Polyvalent Mannuronic Acid-Coated Gold Nanoparticles for Probing Multivalent Lectin–Glycan Interaction and Blocking Virus Infection
Abstract
1. Introduction
2. Experimental Section
2.1. Materials
2.2. Synthesis of [2-(2-Azidoethoxy)ethoxy]-α-d-Mannopyranosiduronic Acid (N3-EG2-ManA] [30,31,32]
2.2.1. Phenyl 2,3,4,6-Tetra-O-Acetyl-1-Thio-α-d-Mannopyranoside (1)
2.2.2. 2,3,4,5-Tetra-O-Acetyl-α-d-[2-(2-Azidoethoxy)ethoxy]mannopyranoside (2)
2.2.3. 2,3,4,5-Tetra-O-Hydroxy-α-d-[2-(2-Azidoethoxy)ethoxy]mannopyranoside (3)
2.2.4. [2-(2-Azidoethoxy)ethoxy] α-d-mannopyranosiduronic Acid (4)
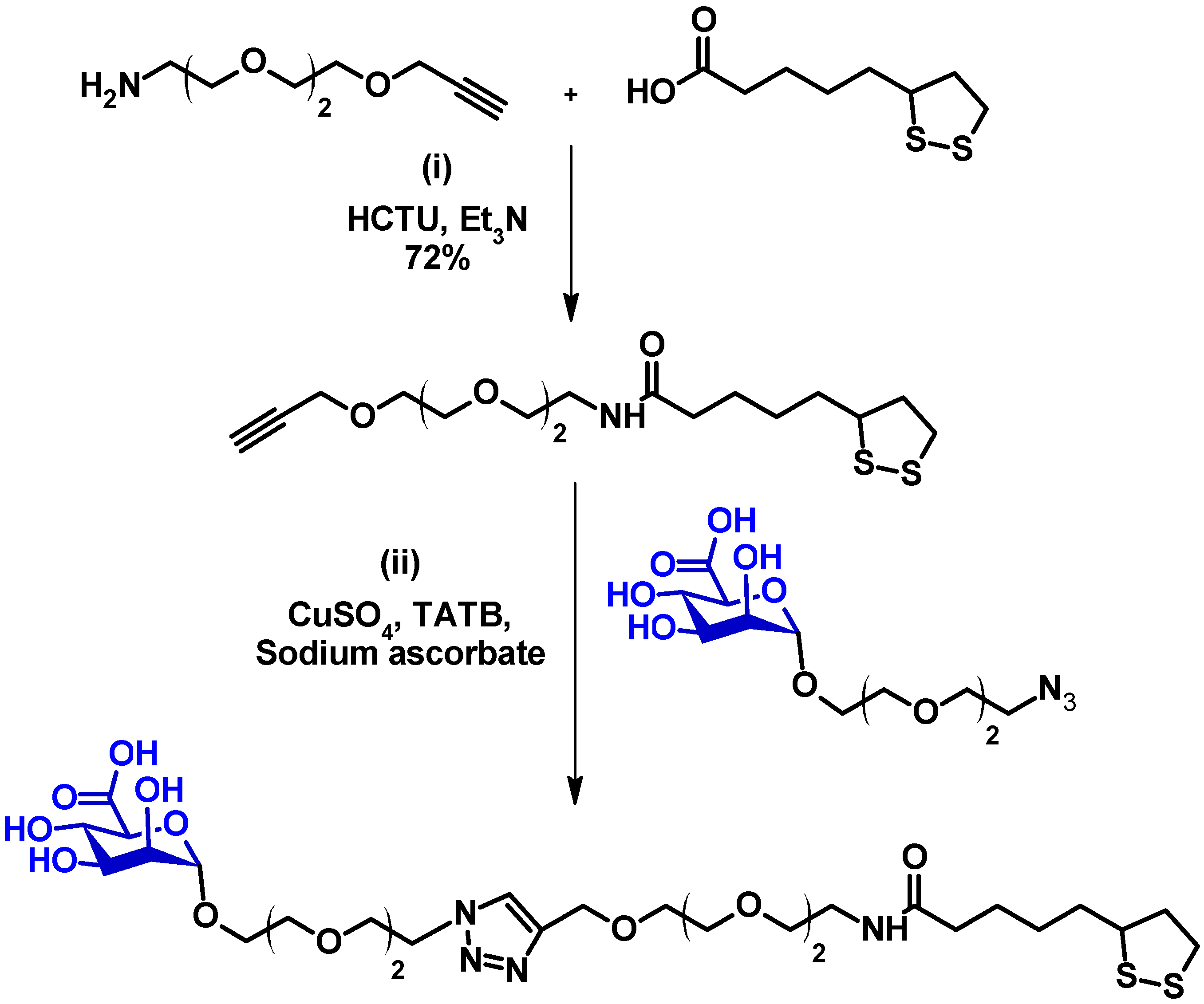
2.3. Synthesis of LA-EG2-ManA [14,19,20,21]
2.4. Preparation of Gx-ManA Conjugates [21]
2.5. Fluorescence Spectra [21]
2.6. Virus Inhibition Studies
2.7. Cytotoxicity Studies (Cell Titer-Glo Assay)
2.8. Data Analysis and Fitting [14,21]
3. Results and Discussion
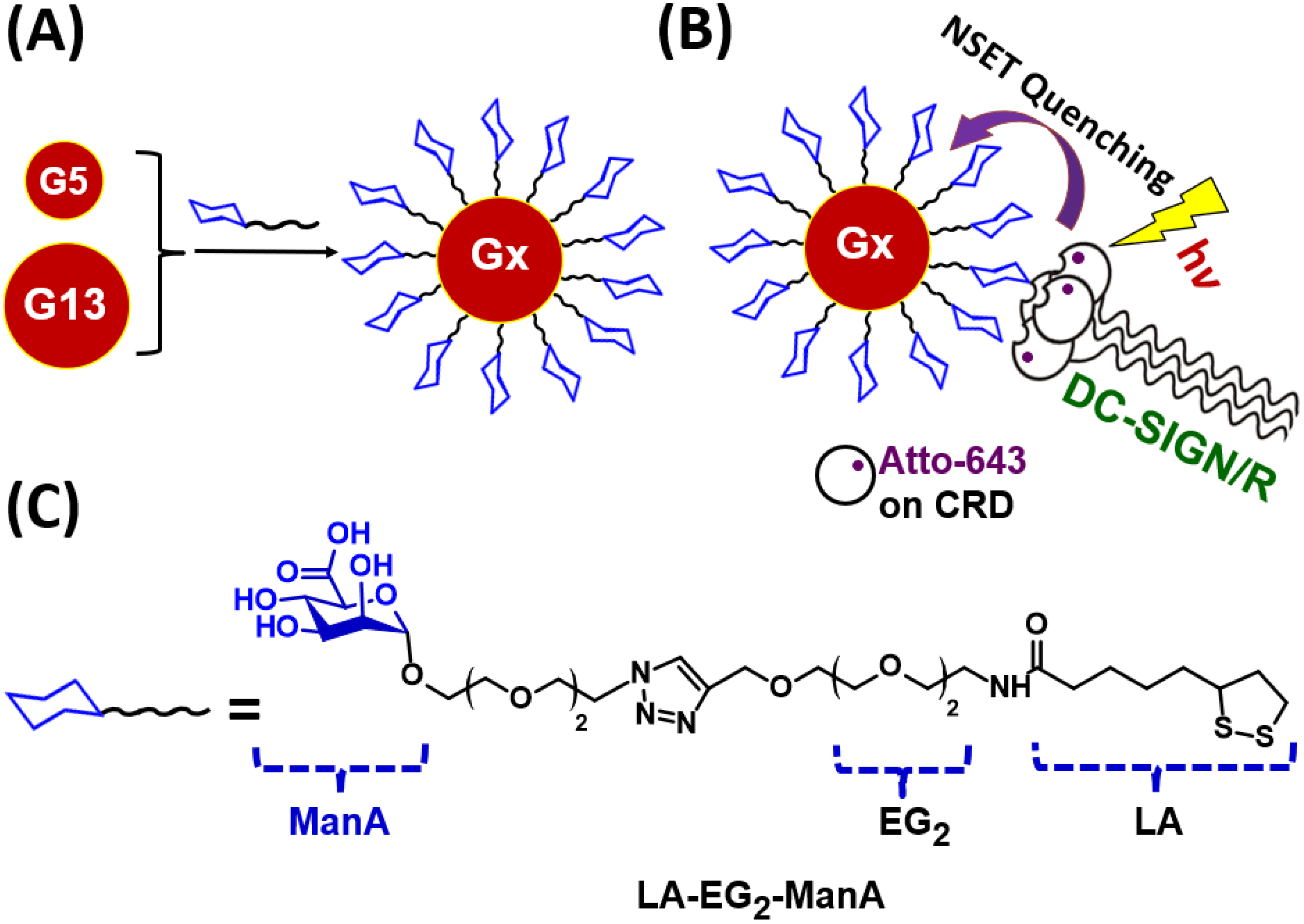
3.1. Ligand Design and Synthesis
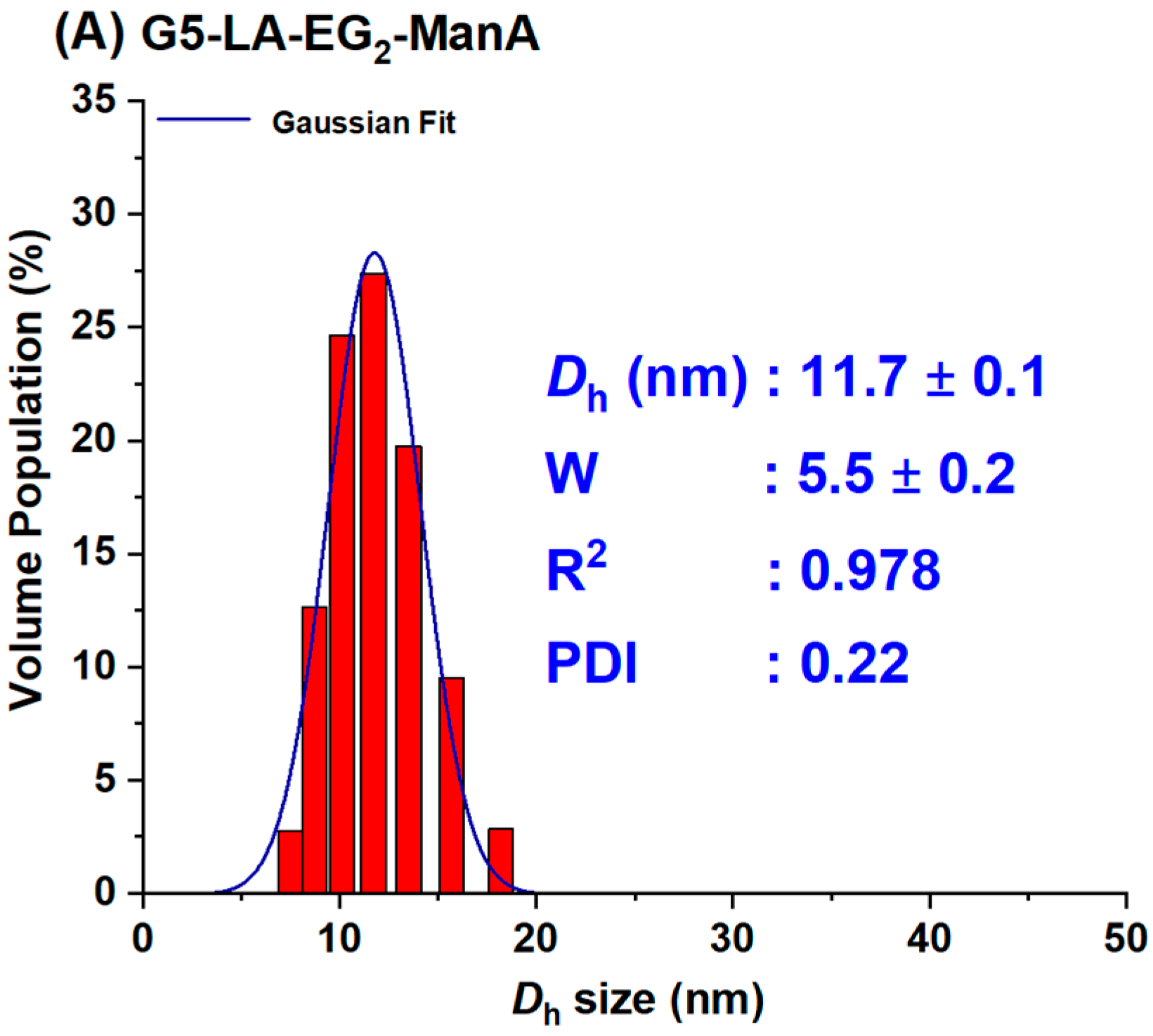
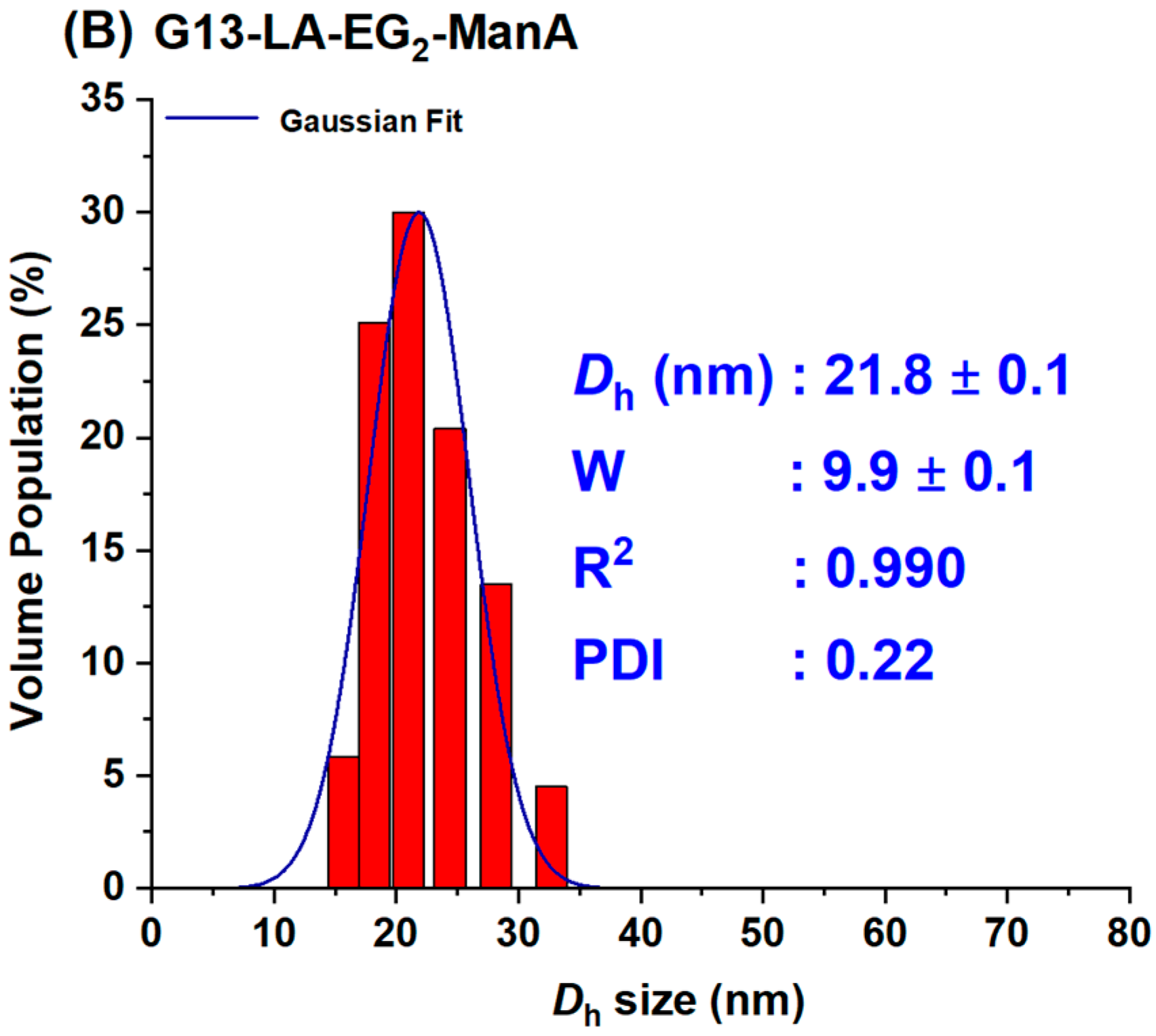
3.2. Preparation of GNP–ManA Conjugates
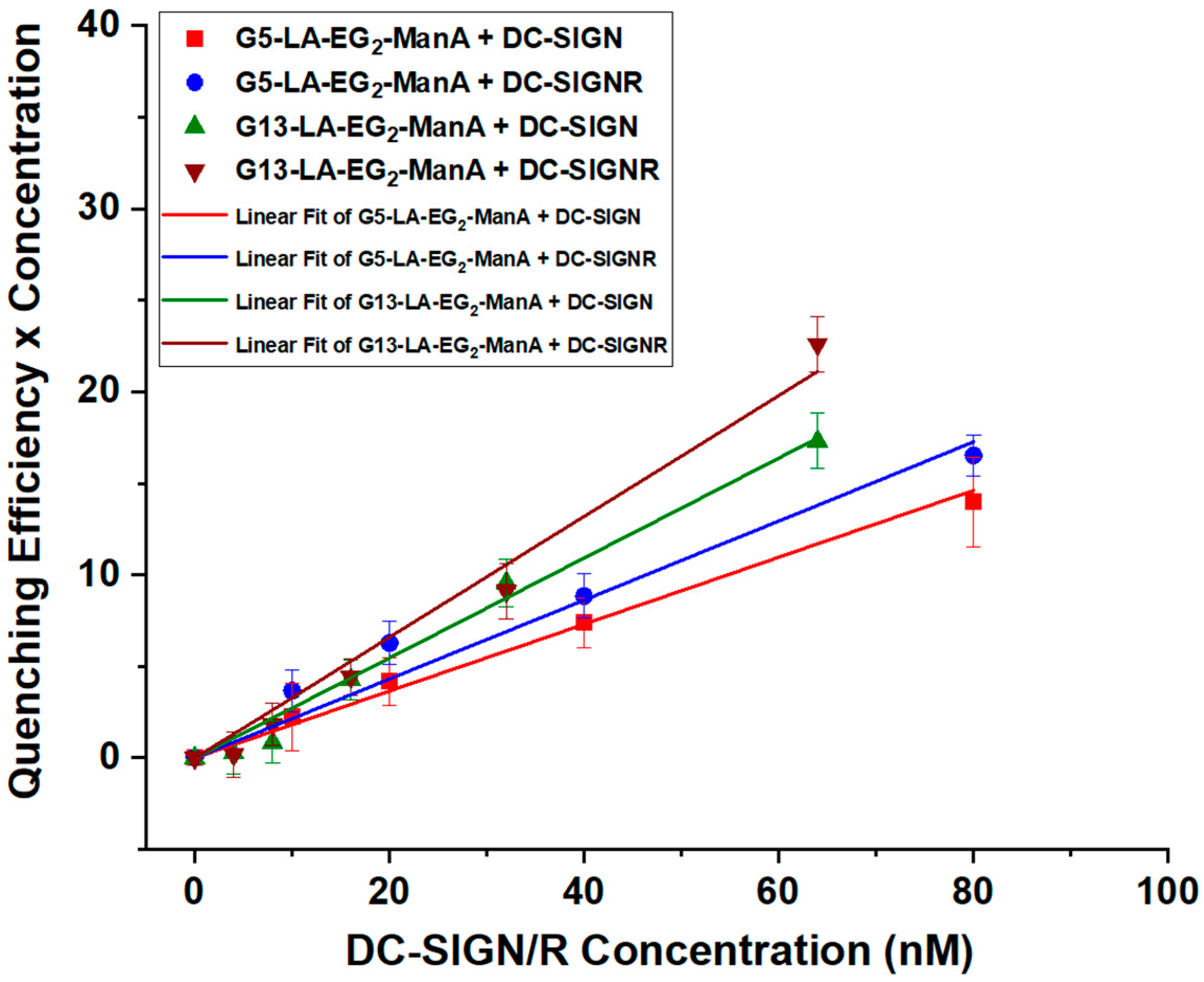
3.3. Quantifying GNP-Glycan-DC-SIGN/R Binding Affinity
3.4. Inhibition of DC-SIGN/R-Promoted EBOVpp Entry into Cells
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- W.H.O. WHO Coronavirus (COVID-19) Dashboard > Deaths [Dashboard]. 2023. Available online: https://data.who.int/dashboards/covid19/deaths (accessed on 2 May 2025).[Green Version]
- Wang, H.; Paulson, K.R.; Pease, S.A.; Watson, S.; Comfort, H.; Zheng, P.; Aravkin, A.Y.; Bisignano, C.; Barber, R.M.; Alam, T.; et al. Estimating excess mortality due to the COVID-19 pandemic: A systematic analysis of COVID-19-related mortality, 2020-21. Lancet 2022, 399, 1513–1536. [Google Scholar] [CrossRef]
- Bhatia, S.; Camacho, L.C.; Haag, R. Pathogen Inhibition by Multivalent Ligand Architectures. J. Am. Chem. Soc. 2016, 138, 8654–8666. [Google Scholar] [CrossRef]
