023, 14, 3629. [Google Scholar] [CrossRef] [PubMed]
- Teney, C.; Poupelin, J.C.; Briot, T.; Le Bouar, M.; Fevre, C.; Brosset, S.; Martin, O.; Valour, F.; Roussel-Gaillard, T.; Leboucher, G.; et al. Phage therapy in a burn patient colonized with extensively drug-resistant Pseudomonas aeruginosa responsible for relapsing ventilator-associated pneumonia and bacteriemia. Viruses 2024, 16, 1080. [Google Scholar] [CrossRef]
- Pirnay, J.P.; Djebara, S.; Steurs, G.; Griselain, J.; Cochez, C.; De Soir, S.; Glonti, T.; Spiessens, A.; Vanden Berghe, E.; Green, S.; et al. Personalized bacteriophage therapy outcomes for 100 consecutive cases: A multicentre, multinational, retrospective observational study. Nat. Microbiol. 2024, 9, 1434–1453. [Google Scholar] [CrossRef]
- Van Nieuwenhuyse, B.; Van der Linden, D.; Chatzis, O.; Lood, C.; Wagemans, J.; Lavigne, R.; Schroven, K.; Paeshuyse, J.; de Magnee, C.; Sokal, E.; et al. Bacteriophage-antibiotic combination therapy against extensively drug-resistant Pseudomonas aeruginosa infection to allow liver transplantation in a toddler. Nat. Commun. 2022, 13, 5725. [Google Scholar] [CrossRef]
- Onsea, J.; Soentjens, P.; Djebara, S.; Merabishvili, M.; Depypere, M.; Spriet, I.; De Munter, P.; Debaveye, Y.; Nijs, S.; Vanderschot, P.; et al. Bacteriophage application for difficult-to-treat musculoskeletal infections: Development of a standardized multidisciplinary treatment protocol. Viruses 2019, 11, 891. [Google Scholar] [CrossRef]
- Jennes, S.; Merabishvili, M.; Soentjens, P.; Pang, K.W.; Rose, T.; Keersebilck, E.; Soete, O.; Francois, P.M.; Teodorescu, S.; Verween, G.; et al. Use of bacteriophages in the treatment of colistin-only-sensitive Pseudomonas aeruginosa septicaemia in a patient with acute kidney injury—A case report. Crit. Care 2017, 21, 129. [Google Scholar] [CrossRef]
- Racenis, K.; Rezevska, D.; Madelane, M.; Lavrinovics, E.; Djebara, S.; Petersons, A.; Kroica, J. Use of phage cocktail bfc 1.10 in combination with ceftazidime-avibactam in the treatment of multidrug-resistant Pseudomonas aeruginosa femur osteomyelitis—A case report. Front. Med. 2022, 9, 851310. [Google Scholar] [CrossRef]
- Chan, B.K.; Turner, P.E.; Kim, S.; Mojibian, H.R.; Elefteriades, J.A.; Narayan, D. Phage treatment of an aortic graft infected with Pseudomonas aeruginosa. Evol. Med. Public Health 2018, 2018, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Liu, Z.; Tan, X.; Wang, H.; Liang, Y.; Kong, Y.; Sun, W.; Sun, L.; Ma, Y.; Lu, H. Bacteriophage therapy for empyema caused by carbapenem-resistant Pseudomonas aeruginosa. Biosci. Trends 2022, 16, 158–162. [Google Scholar] [CrossRef] [PubMed]
- Ferry, T.; Kolenda, C.; Laurent, F.; Leboucher, G.; Merabischvilli, M.; Djebara, S.; Gustave, C.A.; Perpoint, T.; Barrey, C.; Pirnay, J.P.; et al. Personalized bacteriophage therapy to treat pandrug-resistant spinal Pseudomonas aeruginosa infection. Nat. Commun. 2022, 13, 4239. [Google Scholar] [CrossRef] [PubMed]
- Duplessis, C.; Biswas, B.; Hanisch, B.; Perkins, M.; Henry, M.; Quinones, J.; Wolfe, D.; Estrella, L.; Hamilton, T. Refractory pseudomonas bacteremia in a 2-year-old sterilized by bacteriophage therapy. J. Pediatr. Infect. Dis. Soc. 2018, 7, 253–256. [Google Scholar] [CrossRef]
