Mesocricetus auratus (Golden Syrian Hamster) Experimental Model of SARS-CoV-2 Infection Reveals That Lung Injury Is Associated with Phenotypic Differences Between SARS-CoV-2 Variants
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Aspects
2.2. SARS-CoV-2 Inocula
2.3. SARS-CoV-2 Infection in Golden Syrian Hamsters
2.4. SARS-CoV-2 Neutralising Antibodies Detection in Blood Samples
2.5. Quantitative SARS-CoV-2 RNA Detection in Oropharyngeal Swabs and Tissue Samples
2.6. Inflammatory Markers Quantification
2.7. Immunofluorescence
2.8. Histopathology and Transmission Electron Microscopy
2.9. Statistical Analysis
3. Results
3.1. Clinical Manifestations
3.2. Blood Biochemistry and Haematological Findings
3.3. Production of Neutralisation Antibodies
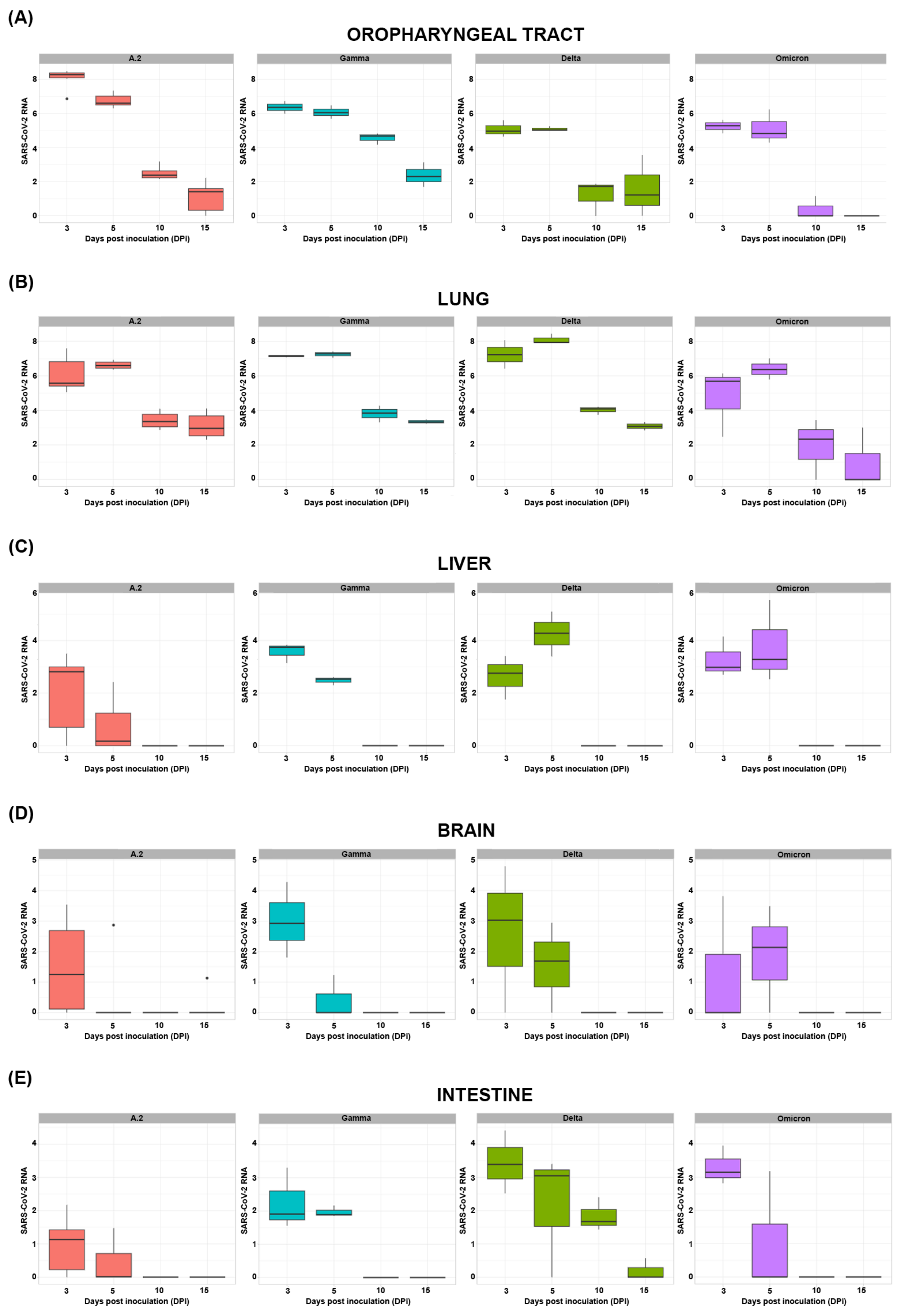
3.4. Quantitative SARS-CoV-2 RNA Detection in Swab and Tissue Samples
3.5. Inflammatory Markers
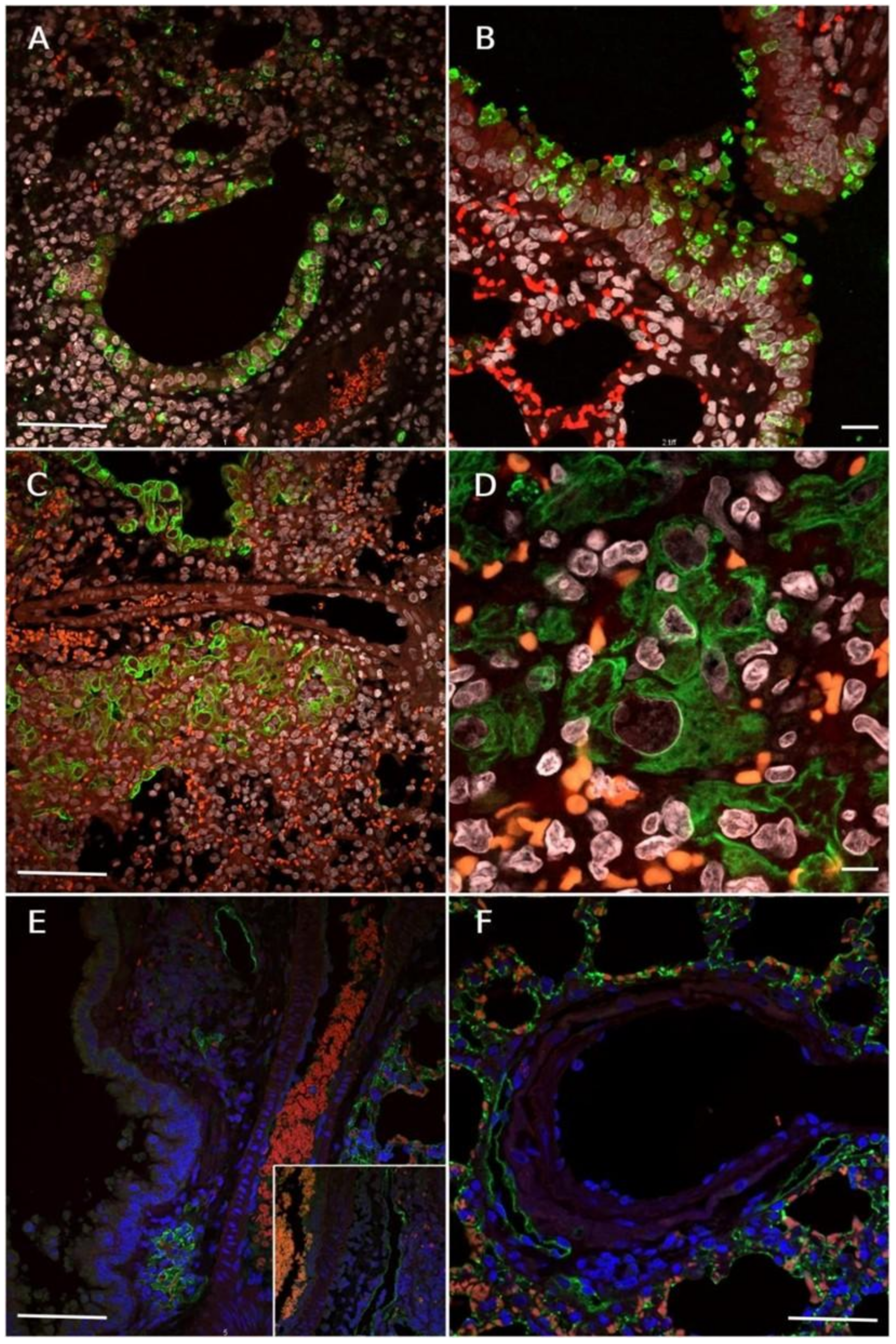
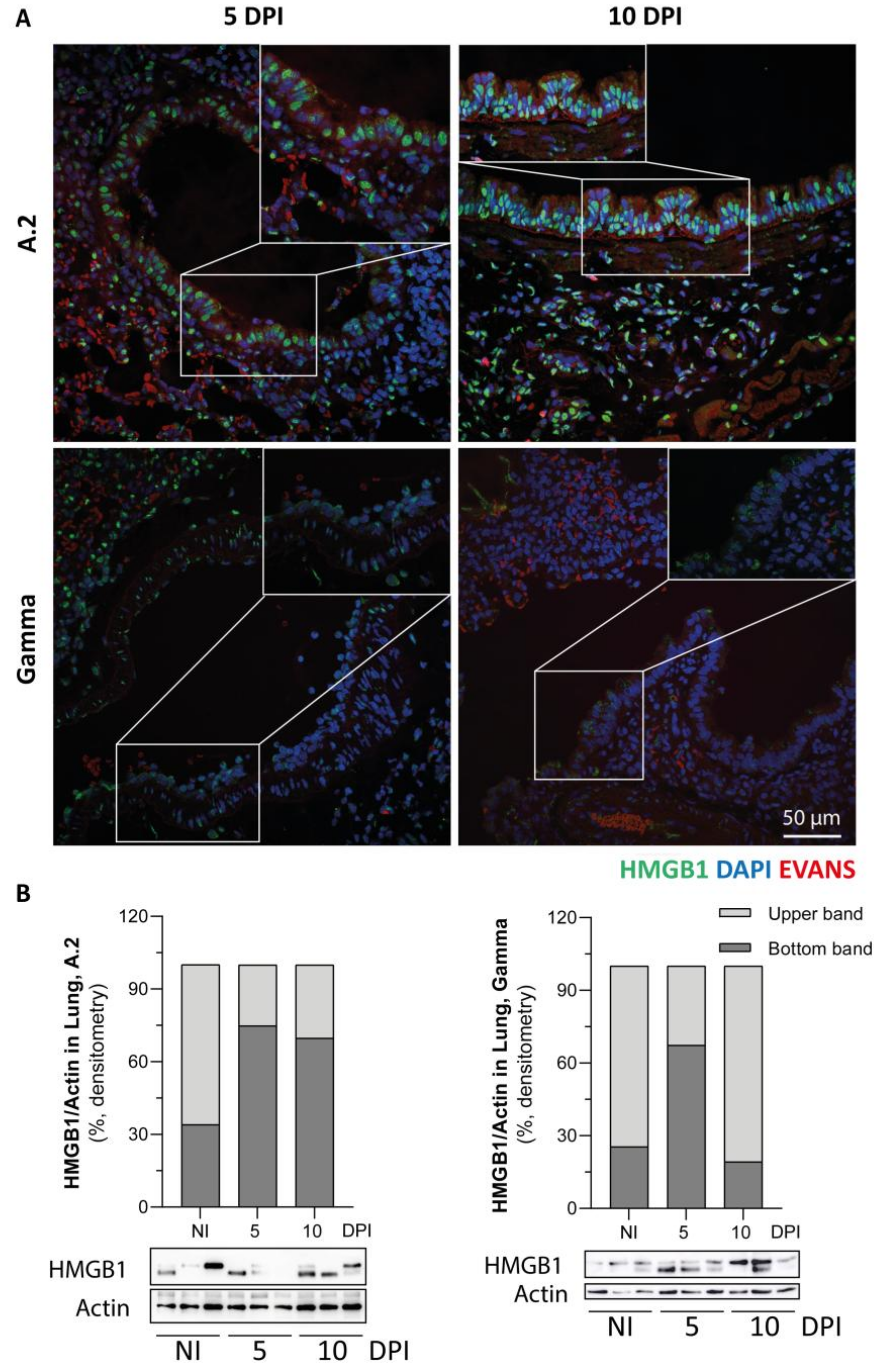
3.6. Immunofluorescence Reaction for SARS-CoV-2 Nucleocapsid, Lymphatic Vessel Endothelial Marker (Podoplanin) and HMGB1 Translocation in Lungs of SARS-CoV-2 Inoculated Hamsters
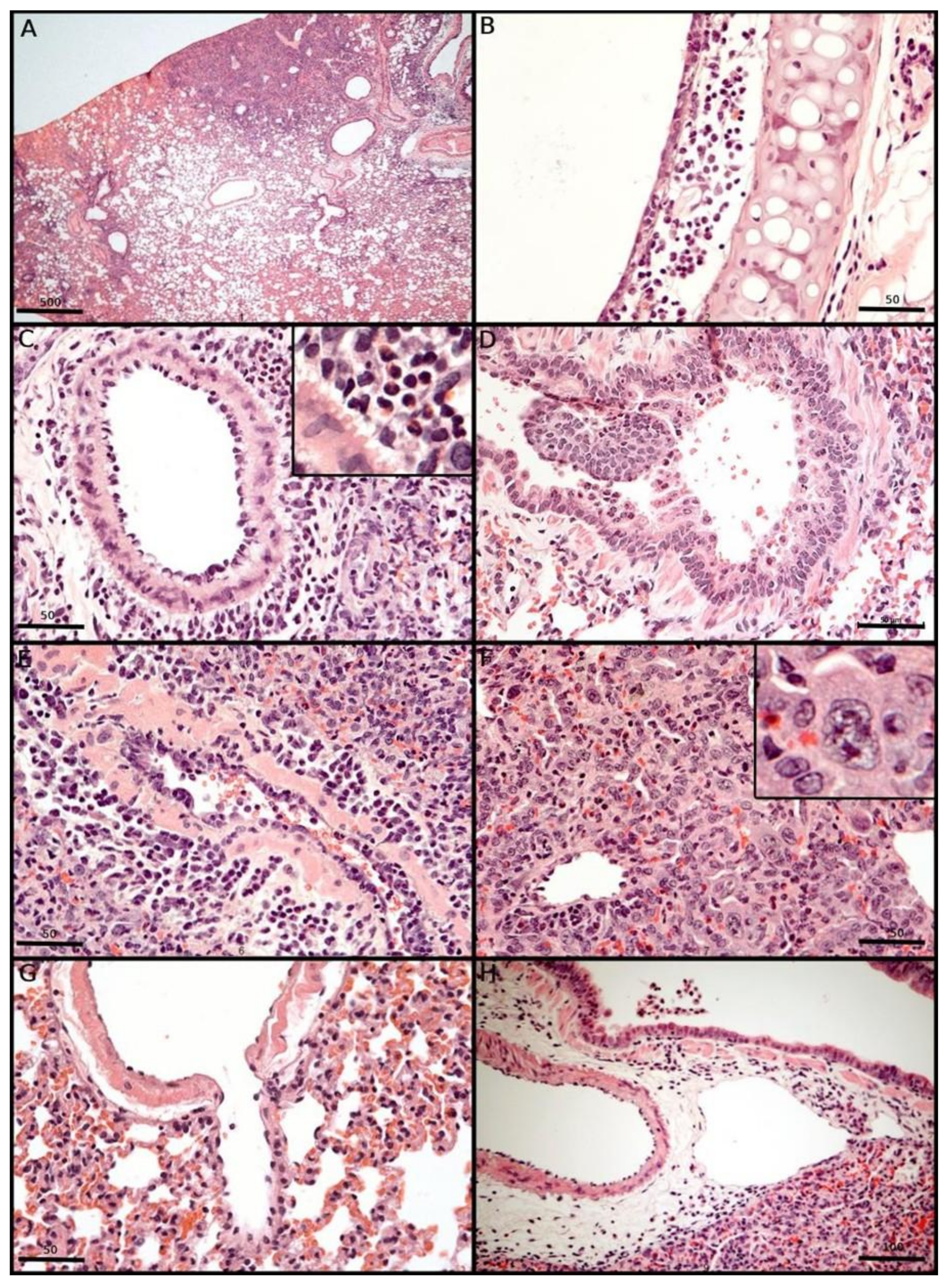
3.7. Morphological Changes Induced in the Respiratory Tract of Syrian Hamsters After Experimental SARS-CoV-2 Inoculation
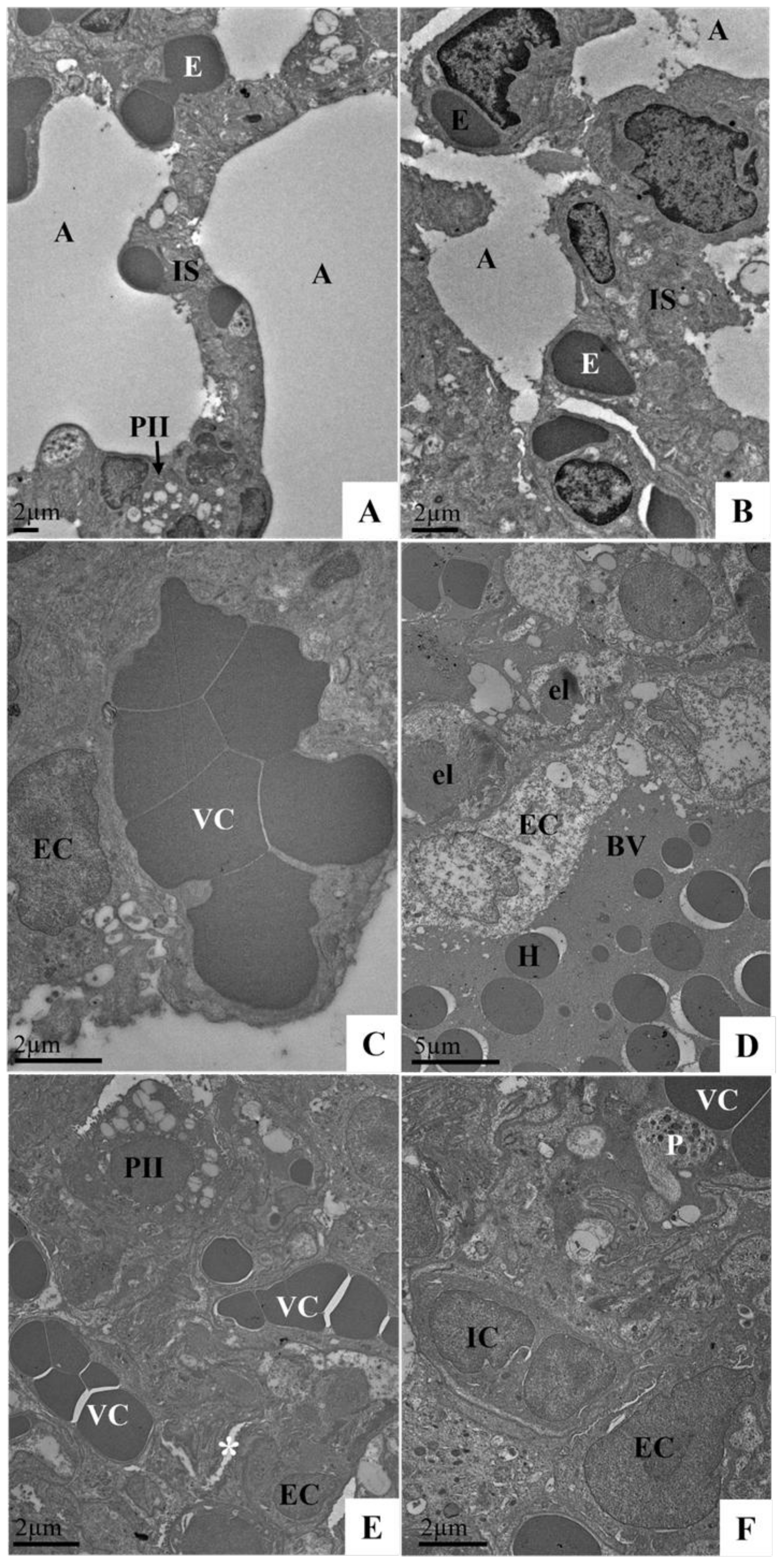
3.8. Ultrastructural Analysis of the Lungs of Syrian Hamster Lung Samples Infected with the SARS-CoV-2 Variant (Delta Variant or Omicron) via TEM
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupta, K.; Toelzer, C.; Williamson, M.K.; Shoemark, D.K.; Oliveira, A.S.F.; Matthews, D.A.; Almuqrin, A.; Staufer, O.; Yadav, S.K.N.; Borucu, U.; et al. Structural Insights in Cell-Type Specific Evolution of Intra-Host Diversity by SARS-CoV-2. Nat. Commun. 2022, 13, 222. [Google Scholar] [CrossRef]
- Meehan, G.R.; Herder, V.; Allan, J.; Huang, X.; Kerr, K.; Mendonca, D.C.; Ilia, G.; Wright, D.W.; Nomikou, K.; Gu, Q.; et al. Phenotyping the Virulence of SARS-CoV-2 Variants in Hamsters by Digital Pathology and Machine Learning. PLoS Pathog. 2023, 19, e1011589. [Google Scholar] [CrossRef]
- Rouzine, I.M. Evolutionary Mechanisms of the Emergence of the Variants of Concern of SARS-CoV-2. Viruses 2025, 17, 197. [Google Scholar] [CrossRef]
- Lima-Martínez, M.M.; Carrera Boada, C.; Madera-Silva, M.D.; Marín, W.; Contreras, M. COVID-19 and Diabetes: A Bidirectional Relationship. Clínica Investig. Arterioscler. (Engl. Ed.) 2021, 33, 151–157. [Google Scholar] [CrossRef]
- Hekman, R.M.; Hume, A.J.; Goel, R.K.; Abo, K.M.; Huang, J.; Blum, B.C.; Werder, R.B.; Suder, E.L.; Paul, I.; Phanse, S.; et al. Actionable Cytopathogenic Host Responses of Human Alveolar Type 2 Cells to SARS-CoV-2. Mol. Cell 2020, 80, 1104–1122.e9. [Google Scholar] [CrossRef]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and Evasion of Type I Interferon Responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.-C.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.-Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef]