- Mammen, M.; Choi, S.K.; Whitesides, G.M. Polyvalent Interactions in Biological Systems: Implications for Design and Use of Multivalent Ligands and Inhibitors. Angew. Chem. Int. Ed. Engl. 1998, 37, 2754–2794. [Google Scholar] [CrossRef]
- Drickamer, K.; Taylor, M.E. Recent insights into structures and functions of C-type lectins in the immune system. Curr. Opin. Struct. Biol. 2015, 34, 26–34. [Google Scholar] [CrossRef]
- Guo, Y.; Feinberg, H.; Conroy, E.; Mitchell, D.A.; Alvarez, R.; Blixt, O.; Taylor, M.E.; Weis, W.I.; Drickamer, K. Structural basis for distinct ligand-binding and targeting properties of the receptors DC-SIGN and DC-SIGNR. Nat. Struct. Mol. Biol. 2004, 11, 591–598. [Google Scholar] [CrossRef]
- Feinberg, H.; Mitchell, D.A.; Drickamer, K.; Weis, W.I. Structural basis for selective recognition of oligosaccharides by DC-SIGN and DC-SIGNR. Science 2001, 294, 2163–2166. [Google Scholar] [CrossRef]
- Bermejo-Jambrina, M.; Eder, J.; Helgers, L.C.; Hertoghs, N.; Nijmeijer, B.M.; Stunnenberg, M.; Geijtenbeek, T.B.H. C-Type Lectin Receptors in Antiviral Immunity and Viral Escape. Front. Immunol. 2018, 9, 590. [Google Scholar] [CrossRef]
- Fasting, C.; Schalley, C.A.; Weber, M.; Seitz, O.; Hecht, S.; Koksch, B.; Dernedde, J.; Graf, C.; Knapp, E.W.; Haag, R. Multivalency as a chemical organization and action principle. Angew. Chem. Int. Ed. Engl. 2012, 51, 10472–10498. [Google Scholar] [CrossRef]
- Yeldell, S.B.; Seitz, O. Nucleic acid constructs for the interrogation of multivalent protein interactions. Chem. Soc. Rev. 2020, 49, 6848–6865. [Google Scholar] [CrossRef]
- Chen, X.; Ramström, O.; Yan, M. Glyconanomaterials: Emerging applications in biomedical research. Nano Res. 2014, 7, 1381–1403. [Google Scholar] [CrossRef]