- Racenis, K.; Lacis, J.; Rezevska, D.; Mukane, L.; Vilde, A.; Putnins, I.; Djebara, S.; Merabishvili, M.; Pirnay, J.P.; Kalnina, M.; et al. Successful bacteriophage-antibiotic combination therapy against multidrug-resistant Pseudomonas aeruginosa left ventricular assist device driveline infection. Viruses 2023, 15, 1210. [Google Scholar] [CrossRef]
- Eiferman, V.; Vion, P.A.; Bleibtreu, A. Phage therapy as a rescue treatment for recurrent Pseudomonas aeruginosa bentall infection. Viruses 2025, 17, 123. [Google Scholar] [CrossRef]
- Blasco, L.; Lopez-Hernandez, I.; Rodriguez-Fernandez, M.; Perez-Florido, J.; Casimiro-Soriguer, C.S.; Djebara, S.; Merabishvili, M.; Pirnay, J.P.; Rodriguez-Bano, J.; Tomas, M.; et al. Case report: Analysis of phage therapy failure in a patient with a Pseudomonas aeruginosa prosthetic vascular graft infection. Front. Med. 2023, 10, 1199657. [Google Scholar] [CrossRef]
- Onallah, H.; Hazan, R.; Nir-Paz, R.; PASA16 Study Group; Brownstein, M.J.; Fackler, J.R.; Horne, B.; Hopkins, R.; Basu, S.; Yerushalmy, O.; et al. Refractory Pseudomonas aeruginosa infections treated with phage PASA16: A compassionate use case series. Med 2023, 4, 600–611.e604. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Freeman, K.G.; Nguyen, J.A.; Bahadirli-Talbott, A.; Smith, B.E.; Wu, A.E.; Ong, A.S.; Lin, C.T.; Ruppel, L.C.; Parrish, N.M.; et al. Potent antibody-mediated neutralization limits bacteriophage treatment of a pulmonary Mycobacterium abscessus infection. Nat. Med. 2021, 27, 1357–1361. [Google Scholar] [CrossRef]
- Bao, J.; Wu, N.; Zeng, Y.; Chen, L.; Li, L.; Yang, L.; Zhang, Y.; Guo, M.; Li, L.; Li, J.; et al. Non-active antibiotic and bacteriophage synergism to successfully treat recurrent urinary tract infection caused by extensively drug-resistant Klebsiella pneumoniae. Emerg. Microbes Infect. 2020, 9, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Zhvania, P.; Hoyle, N.S.; Nadareishvili, L.; Nizharadze, D.; Kutateladze, M. Phage therapy in a 16-year-old boy with netherton syndrome. Front. Med. 2017, 4, 94. [Google Scholar] [CrossRef]
- Jault, P.; Leclerc, T.; Jennes, S.; Pirnay, J.P.; Que, Y.A.; Resch, G.; Rousseau, A.F.; Ravat, F.; Carsin, H.; Le Floch, R.; et al. Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): A randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 2019, 19, 35–45. [Google Scholar] [CrossRef]
- Hampton, H.G.; Watson, B.N.J.; Fineran, P.C. The arms race between bacteria and their phage foes. Nature 2020, 577, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Jdeed, G.; Kravchuk, B.; Tikunova, N.V. Factors affecting phage-bacteria coevolution dynamics. Viruses 2025, 17, 235. [Google Scholar] [CrossRef] [PubMed]
- Borin, J.M.; Lee, J.J.; Lucia-Sanz, A.; Gerbino, K.R.; Weitz, J.S.; Meyer, J.R. Rapid bacteria-phage coevolution drives the emergence of multiscale networks. Science 2023, 382, 674–678. [Google Scholar] [CrossRef]
- Goldhill, D.H.; Turner, P.E. The evolution of life history trade-offs in viruses. Curr. Opin. Virol. 2014, 8, 79–84. [Google Scholar] [CrossRef]
- Oechslin, F. Resistance development to bacteriophages occurring during bacteriophage therapy. Viruses 2018, 10, 351. [Google Scholar] [CrossRef]
- Mangalea, M.R.; Duerkop, B.A. Fitness trade-offs resulting from bacteriophage resistance potentiate synergistic antibacterial strategies. Infect. Immun. 2020, 88, e00926-19. [Google Scholar] [CrossRef]