- Habashi, N.M.; Camporota, L.; Gatto, L.A.; Nieman, G. Functional Pathophysiology of SARS-CoV-2-Induced Acute Lung Injury and Clinical Implications. J. Appl. Physiol. 2021, 130, 877–891. [Google Scholar] [CrossRef]
- Sastry, S.; Cuomo, F.; Muthusamy, J. COVID-19 and Thrombosis: The Role of Hemodynamics. Thromb. Res. 2022, 212, 51–57. [Google Scholar] [CrossRef]
- Muñoz-Fontela, C.; Dowling, W.E.; Funnell, S.G.P.; Gsell, P.-S.; Riveros-Balta, A.X.; Albrecht, R.A.; Andersen, H.; Baric, R.S.; Carroll, M.W.; Cavaleri, M.; et al. Animal Models for COVID-19. Nature 2020, 586, 509–515. [Google Scholar] [CrossRef]
- Zhai, C.; Wang, M.; Chung, H.-J.; Hassan, M.; Lee, S.; Kim, H.-J.; Hong, S.-T. Roborovski Hamster (Phodopus roborovskii) Strain SH101 as a Systemic Infection Model of SARS-CoV-2. Virulence 2021, 12, 2430–2442. [Google Scholar] [CrossRef]
- McCray, P.B.; Pewe, L.; Wohlford-Lenane, C.; Hickey, M.; Manzel, L.; Shi, L.; Netland, J.; Jia, H.P.; Halabi, C.; Sigmund, C.D.; et al. Lethal Infection of K18-hACE2 Mice Infected with Severe Acute Respiratory Syndrome Coronavirus. J. Virol. 2007, 81, 813–821. [Google Scholar] [CrossRef]
- Gruber, A.D.; Firsching, T.C.; Trimpert, J.; Dietert, K. Hamster Models of COVID-19 Pneumonia Reviewed: How Human Can They Be? Vet. Pathol. 2022, 59, 528–545. [Google Scholar] [CrossRef]
- Bjornson-Hooper, Z.B.; Fragiadakis, G.K.; Spitzer, M.H.; Chen, H.; Madhireddy, D.; Hu, K.; Lundsten, K.; McIlwain, D.R.; Nolan, G.P. A Comprehensive Atlas of Immunological Differences Between Humans, Mice, and Non-Human Primates. Front. Immunol. 2022, 13, 867015. [Google Scholar] [CrossRef]
- Reczyńska, K.; Tharkar, P.; Kim, S.Y.; Wang, Y.; Pamuła, E.; Chan, H.-K.; Chrzanowski, W. Animal Models of Smoke Inhalation Injury and Related Acute and Chronic Lung Diseases. Adv. Drug Deliv. Rev. 2018, 123, 107–134. [Google Scholar] [CrossRef]
- Rosenke, K.; Meade-White, K.; Letko, M.; Clancy, C.; Hansen, F.; Liu, Y.; Okumura, A.; Tang-Huau, T.-L.; Li, R.; Saturday, G.; et al. Defining the Syrian Hamster as a Highly Susceptible Preclinical Model for SARS-CoV-2 Infection. Emerg. Microbes Infect. 2020, 9, 2673–2684. [Google Scholar] [CrossRef]
- Pach, S.; Nguyen, T.N.; Trimpert, J.; Kunec, D.; Osterrieder, N.; Wolber, G. ACE2-Variants Indicate Potential SARS-CoV-2-Susceptibility in Animals: A Molecular Dynamics Study. Mol. Inform. 2021, 40, 2100031. [Google Scholar] [CrossRef]
- Tomris, I.; Bouwman, K.M.; Adolfs, Y.; Noack, D.; van der Woude, R.; Kerster, G.; Herfst, S.; Sanders, R.W.; van Gils, M.J.; Boons, G.-J.; et al. Distinct Spatial Arrangements of ACE2 and TMPRSS2 Expression in Syrian Hamster Lung Lobes Dictates SARS-CoV-2 Infection Patterns. PLoS Pathog. 2022, 18, e1010340. [Google Scholar] [CrossRef]