- Bernardi, A.; Jiménez-Barbero, J.; Casnati, A.; De Castro, C.; Darbre, T.; Fieschi, F.; Finne, J.; Funken, H.; Jaeger, K.-E.; Lahmann, M.; et al. Multivalent glycoconjugates as anti-pathogenic agents. Chem. Soc. Rev. 2013, 42, 4709–4727. [Google Scholar] [CrossRef]
- Illescas, B.M.; Rojo, J.; Delgado, R.; Martín, N. Multivalent Glycosylated Nanostructures To Inhibit Ebola Virus Infection. J. Am. Chem. Soc. 2017, 139, 6018–6025. [Google Scholar] [CrossRef]
- Budhadev, D.; Poole, E.; Nehlmeier, I.; Liu, Y.; Hooper, J.; Kalverda, E.; Akshath, U.S.; Hondow, N.; Turnbull, W.B.; Pöhlmann, S.; et al. Glycan-Gold Nanoparticles as Multifunctional Probes for Multivalent Lectin-Carbohydrate Binding: Implications for Blocking Virus Infection and Nanoparticle Assembly. J. Am. Chem. Soc. 2020, 142, 18022–18034. [Google Scholar] [CrossRef]
- Chien, Y.-Y.; Jan, M.-D.; Adak, A.K.; Tzeng, H.-C.; Lin, Y.-P.; Chen, Y.-J.; Wang, K.-T.; Chen, C.-T.; Chen, C.-C.; Lin, C.-C. Globotriose-Functionalized Gold Nanoparticles as Multivalent Probes for Shiga-like Toxin. ChemBioChem 2008, 9, 1100–1109. [Google Scholar] [CrossRef]
- Schofield, C.L.; Field, R.A.; Russell, D.A. Glyconanoparticles for the Colorimetric Detection of Cholera Toxin. Anal. Chem. 2007, 79, 1356–1361. [Google Scholar] [CrossRef]
- Giljohann, D.A.; Seferos, D.S.; Daniel, W.L.; Massich, M.D.; Patel, P.C.; Mirkin, C.A. Gold Nanoparticles for Biology and Medicine. Angew. Chem. Int. Ed. 2010, 49, 3280–3294. [Google Scholar] [CrossRef]
- Daraee, H.; Eatemadi, A.; Abbasi, E.; Fekri Aval, S.; Kouhi, M.; Akbarzadeh, A. Application of gold nanoparticles in biomedical and drug delivery. Artif. Cells Nanomed. Biotechnol. 2016, 44, 410–422. [Google Scholar] [CrossRef]
- Guo, Y.; Nehlmeier, I.; Poole, E.; Sakonsinsiri, C.; Hondow, N.; Brown, A.; Li, Q.; Li, S.; Whitworth, J.; Li, Z.; et al. Dissecting Multivalent Lectin-Carbohydrate Recognition Using Polyvalent Multifunctional Glycan-Quantum Dots. J. Am. Chem. Soc. 2017, 139, 11833–11844. [Google Scholar] [CrossRef]