- Li, N.; Zeng, Y.; Wang, M.; Bao, R.; Chen, Y.; Li, X.; Pan, J.; Zhu, T.; Hu, B.; Tan, D. Characterization of phage resistance and their impacts on bacterial fitness in Pseudomonas aeruginosa. Microbiol. Spectr. 2022, 10, e0207222. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, T.; Li, L.; Zheng, C.; Tan, D.; Wu, N.; Wang, M.; Zhu, T. Characterization of Pseudomonas aeruginosa bacteriophage l5 which requires type iv pili for infection. Front. Microbiol. 2022, 13, 907958. [Google Scholar] [CrossRef]
- Thongchol, J.; Yu, Z.; Harb, L.; Lin, Y.; Koch, M.; Theodore, M.; Narsaria, U.; Shaevitz, J.; Gitai, Z.; Wu, Y.; et al. Removal of Pseudomonas type iv pili by a small rna virus. Science 2024, 384, eadl0635. [Google Scholar] [CrossRef] [PubMed]
- Nordstrom, H.R.; Evans, D.R.; Finney, A.G.; Westbrook, K.J.; Zamora, P.F.; Hofstaedter, C.E.; Yassin, M.H.; Pradhan, A.; Iovleva, A.; Ernst, R.K.; et al. Genomic characterization of lytic bacteriophages targeting genetically diverse Pseudomonas aeruginosa clinical isolates. iScience 2022, 25, 104372. [Google Scholar] [CrossRef] [PubMed]
- Mi, Y.; He, Y.; Mi, J.; Huang, Y.; Fan, H.; Song, L.; An, X.; Xu, S.; Li, M.; Tong, Y. Genetic and phenotypic analysis of phage-resistant mutant fitness triggered by phage-host interactions. Int. J. Mol. Sci. 2023, 24, 15594. [Google Scholar] [CrossRef]
- Floyd, M.; Winn, M.; Cullen, C.; Sil, P.; Chassaing, B.; Yoo, D.G.; Gewirtz, A.T.; Goldberg, J.B.; McCarter, L.L.; Rada, B. Swimming motility mediates the formation of neutrophil extracellular traps induced by flagellated Pseudomonas aeruginosa. PLoS Pathog. 2016, 12, e1005987. [Google Scholar] [CrossRef]
- Demirdjian, S.; Schutz, K.; Wargo, M.J.; Lam, J.S.; Berwin, B. The effect of loss of o-antigen ligase on phagocytic susceptibility of motile and non-motile Pseudomonas aeruginosa. Mol. Immunol. 2017, 92, 106–115. [Google Scholar] [CrossRef]
- Wannasrichan, W.; Htoo, H.H.; Suwansaeng, R.; Pogliano, J.; Nonejuie, P.; Chaikeeratisak, V. Phage-resistant Pseudomonas aeruginosa against a novel lytic phage jj01 exhibits hypersensitivity to colistin and reduces biofilm production. Front. Microbiol. 2022, 13, 1004733. [Google Scholar] [CrossRef]
- Roach, D.R.; Leung, C.Y.; Henry, M.; Morello, E.; Singh, D.; Di Santo, J.P.; Weitz, J.S.; Debarbieux, L. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host Microbe 2017, 22, 38–47.e4. [Google Scholar] [CrossRef] [PubMed]
- Masuda, N.; Sakagawa, E.; Ohya, S.; Gotoh, N.; Tsujimoto, H.; Nishino, T. Substrate specificities of MexAB-OprM, MexCD-OprJ, and MexXY-oprM efflux pumps in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2000, 44, 3322–3327. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Kimura, N.; Mima, T.; Mizushima, T.; Tsuchiya, T. Roles of MexXY- and MexAB-multidrug efflux pumps in intrinsic multidrug resistance of Pseudomonas aeruginosa PAO1. J. Gen. Appl. Microbiol. 2001, 47, 27–32. [Google Scholar] [CrossRef]
- Gurney, J.; Pradier, L.; Griffin, J.S.; Gougat-Barbera, C.; Chan, B.K.; Turner, P.E.; Kaltz, O.; Hochberg, M.E. Phage steering of antibiotic-resistance evolution in the bacterial pathogen, Pseudomonas aeruginosa. Evol. Med. Public Health 2020, 2020, 148–157. [Google Scholar] [CrossRef]