- Carossino, M.; Izadmehr, S.; Trujillo, J.D.; Gaudreault, N.N.; Dittmar, W.; Morozov, I.; Balasuriya, U.B.R.; Cordon-Cardo, C.; García-Sastre, A.; Richt, J.A. ACE2 and TMPRSS2 Distribution in the Respiratory Tract of Different Animal Species and Its Correlation with SARS-CoV-2 Tissue Tropism. Microbiol. Spectr. 2024, 12, e0327023. [Google Scholar] [CrossRef]
- Ishige, T. Molecular Biology of SARS-CoV-2 and Techniques of Diagnosis and Surveillance. Adv. Clin. Chem. 2024, 118, 35–85. [Google Scholar] [CrossRef]
- Abdelnabi, R.; Lassaunière, R.; Maes, P.; Weynand, B.; Neyts, J. Comparing the Infectivity of Recent SARS-CoV-2 Omicron Sub-Variants in Syrian Hamsters. Viruses 2024, 16, 122. [Google Scholar] [CrossRef]
- Coussement, L.; Oosterhof, M.M.; Guryev, V.; Reitsema, V.A.; Bruintjes, J.J.; Goris, M.; Bouma, H.R.; de Meyer, T.; Rots, M.G.; Henning, R.H. Liver Transcriptomic and Methylomic Analyses Identify Transcriptional Mitogen-Activated Protein Kinase Regulation in Facultative Hibernation of Syrian Hamster. Proc. R. Soc. B 2023, 290, 20230368. [Google Scholar] [CrossRef]
- Plunkard, J.; Mulka, K.; Zhou, R.; Tarwater, P.; Zhong, W.; Lowman, M.; Wong, A.; Pekosz, A.; Villano, J. SARS-CoV-2 Variant Pathogenesis Following Primary Infection and Reinfection in Syrian Hamsters. mBio 2023, 14, e0007823. [Google Scholar] [CrossRef]
- Shiwa-Sudo, N.; Sakai, Y.; Iwata-Yoshikawa, N.; Watanabe, S.; Yamada, S.; Kuroda, Y.; Yamamoto, T.; Shirakura, M.; Fujisaki, S.; Miyazaki, K.; et al. Impact of Reinfection with SARS-CoV-2 Omicron Variants in Previously Infected Hamsters. J. Virol. 2023, 97, e0136622. [Google Scholar] [CrossRef]
- Abdelnabi, R.; Boudewijns, R.; Foo, C.S.; Seldeslachts, L.; Sanchez-Felipe, L.; Zhang, X.; Delang, L.; Maes, P.; Kaptein, S.J.F.; Weynand, B.; et al. Comparing Infectivity and Virulence of Emerging SARS-CoV-2 Variants in Syrian Hamsters. EBioMedicine 2021, 68, 103403. [Google Scholar] [CrossRef]
- Wickenhagen, A.; Flagg, M.; Port, J.R.; Yinda, C.K.; Goldin, K.; Gallogly, S.; Schulz, J.E.; Lutterman, T.; Williamson, B.N.; Kaiser, F.; et al. Evolution of Omicron Lineage towards Increased Fitness in the Upper Respiratory Tract in the Absence of Severe Lung Pathology. Nat. Commun. 2025, 16, 594. [Google Scholar] [CrossRef]
- Percie du Sert, N.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; et al. The ARRIVE Guidelines 2.0: Updated Guidelines for Reporting Animal Research. BMC Vet. Res. 2020, 16, 242. [Google Scholar] [CrossRef]
- Matute-Bello, G.; Downey, G.; Moore, B.B.; Groshong, S.D.; Matthay, M.A.; Slutsky, A.S.; Kuebler, W.M. An Official American Thoracic Society Workshop Report: Features and Measurements of Experimental Acute Lung Injury in Animals. Am. J. Respir. Cell Mol. Biol. 2011, 44, 725–738. [Google Scholar] [CrossRef]
- Klopfleisch, R. Multiparametric and Semiquantitative Scoring Systems for the Evaluation of Mouse Model Histopathology—A Systematic Review. BMC Vet. Res. 2013, 9, 123. [Google Scholar] [CrossRef]
- Dietert, K.; Gutbier, B.; Wienhold, S.M.; Reppe, K.; Jiang, X.; Yao, L.; Chaput, C.; Naujoks, J.; Brack, M.; Kupke, A.; et al. Spectrum of Pathogen- and Model-Specific Histopathologies in Mouse Models of Acute Pneumonia. PLoS ONE 2017, 12, e0188251. [Google Scholar] [CrossRef]