- Budhadev, D.; Hooper, J.; Rocha, C.; Nehlmeier, I.; Kempf, A.M.; Hoffmann, M.; Krüger, N.; Zhou, D.; Pöhlmann, S.; Guo, Y. Polyvalent Nano-Lectin Potently Neutralizes SARS-CoV-2 by Targeting Glycans on the Viral Spike Protein. JACS Au 2023, 3, 1755–1766. [Google Scholar] [CrossRef]
- Basaran, R.; Budhadev, D.; Kempf, A.; Nehlmeier, I.; Hondow, N.; Pöhlmann, S.; Guo, Y.; Zhou, D. Probing scaffold size effects on multivalent lectin–glycan binding affinity, thermodynamics and antiviral properties using polyvalent glycan-gold nanoparticles. Nanoscale 2024, 16, 13962–13978. [Google Scholar] [CrossRef]
- Hooper, J.; Budhadev, D.; Fernandez Ainaga, D.L.; Hondow, N.; Zhou, D.; Guo, Y. Polyvalent Glycan Functionalized Quantum Nanorods as Mechanistic Probes for Shape-Selective Multivalent Lectin-Glycan Recognition. ACS Appl. Nano Mater. 2023, 6, 4201–4213. [Google Scholar] [CrossRef] [PubMed]
- Hooper, J.; Liu, Y.; Budhadev, D.; Ainaga, D.F.; Hondow, N.; Zhou, D.; Guo, Y. Polyvalent Glycan Quantum Dots as a Multifunctional Tool for Revealing Thermodynamic, Kinetic, and Structural Details of Multivalent Lectin-Glycan Interactions. ACS Appl. Mater. Interfaces 2022, 14, 47385–47396. [Google Scholar] [CrossRef]
- Porkolab, V.; Lepšík, M.; Ordanini, S.; St John, A.; Le Roy, A.; Thépaut, M.; Paci, E.; Ebel, C.; Bernardi, A.; Fieschi, F. Powerful Avidity with a Limited Valency for Virus-Attachment Blockers on DC-SIGN: Combining Chelation and Statistical Rebinding with Structural Plasticity of the Receptor. ACS Cent. Sci. 2023, 9, 709–718. [Google Scholar] [CrossRef]
- Geijtenbeek, T.B.; Kwon, D.S.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.; Middel, J.; Cornelissen, I.L.; Nottet, H.S.; KewalRamani, V.N.; Littman, D.R.; et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 2000, 100, 587–597. [Google Scholar] [CrossRef]