- Koderi Valappil, S.; Shetty, P.; Deim, Z.; Terhes, G.; Urban, E.; Vaczi, S.; Patai, R.; Polgar, T.; Pertics, B.Z.; Schneider, G.; et al. Survival comes at a cost: A coevolution of phage and its host leads to phage resistance and antibiotic sensitivity of Pseudomonas aeruginosa multidrug resistant strains. Front. Microbiol. 2021, 12, 783722. [Google Scholar] [CrossRef]
- Nakamura, K.; Fujiki, J.; Nakamura, T.; Furusawa, T.; Gondaira, S.; Usui, M.; Higuchi, H.; Tamura, Y.; Iwano, H. Fluctuating bacteriophage-induced galU deficiency region is involved in trade-off effects on the phage and fluoroquinolone sensitivity in Pseudomonas aeruginosa. Virus Res. 2021, 306, 198596. [Google Scholar] [CrossRef]
- Fujiki, J.; Nakamura, K.; Ishiguro, Y.; Iwano, H. Using phage to drive selections toward restoring antibiotic sensitivity in Pseudomonas aeruginosa via chromosomal deletions. Front. Microbiol. 2024, 15, 1401234. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Chen, H.; Qian, C.; Zhao, Y.; Wang, W.; Liu, Y.; Xu, M.; Cao, J.; Zhou, T.; Wu, Q. Resistance, mechanism, and fitness cost of specific bacteriophages for Pseudomonas aeruginosa. mSphere 2024, 9, e0055323. [Google Scholar] [CrossRef]
- Menon, N.D.; Penziner, S.; Montano, E.T.; Zurich, R.; Pride, D.T.; Nair, B.G.; Kumar, G.B.; Nizet, V. Increased innate immune susceptibility in hyperpigmented bacteriophage-resistant mutants of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2022, 66, e0023922. [Google Scholar] [CrossRef]
- Chan, B.K.; Stanley, G.L.; Kortright, K.E.; Vill, A.C.; Modak, M.; Ott, I.M.; Sun, Y.; Wurstle, S.; Grun, C.N.; Kazmierczak, B.I.; et al. Personalized inhaled bacteriophage therapy for treatment of multidrug-resistant Pseudomonas aeruginosa in cystic fibrosis. Nat. Med. 2025, 31, 1494–1501. [Google Scholar] [CrossRef]
- Kortright, K.E.; Doss-Gollin, S.; Chan, B.K.; Turner, P.E. Evolution of bacterial cross-resistance to lytic phages and albicidin antibiotic. Front. Microbiol. 2021, 12, 658374. [Google Scholar] [CrossRef]
- Burmeister, A.R.; Fortier, A.; Roush, C.; Lessing, A.J.; Bender, R.G.; Barahman, R.; Grant, R.; Chan, B.K.; Turner, P.E. Pleiotropy complicates a trade-off between phage resistance and antibiotic resistance. Proc. Natl. Acad. Sci. USA 2020, 117, 11207–11216. [Google Scholar] [CrossRef] [PubMed]
- Nambu, Y.; Jona-Lasinio, G. Dynamical model of elementary particles based on an analogy with superconductivity. I. Phys. Rev. 1961, 122, 345–358. [Google Scholar] [CrossRef]
- Nambu, Y.; Jona-Lasinio, G. Dynamical model of elementary particles based on an analogy with superconductivity. II. Phys. Rev. 1961, 124, 246–254. [Google Scholar] [CrossRef]
- Shen, M.; Zhang, H.; Shen, W.; Zou, Z.; Lu, S.; Li, G.; He, X.; Agnello, M.; Shi, W.; Hu, F.; et al. Pseudomonas aeruginosa MutL promotes large chromosomal deletions through non-homologous end joining to prevent bacteriophage predation. Nucleic Acids Res. 2018, 46, 4505–4514. [Google Scholar] [CrossRef]
- Le, S.; Yao, X.; Lu, S.; Tan, Y.; Rao, X.; Li, M.; Jin, X.; Wang, J.; Zhao, Y.; Wu, N.C.; et al. Chromosomal DNA deletion confers phage resistance to Pseudomonas aeruginosa. Sci. Rep. 2014, 4, 4738. [Google Scholar] [CrossRef] [PubMed]