- Gruber, A.D.; Osterrieder, N.; Bertzbach, L.D.; Vladimirova, D.; Greuel, S.; Ihlow, J.; Horst, D.; Trimpert, J.; Dietert, K. Standardization of Reporting Criteria for Lung Pathology in SARS-CoV-2–Infected Hamsters: What Matters? Am. J. Respir. Cell Mol. Biol. 2020, 63, 856–859. [Google Scholar] [CrossRef]
- Meyerholz, D.K.; Beck, A.P. Histopathologic Evaluation and Scoring of Viral Lung Infection. In MERS Coronavirus: Methods and Protocols; Vijay, R., Ed.; Springer: New York, NY, USA, 2020; pp. 205–220. ISBN 978-1-0716-0211-9. [Google Scholar]
- Barth, O.M.; da Silva, M.A.N.; Barreto-Vieira, D.F. Low Impact to Fixed Cell Processing Aiming Transmission Electron Microscopy. Mem. Inst. Oswaldo Cruz 2016, 111, 411–413. [Google Scholar] [CrossRef]
- Hoppe, G.; Talcott, K.E.; Bhattacharya, S.K.; Crabb, J.W.; Sears, J.E. Molecular Basis for the Redox Control of Nuclear Transport of the Structural Chromatin Protein Hmgb1. Exp. Cell Res. 2006, 312, 3526–3538. [Google Scholar] [CrossRef]
- Davies, E.R.; Ryan, K.A.; Bewley, K.R.; Coombes, N.S.; Salguero, F.J.; Carnell, O.T.; Biddlecombe, S.; Charlton, M.; Challis, A.; Cross, E.S.; et al. The Omicron Sub-Variant BA.4 Displays a Remarkable Lack of Clinical Signs in a Golden Syrian Hamster Model of SARS-CoV-2 Infection. Viruses 2023, 15, 1133. [Google Scholar] [CrossRef]
- Voss, J.D.; Skarzynski, M.; McAuley, E.M.; Maier, E.J.; Gibbons, T.; Fries, A.C.; Chapleau, R.R. Variants in SARS-CoV-2 Associated with Mild or Severe Outcome. Evol. Med. Public Health 2021, 9, 267–275. [Google Scholar] [CrossRef]
- Armando, F.; Beythien, G.; Kaiser, F.K.; Allnoch, L.; Heydemann, L.; Rosiak, M.; Becker, S.; Gonzalez-Hernandez, M.; Lamers, M.M.; Haagmans, B.L.; et al. SARS-CoV-2 Omicron Variant Causes Mild Pathology in the Upper and Lower Respiratory Tract of Hamsters. Nat. Commun. 2022, 13, 3519. [Google Scholar] [CrossRef]
- Abdelnabi, R.; Foo, C.S.; Zhang, X.; Lemmens, V.; Maes, P.; Slechten, B.; Raymenants, J.; André, E.; Weynand, B.; Dallmeier, K.; et al. The Omicron (B.1.1.529) SARS-CoV-2 Variant of Concern Does Not Readily Infect Syrian Hamsters. Antivir. Res. 2022, 198, 105253. [Google Scholar] [CrossRef]
- Zheng, J.; Roy Wong, L.-Y.; Li, K.; Verma, A.K.; Ortiz, M.; Wohlford-Lenane, C.; Leidinger, M.R.; Knudson, C.M.; Meyerholz, D.K.; McCray, P.B.; et al. K18-hACE2 Mice for Studies of COVID-19 Treatments and Pathogenesis Including Anosmia. BioRxiv 2020, unpublished. [Google Scholar] [CrossRef]
- Zheng, J.; Wong, L.-Y.R.; Li, K.; Verma, A.K.; Ortiz, M.E.; Wohlford-Lenane, C.; Leidinger, M.R.; Knudson, C.M.; Meyerholz, D.K.; McCray, P.B.; et al. COVID-19 Treatments and Pathogenesis Including Anosmia in K18-hACE2 Mice. Nature 2021, 589, 603–607. [Google Scholar] [CrossRef]