- Pöhlmann, S.; Soilleux, E.J.; Baribaud, F.; Leslie, G.J.; Morris, L.S.; Trowsdale, J.; Lee, B.; Coleman, N.; Doms, R.W. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc. Natl. Acad. Sci. USA 2001, 98, 2670–2675. [Google Scholar] [CrossRef]
- Amraei, R.; Yin, W.; Napoleon, M.A.; Suder, E.L.; Berrigan, J.; Zhao, Q.; Olejnik, J.; Chandler, K.B.; Xia, C.; Feldman, J.; et al. CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2. ACS Cent. Sci. 2021, 7, 1156–1165. [Google Scholar] [CrossRef]
- Davis, C.W.; Nguyen, H.-Y.; Hanna, S.L.; Sánchez, M.D.; Doms, R.W.; Pierson, T.C. West Nile Virus Discriminates between DC-SIGN and DC-SIGNR for Cellular Attachment and Infection. J. Virol. 2006, 80, 1290–1301. [Google Scholar] [CrossRef]
- Guo, Y.; Bruce Turnbull, W.; Zhou, D. Probing Multivalent Protein-Carbohydrate Interactions by Quantum Dot-Förster Resonance Energy Transfer. Methods Enzym. 2018, 598, 71–100. [Google Scholar] [CrossRef]
- Beswick, L.; Dimitriou, E.; Ahmadipour, S.; Zafar, A.; Rejzek, M.; Reynisson, J.; Field, R.A.; Miller, G.J. Inhibition of the GDP-d-mannose dehydrogenase from Pseudomonas aeruginosa using targeted sugar nucleotide probes. ACS Chem. Biol. 2020, 15, 3086–3092. [Google Scholar] [CrossRef]
- Ahmadipour, S.; Pergolizzi, G.; Rejzek, M.; Field, R.A.; Miller, G.J. Chemoenzymatic synthesis of C6-modified sugar nucleotides to probe the GDP-d-mannose dehydrogenase from Pseudomonas aeruginosa. Org. Lett. 2019, 21, 4415–4419. [Google Scholar] [CrossRef]
- Beswick, L.; Ahmadipour, S.; Dolan, J.P.; Rejzek, M.; Field, R.A.; Miller, G.J. Chemical and enzymatic synthesis of the alginate sugar nucleotide building block: GDP-d-mannuronic acid. Carbohydr. Res. 2019, 485, 107819. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Sakonsinsiri, C.; Nehlmeier, I.; Fascione, M.A.; Zhang, H.; Wang, W.; Pöhlmann, S.; Turnbull, W.B.; Zhou, D. Compact, Polyvalent Mannose Quantum Dots as Sensitive, Ratiometric FRET Probes for Multivalent Protein-Ligand Interactions. Angew. Chem. Int. Ed. Engl. 2016, 55, 4738–4742. [Google Scholar] [CrossRef] [PubMed]