- Beard, W.A.; Wilson, S.H. Structural design of a eukaryotic DNA repair polymerase: DNA polymerase beta. Mutat. Res. 2000, 460, 231–244. [Google Scholar] [CrossRef]
- Ortega, J.; Lee, G.S.; Gu, L.; Yang, W.; Li, G.M. Mispair-bound human MutS-MutL complex triggers DNA incisions and activates mismatch repair. Cell Res. 2021, 31, 542–553. [Google Scholar] [CrossRef]
- Prunier, A.L.; Leclercq, R. Role of mutS and mutL genes in hypermutability and recombination in Staphylococcus aureus. J. Bacteriol. 2005, 187, 3455–3464. [Google Scholar] [CrossRef]
- Deriano, L.; Roth, D.B. Modernizing the nonhomologous end-joining repertoire: Alternative and classical nhej share the stage. Annu. Rev. Genet. 2013, 47, 433–455. [Google Scholar] [CrossRef]
- Lieber, M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010, 79, 181–211. [Google Scholar] [CrossRef]
- Zhu, H.; Shuman, S. Gap filling activities of Pseudomonas DNA ligase d (ligd) polymerase and functional interactions of ligd with the DNA end-binding ku protein. J. Biol. Chem. 2010, 285, 4815–4825. [Google Scholar] [CrossRef]
- Stephanou, N.C.; Gao, F.; Bongiorno, P.; Ehrt, S.; Schnappinger, D.; Shuman, S.; Glickman, M.S. Mycobacterial nonhomologous end joining mediates mutagenic repair of chromosomal double-strand DNA breaks. J. Bacteriol. 2007, 189, 5237–5246. [Google Scholar] [CrossRef]
- Bianco, P.R.; Kowalczykowski, S.C. The recombination hotspot chi is recognized by the translocating RecBCD enzyme as the single strand of DNA containing the sequence 5′-GCTGGTGG-3′. Proc. Natl. Acad. Sci. USA 1997, 94, 6706–6711. [Google Scholar] [CrossRef]
- Canfield, G.S.; Duerkop, B.A. Molecular mechanisms of enterococcal-bacteriophage interactions and implications for human health. Curr. Opin. Microbiol. 2020, 56, 38–44. [Google Scholar] [CrossRef]
- Uemura, K.; Sato, T.; Yamamoto, S.; Ogasawara, N.; Toyting, J.; Aoki, K.; Takasawa, A.; Koyama, M.; Saito, A.; Wada, T.; et al. Rapid and integrated bacterial evolution analysis unveils gene mutations and clinical risk of Klebsiella pneumoniae. Nat. Commun. 2025, 16, 2917. [Google Scholar] [CrossRef] [PubMed]
- Chu, N.D.; Clarke, S.A.; Timberlake, S.; Polz, M.F.; Grossman, A.D.; Alm, E.J. A mobile element in mutS drives hypermutation in a marine vibrio. mBio 2017, 8, e02045-16. [Google Scholar] [CrossRef] [PubMed]
- Little, J.W. Autodigestion of lexA and phage lambda repressors. Proc. Natl. Acad. Sci. USA 1984, 81, 1375–1379. [Google Scholar] [CrossRef] [PubMed]
- Fernandez De Henestrosa, A.R.; Ogi, T.; Aoyagi, S.; Chafin, D.; Hayes, J.J.; Ohmori, H.; Woodgate, R. Identification of additional genes belonging to the LexA regulon in Escherichia coli. Mol. Microbiol. 2000, 35, 1560–1572. [Google Scholar] [CrossRef]
- Little, J.W. Mechanism of specific LexA cleavage: Autodigestion and the role of RecA coprotease. Biochimie 1991, 73, 411–421. [Google Scholar] [CrossRef]
- Fornelos, N.; Browning, D.F.; Butala, M. The use and abuse of LexA by mobile genetic elements. Trends Microbiol. 2016, 24, 391–401. [Google Scholar] [CrossRef]
- Tippin, B.; Pham, P.; Goodman, M.F. Error-prone replication for better or worse. Trends Microbiol. 2004, 12, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Cirz, R.T.; Romesberg, F.E. Controlling mutation: Intervening in evolution as a therapeutic strategy. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Ennis, D.G.; Fisher, B.; Edmiston, S.; Mount, D.W. Dual role for Escherichia coli RecA protein in sos mutagenesis. Proc. Natl. Acad. Sci. USA 1985, 82, 3325–3329. [Google Scholar] [CrossRef] [PubMed]