- de Melo, G.D.; Lazarini, F.; Levallois, S.; Hautefort, C.; Michel, V.; Larrous, F.; Verillaud, B.; Aparicio, C.; Wagner, S.; Gheusi, G.; et al. COVID-19-Related Anosmia Is Associated with Viral Persistence and Inflammation in Human Olfactory Epithelium and Brain Infection in Hamsters. Sci. Transl. Med. 2021, 13, eabf8396. [Google Scholar] [CrossRef]
- Tsukahara, T.; Brann, D.H.; Datta, S.R. Mechanisms of SARS-CoV-2-Associated Anosmia. Physiol. Rev. 2023, 103, 2759–2766. [Google Scholar] [CrossRef]
- Halfmann, P.J.; Iida, S.; Iwatsuki-Horimoto, K.; Maemura, T.; Kiso, M.; Scheaffer, S.M.; Darling, T.L.; Joshi, A.; Loeber, S.; Singh, G.; et al. SARS-CoV-2 Omicron Virus Causes Attenuated Disease in Mice and Hamsters. Nature 2022, 603, 687–692. [Google Scholar] [CrossRef]
- Kozlov, M. Omicron’s Feeble Attack on the Lungs Could Make It Less Dangerous. Nature 2022, 601, 177. [Google Scholar] [CrossRef]
- Brüssow, H. COVID-19: Omicron—The Latest, the Least Virulent, but Probably Not the Last Variant of Concern of SARS-CoV-2. Microb. Biotechnol. 2022, 15, 1927–1939. [Google Scholar] [CrossRef]
- Diamond, M.S.; Kanneganti, T.-D. Innate Immunity: The First Line of Defense against SARS-CoV-2. Nat. Immunol. 2022, 23, 165–176. [Google Scholar] [CrossRef]
- Martins, M.; do Nascimento, G.M.; Nooruzzaman, M.; Yuan, F.; Chen, C.; Caserta, L.C.; Miller, A.D.; Whittaker, G.R.; Fang, Y.; Diel, D.G. The Omicron Variant BA.1.1 Presents a Lower Pathogenicity than B.1 D614G and Delta Variants in a Feline Model of SARS-CoV-2 Infection. J. Virol. 2022, 96, e0096122. [Google Scholar] [CrossRef]
- Czura, C.J.; Bikson, M.; Charvet, L.; Chen, J.D.Z.; Franke, M.; Fudim, M.; Grigsby, E.; Hamner, S.; Huston, J.M.; Khodaparast, N.; et al. Neuromodulation Strategies to Reduce Inflammation and Improve Lung Complications in COVID-19 Patients. Front. Neurol. 2022, 13, 897124. [Google Scholar] [CrossRef]
- Ode, Y.; Aziz, M.; Jin, H.; Arif, A.; Nicastro, J.G.; Wang, P. Cold-Inducible RNA-Binding Protein Induces Neutrophil Extracellular Traps in the Lungs during Sepsis. Sci. Rep. 2019, 9, 6252. [Google Scholar] [CrossRef]
- Scaffidi, P.; Misteli, T.; Bianchi, M.E. Release of Chromatin Protein HMGB1 by Necrotic Cells Triggers Inflammation. Nature 2002, 418, 191–195. [Google Scholar] [CrossRef]
- Malavige, G.N.; Jeewandara, C.; Ogg, G.S. Dengue and COVID-19: Two Sides of the Same Coin. J. Biomed. Sci. 2022, 29, 48. [Google Scholar] [CrossRef]
- Calabrese, F.; Lunardi, F.; Baldasso, E.; Pezzuto, F.; Kilitci, A.; Olteanu, G.-E.; Del Vecchio, C.; Fortarezza, F.; Boscolo, A.; Schiavon, M.; et al. Comprehensive Bronchoalveolar Lavage Characterization in COVID-19 Associated Acute Respiratory Distress Syndrome Patients: A Prospective Cohort Study. Respir. Res. 2023, 24, 152. [Google Scholar] [CrossRef]
- Bednash, J.S.; Kagan, V.E.; Englert, J.A.; Farkas, D.; Tyurina, Y.Y.; Tyurin, V.A.; Samovich, S.N.; Farkas, L.; Elhance, A.; Johns, F.; et al. Syrian Hamsters as a Model of Lung Injury with SARS-CoV-2 Infection: Pathologic, Physiologic, and Detailed Molecular Profiling. Transl. Res. 2022, 240, 1–16. [Google Scholar] [CrossRef]