- Susumu, K.; Uyeda, H.T.; Medintz, I.L.; Pons, T.; Delehanty, J.B.; Mattoussi, H. Enhancing the stability and biological functionalities of quantum dots via compact multifunctional ligands. J. Am. Chem. Soc. 2007, 129, 13987–13996. [Google Scholar] [CrossRef]
- Zhou, D.; Bruckbauer, A.; Abell, C.; Klenerman, D.; Kang, D.J. Fabrication of Three-Dimensional Surface Structures with Highly Fluorescent Quantum Dots by Surface-Templated Layer-by-Layer Assembly. Adv. Mater. 2005, 17, 1243–1248. [Google Scholar] [CrossRef]
- Zhou, D.; Bruckbauer, A.; Ying; Abell, C.; Klenerman, D. Building three-dimensional surface biological assemblies on the nanometer scale. Nano Lett. 2003, 3, 1517–1520. [Google Scholar] [CrossRef]
- Lee, J.H.; Ozorowski, G.; Ward, A.B. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 2016, 351, 1043–1048. [Google Scholar] [CrossRef]
- Stewart-Jones, G.B.; Soto, C.; Lemmin, T.; Chuang, G.Y.; Druz, A.; Kong, R.; Thomas, P.V.; Wagh, K.; Zhou, T.; Behrens, A.J.; et al. Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell 2016, 165, 813–826. [Google Scholar] [CrossRef]
- Saha, S.K.; Brewer, C.F. Determination of the concentrations of oligosaccharides, complex type carbohydrates, and glycoproteins using the phenol-sulfuric acid method. Carbohydr. Res. 1994, 254, 157–167. [Google Scholar] [CrossRef]
- Ning, X.; Budhadev, D.; Pollastri, S.; Nehlmeier, I.; Kempf, A.; Manfield, I.; Turnbull, W.B.; Pöhlmann, S.; Bernardi, A.; Li, X. Polyvalent Glycomimetic-Gold Nanoparticles Revealing Critical Roles of Glycan Display on Multivalent Lectin–Glycan Interaction Biophysics and Antiviral Properties. JACS Au 2024, 4, 3295–3309. [Google Scholar] [CrossRef] [PubMed]