- Sagi, D.; Tlusty, T.; Stavans, J. High fidelity of RecA-catalyzed recombination: A watchdog of genetic diversity. Nucleic Acids Res. 2006, 34, 5021–5031. [Google Scholar] [CrossRef]
- Shibata, T.; Ikawa, S.; Iwasaki, W.; Sasanuma, H.; Masai, H.; Hirota, K. Homology recognition without double-stranded DNA-strand separation in d-loop formation by RecA. Nucleic Acids Res. 2024, 52, 2565–2577. [Google Scholar] [CrossRef]
- Smith, G.R. How RecBCD enzyme and Chi promote DNA break repair and recombination: A molecular biologist’s view. Microbiol. Mol. Biol. Rev. 2012, 76, 217–228. [Google Scholar] [CrossRef]
- Dillingham, M.S.; Kowalczykowski, S.C. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008, 72, 642–671, Table of Contents. [Google Scholar] [CrossRef]
- Levy, A.; Goren, M.G.; Yosef, I.; Auster, O.; Manor, M.; Amitai, G.; Edgar, R.; Qimron, U.; Sorek, R. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature 2015, 520, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Bobay, L.M.; Touchon, M.; Rocha, E.P. Manipulating or superseding host recombination functions: A dilemma that shapes phage evolvability. PLoS Genet. 2013, 9, e1003825. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.; Wilkinson, O.J.; Feyerherm, C.; Fletcher, E.E.; Wigley, D.B.; Dillingham, M.S. Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub. Elife 2022, 11, e83409. [Google Scholar] [CrossRef]
- Tabor, S.; Huber, H.E.; Richardson, C.C. Escherichia coli thioredoxin confers processivity on the DNA polymerase activity of the gene 5 protein of bacteriophage T7. J. Biol. Chem. 1987, 262, 16212–16223. [Google Scholar] [CrossRef] [PubMed]
- Savalia, D.; Robins, W.; Nechaev, S.; Molineux, I.; Severinov, K. The role of the T7 Gp2 inhibitor of host RNA polymerase in phage development. J. Mol. Biol. 2010, 402, 118–126. [Google Scholar] [CrossRef]
- Tabib-Salazar, A.; Liu, B.; Barker, D.; Burchell, L.; Qimron, U.; Matthews, S.J.; Wigneshweraraj, S. T7 phage factor required for managing RpoS in Escherichia coli. Proc. Natl. Acad. Sci. USA 2018, 115, E5353–E5362. [Google Scholar] [CrossRef]
- Zhang, X.; Studier, F.W. Multiple roles of T7 RNA polymerase and T7 lysozyme during bacteriophage T7 infection. J. Mol. Biol. 2004, 340, 707–730. [Google Scholar] [CrossRef]
- Yamashita, W.; Chihara, K.; Azam, A.H.; Kondo, K.; Ojima, S.; Tamura, A.; Imanaka, M.; Nobrega, F.L.; Takahashi, Y.; Watashi, K.; et al. Phage engineering to overcome bacterial Tmn immunity in Dhillonvirus. Commun. Biol. 2025, 8, 290. [Google Scholar] [CrossRef]
- Kiga, K.; Tan, X.E.; Ibarra-Chavez, R.; Watanabe, S.; Aiba, Y.; Sato’o, Y.; Li, F.Y.; Sasahara, T.; Cui, B.; Kawauchi, M.; et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat. Commun. 2020, 11, 2934. [Google Scholar] [CrossRef]
- Mitsunaka, S.; Yamazaki, K.; Pramono, A.K.; Ikeuchi, M.; Kitao, T.; Ohara, N.; Kubori, T.; Nagai, H.; Ando, H. Synthetic engineering and biological containment of bacteriophages. Proc. Natl. Acad. Sci. USA 2022, 119, e2206739119. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).