- Guo, T.; Wang, X.; Qu, Y.; Yin, Y.; Jing, T.; Zhang, Q. Megakaryopoiesis and Platelet Production: Insight into Hematopoietic Stem Cell Proliferation and Differentiation. Stem Cell Investig. 2015, 2, 3. [Google Scholar] [CrossRef]
- Pannone, G.; Pedicillo, M.C.; De Stefano, I.S.; Angelillis, F.; Barile, R.; Pannone, C.; Villani, G.; Miele, F.; Municinò, M.; Ronchi, A.; et al. The Role of TLR-2 in Lethal COVID-19 Disease Involving Medullary and Resident Lung Megakaryocyte Up-Regulation in the Microthrombosis Mechanism. Cells 2024, 13, 854. [Google Scholar] [CrossRef]
- Rolla, R.; Puricelli, C.; Bertoni, A.; Boggio, E.; Gigliotti, C.L.; Chiocchetti, A.; Cappellano, G.; Dianzani, U. Platelets: “Multiple Choice” Effectors in the Immune Response and Their Implication in COVID-19 Thromboinflammatory Process. Int. J. Lab. Hematol. 2021, 43, 895–906. [Google Scholar] [CrossRef]
- Greve, F.; Mair, O.; Aulbach, I.; Biberthaler, P.; Hanschen, M. Correlation between Platelet Count and Lung Dysfunction in Multiple Trauma Patients—A Retrospective Cohort Analysis. J. Clin. Med. 2022, 11, 1400. [Google Scholar] [CrossRef]
- Caillon, A.; Trimaille, A.; Favre, J.; Jesel, L.; Morel, O.; Kauffenstein, G. Role of Neutrophils, Platelets, and Extracellular Vesicles and Their Interactions in COVID-19-associated Thrombopathy. J. Thromb. Haemost. 2022, 20, 17–31. [Google Scholar] [CrossRef]
- De Vrij, E.L.; Bouma, H.R.; Henning, R.H.; Cooper, S.T. Hibernation and Hemostasis. Front. Physiol. 2023, 14, 1207003. [Google Scholar] [CrossRef]
- Ackermann, M.; Verleden, S.E.; Kuehnel, M.; Haverich, A.; Welte, T.; Laenger, F.; Vanstapel, A.; Werlein, C.; Stark, H.; Tzankov, A.; et al. Pulmonary Vascular Endothelialitis, Thrombosis, and Angiogenesis in COVID-19. N. Engl. J. Med. 2020, 383, 120–128. [Google Scholar] [CrossRef]
- Kulkarni, R.M.; Greenberg, J.M.; Akeson, A.L. NFATc1 Regulates Lymphatic Endothelial Development. Mech. Dev. 2009, 126, 350–365. [Google Scholar] [CrossRef]
- Borba-Junior, I.T.; Lima, F.; Sidarta-Oliveira, D.; Moraes, C.R.P.; Annichino-Bizzacchi, J.M.; Bombassaro, B.; Palma, A.C.; Costa, F.T.M.; Moretti, M.L.; Mansour, E.; et al. Podoplanin and CLEC-2 Levels in Patients with COVID-19. Res. Pract. Thromb. Haemost. 2023, 7, 100282. [Google Scholar] [CrossRef]
- Hanff, T.C.; Mohareb, A.M.; Giri, J.; Cohen, J.B.; Chirinos, J.A. Thrombosis in COVID-19. Am. J. Hematol. 2020, 95, 1578–1589. [Google Scholar] [CrossRef]
- Xu, S.; Ilyas, I.; Weng, J. Endothelial Dysfunction in COVID-19: An Overview of Evidence, Biomarkers, Mechanisms and Potential Therapies. Acta Pharmacol. Sin. 2023, 44, 695–709. [Google Scholar] [CrossRef]
- Perico, L.; Morigi, M.; Pezzotta, A.; Locatelli, M.; Imberti, B.; Corna, D.; Cerullo, D.; Benigni, A.; Remuzzi, G. SARS-CoV-2 Spike Protein Induces Lung Endothelial Cell Dysfunction and Thrombo-Inflammation Depending on the C3a/C3a Receptor Signalling. Sci. Rep. 2023, 13, 11392. [Google Scholar] [CrossRef]