- Hill, H.D.; Millstone, J.E.; Banholzer, M.J.; Mirkin, C.A. The Role Radius of Curvature Plays in Thiolated Oligonucleotide Loading on Gold Nanoparticles. ACS Nano 2009, 3, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Dubertret, B.; Calame, M.; Libchaber, A.J. Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat. Biotechnol. 2001, 19, 365–370. [Google Scholar] [CrossRef] [PubMed]
- Dulkeith, E.; Ringler, M.; Klar, T.A.; Feldmann, J.; Muñoz Javier, A.; Parak, W.J. Gold nanoparticles quench fluorescence by phase induced radiative rate suppression. Nano Lett. 2005, 5, 585–589. [Google Scholar] [CrossRef]
- Jennings, T.L.; Singh, M.P.; Strouse, G.F. Fluorescent lifetime quenching near d = 1.5 nm gold nanoparticles: Probing NSET validity. J. Am. Chem. Soc. 2006, 128, 5462–5467. [Google Scholar] [CrossRef]
- Seferos, D.S.; Giljohann, D.A.; Hill, H.D.; Prigodich, A.E.; Mirkin, C.A. Nano-flares: Probes for transfection and mRNA detection in living cells. J. Am. Chem. Soc. 2007, 129, 15477–15479. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Liang, Z.; Zhang, J.; Wang, L.; Li, G.; Fan, C. Gold-nanoparticle-based multicolor nanobeacons for sequence-specific DNA analysis. Angew. Chem. Int. Ed. Engl. 2009, 48, 8670–8674. [Google Scholar] [CrossRef]
- Liu, X.; Atwater, M.; Wang, J.; Huo, Q. Extinction coefficient of gold nanoparticles with different sizes and different capping ligands. Colloids. Surf. B Biointerfaces 2007, 58, 3–7. [Google Scholar] [CrossRef]
- Wang, X.; Ramstrom, O.; Yan, M. Quantitative analysis of multivalent ligand presentation on gold glyconanoparticles and the impact on lectin binding. Anal. Chem. 2010, 82, 9082–9089. [Google Scholar] [CrossRef]
- Liyanage, S.H.; Yan, M. Quantification of binding affinity of glyconanomaterials with lectins. Chem. Commun. 2020, 56, 13491–13505. [Google Scholar] [CrossRef] [PubMed]
| Gx-ManA | Lectin Receptor | QE | Kd (nM) |
|---|---|---|---|
| G5-ManA | DC-SIGN | 0.125 ± 0.006 | 61 ± 3 |
| G13-ManA | DC-SIGN | 0.213 ± 0.014 | 11.5 ± 0.5 |
| G5-ManA | DC-SIGNR | 0.150 ± 0.016 | 48 ± 4 |
| G13-ManA | DC-SIGNR | 0.260 ± 0.019 | 8.4 ± 1.1 |
| Gx-ManA | Lectin Receptor | n | EC50 (nM) | R2 |
|---|---|---|---|---|
| G5-ManA | DC-SIGN | 0.64 ± 0.21 | 10.4 ± 4.7 | 0.839 |
| G13-ManA | DC-SIGN | 0.35 ± 0.09 | 0.53 ± 0.19 | 0.825 |
| G5-ManA | DC-SIGNR | 0.95 ± 0.40 | 18.9 ± 4.9 | 0.790 |
| G13-ManA | DC-SIGNR | 0.43 ± 0.09 | 0.29 ± 0.12 | 0.869 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Basaran, R.; Budhadev, D.; Dimitriou, E.; Wootton, H.S.; Miller, G.J.; Kempf, A.; Nehlmeier, I.; Pöhlmann, S.; Guo, Y.; Zhou, D. Polyvalent Mannuronic Acid-Coated Gold Nanoparticles for Probing Multivalent Lectin–Glycan Interaction and Blocking Virus Infection. Viruses 2025, 17, 1066. https://doi.org/10.3390/v17081066
Basaran R, Budhadev D, Dimitriou E, Wootton HS, Miller GJ, Kempf A, Nehlmeier I, Pöhlmann S, Guo Y, Zhou D. Polyvalent Mannuronic Acid-Coated Gold Nanoparticles for Probing Multivalent Lectin–Glycan Interaction and Blocking Virus Infection. Viruses. 2025; 17(8):1066. https://doi.org/10.3390/v17081066
Chicago/Turabian StyleBasaran, Rahman, Darshita Budhadev, Eleni Dimitriou, Hannah S. Wootton, Gavin J. Miller, Amy Kempf, Inga Nehlmeier, Stefan Pöhlmann, Yuan Guo, and Dejian Zhou. 2025. "Polyvalent Mannuronic Acid-Coated Gold Nanoparticles for Probing Multivalent Lectin–Glycan Interaction and Blocking Virus Infection" Viruses 17, no. 8: 1066. https://doi.org/10.3390/v17081066
APA StyleBasaran, R., Budhadev, D., Dimitriou, E., Wootton, H. S., Miller, G. J., Kempf, A., Nehlmeier, I., Pöhlmann, S., Guo, Y., & Zhou, D. (2025). Polyvalent Mannuronic Acid-Coated Gold Nanoparticles for Probing Multivalent Lectin–Glycan Interaction and Blocking Virus Infection. Viruses, 17(8), 1066. https://doi.org/10.3390/v17081066