- McFadyen, J.D.; Stevens, H.; Peter, K. The Emerging Threat of (Micro)Thrombosis in COVID-19 and Its Therapeutic Implications. Circ. Res. 2020, 127, 571–587. [Google Scholar] [CrossRef]
- Trivedi, A.; Lu, T.M.; Summers, B.; Kim, K.; Rhee, A.J.; Houghton, S.; Byers, D.E.; Lis, R.; Reed, H.O. Lung Lymphatic Endothelial Cells Undergo Inflammatory and Prothrombotic Changes in a Model of Chronic Obstructive Pulmonary Disease. Front. Cell Dev. Biol. 2024, 12, 1344070. [Google Scholar] [CrossRef]
- MacDonald, M.E.; Weathered, R.K.; Stewart, E.C.; Magold, A.I.; Mukherjee, A.; Gurbuxani, S.; Smith, H.; McMullen, P.; Mueller, J.; Husain, A.N.; et al. Lymphatic Coagulation and Neutrophil Extracellular Traps in Lung-Draining Lymph Nodes of COVID-19 Decedents. Blood Adv. 2022, 6, 6249–6262. [Google Scholar] [CrossRef]
- Ball, E.E.; Weiss, C.M.; Liu, H.; Jackson, K.; Keel, M.K.; Miller, C.J.; Van Rompay, K.K.A.; Coffey, L.L.; Pesavento, P.A. Severe Acute Respiratory Syndrome Coronavirus 2 Vasculopathy in a Syrian Golden Hamster Model. Am. J. Pathol. 2023, 193, 690–701. [Google Scholar] [CrossRef]
- Horii, Y.; Shiina, T.; Shimizu, Y. The Mechanism Enabling Hibernation in Mammals. Adv. Exp. Med. Biol. 2018, 1081, 45–60. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, Y.; Li, C.; Xu, Y.; Wang, X.; Wu, D.; Gao, Z.; Qian, H.; You, Z.; Zhang, Z.; et al. Extracellular CIRP-Impaired Rab26 Restrains EPOR-Mediated Macrophage Polarization in Acute Lung Injury. Front. Immunol. 2021, 12, 768435. [Google Scholar] [CrossRef]
- Zhong, P.; Zhou, M.; Zhang, J.; Peng, J.; Zeng, G.; Huang, H. The Role of Cold-Inducible RNA-Binding Protein in Respiratory Diseases. J. Cell. Mol. Med. 2022, 26, 957–965. [Google Scholar] [CrossRef]
- Ito, J.T.; Lourenço, J.D.; Righetti, R.F.; Tibério, I.F.L.C.; Prado, C.M.; Lopes, F.D.T.Q.S. Extracellular Matrix Component Remodeling in Respiratory Diseases: What Has Been Found in Clinical and Experimental Studies? Cells 2019, 8, 342. [Google Scholar] [CrossRef]
- Sano, Y.; Shiina, T.; Naitou, K.; Nakamori, H.; Shimizu, Y. Hibernation-Specific Alternative Splicing of the mRNA Encoding Cold-Inducible RNA-Binding Protein in the Hearts of Hamsters. Biochem. Biophys. Res. Commun. 2015, 462, 322–325. [Google Scholar] [CrossRef]
- Logan, S.M.; Storey, K.B. Cold-Inducible RNA-Binding Protein Cirp, but Not Rbm3, May Regulate Transcript Processing and Protection in Tissues of the Hibernating Ground Squirrel. Cell Stress Chaperones 2020, 25, 857–868. [Google Scholar] [CrossRef]



| Group | 3 DPI | 5 DPI | 10 DPI | 15 DPI |
|---|---|---|---|---|
| Negative Control | Negative | Negative | Negative | Negative |
| A.2 | Negative | Negative | 86.75% | 93.08% |
| Gamma | Negative | 30.18% | 89.46% | 92.87% |
| Delta | Negative | 54.81% | 90.42% | 95.90% |
| Omicron | Negative | Negative | 60.86% | 57.43% |
| Oropharyngeal Tract RNA | Lung RNA | Liver RNA | Brain RNA | Intestine RNA | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predictors | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p | Estimates | CI | p |
| 5 DPI | −0.61 | −1.30–0.09 | 0.086 | 0.74 | 0.13–1.35 | 0.018 | −0.32 | −1.03–0.39 | 0.377 | −1.02 | −1.82–−0.21 | 0.013 | −0.99 | −1.60–−0.39 | 0.001 |
| 10 DPI | −4.34 | −5.03–−3.65 | <0.001 | −2.94 | −3.55–−2.32 | <0.001 | −2.71 | −3.42–−2.00 | <0.001 | −1.97 | −2.78–−1.17 | <0.001 | −1.82 | −2.43–−1.22 | <0.001 |
| 15 DPI | −5.33 | −6.02–−4.64 | <0.001 | −3.52 | −4.14–−2.91 | <0.001 | −2.71 | −3.42–−2.00 | <0.001 | −1.90 | −2.70–−1.09 | <0.001 | −2.15 | −2.76–−1.55 | <0.001 |
| Gamma | 0.24 | −0.43–0.91 | 0.480 | 0.58 | −0.01–1.18 | 0.053 | 0.82 | 0.14–1.51 | 0.019 | 0.31 | −0.46–1.09 | 0.429 | 0.71 | 0.12–1.30 | 0.018 |
| Delta | −1.37 | −2.04–−0.69 | <0.001 | 0.81 | 0.22–1.40 | 0.008 | 1.04 | 0.35–1.72 | 0.003 | 0.50 | −0.28–1.28 | 0.210 | 1.56 | 0.97–2.15 | <0.001 |
| Omicron | −1.92 | −2.59–−1.24 | <0.001 | −1.28 | −1.88–−0.69 | <0.001 | 1.08 | 0.39–1.76 | 0.002 | 0.25 | −0.53–1.03 | 0.533 | 0.75 | 0.16–1.33 | 0.013 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, D.d.R.F.; da Silva, A.d.S.; Alves, A.D.R.; Rossi, B.A.; Lima, R.d.A.; Faria, S.B.S.C.; Cruz, O.G.; Muller, R.; Scharfstein, J.; Vicentino, A.R.R.; et al. Mesocricetus auratus (Golden Syrian Hamster) Experimental Model of SARS-CoV-2 Infection Reveals That Lung Injury Is Associated with Phenotypic Differences Between SARS-CoV-2 Variants. Viruses 2025, 17, 1048. https://doi.org/10.3390/v17081048
Rodrigues DdRF, da Silva AdS, Alves ADR, Rossi BA, Lima RdA, Faria SBSC, Cruz OG, Muller R, Scharfstein J, Vicentino ARR, et al. Mesocricetus auratus (Golden Syrian Hamster) Experimental Model of SARS-CoV-2 Infection Reveals That Lung Injury Is Associated with Phenotypic Differences Between SARS-CoV-2 Variants. Viruses. 2025; 17(8):1048. https://doi.org/10.3390/v17081048
Chicago/Turabian StyleRodrigues, Daniela del Rosario Flores, Alexandre dos Santos da Silva, Arthur Daniel Rocha Alves, Bárbara Araujo Rossi, Richard de Almeida Lima, Sarah Beatriz Salvador Castro Faria, Oswaldo Gonçalves Cruz, Rodrigo Muller, Julio Scharfstein, Amanda Roberta Revoredo Vicentino, and et al. 2025. "Mesocricetus auratus (Golden Syrian Hamster) Experimental Model of SARS-CoV-2 Infection Reveals That Lung Injury Is Associated with Phenotypic Differences Between SARS-CoV-2 Variants" Viruses 17, no. 8: 1048. https://doi.org/10.3390/v17081048
APA StyleRodrigues, D. d. R. F., da Silva, A. d. S., Alves, A. D. R., Rossi, B. A., Lima, R. d. A., Faria, S. B. S. C., Cruz, O. G., Muller, R., Scharfstein, J., Vicentino, A. R. R., Matos, A. d. R., dos Santos, J. P. R., Manso, P. P. A., Paiva, M. B., Barreto-Vieira, D. F., Caldas, G. C., Machado, M. P., & Pinto, M. A. (2025). Mesocricetus auratus (Golden Syrian Hamster) Experimental Model of SARS-CoV-2 Infection Reveals That Lung Injury Is Associated with Phenotypic Differences Between SARS-CoV-2 Variants. Viruses, 17(8), 1048. https://doi.org/10.3390/v17081048






