Genetic and Immunological Profiling of Recent SARS-CoV-2 Omicron Subvariants: Insights into Immune Evasion and Infectivity in Monoinfections and Coinfections
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Viral Infections, Cytopathic Effect and Stock Preparation
2.2.1. SARS-CoV-2
2.2.2. Influenza A H1N1
2.2.3. Respiratory Syncytial Virus
2.3. Plaque Forming Unit Assay (PFU)
2.4. Antibodies and Human Convalescent Plasma
2.5. Neutralization Assay
2.6. Spike Protein Target-Based Sequencing
2.7. Ex Vivo Air–Liquid Interface Model (ALI)
2.7.1. Human Bronchial Airway Epithelial Cells (hBAECs)
2.7.2. Monoinfection and Coinfection Assays in Human Bronchial Airway Epithelial Cells
2.7.3. Viral Quantification by Quantitative Reverse Transcription-PCR (RT-qPCR)
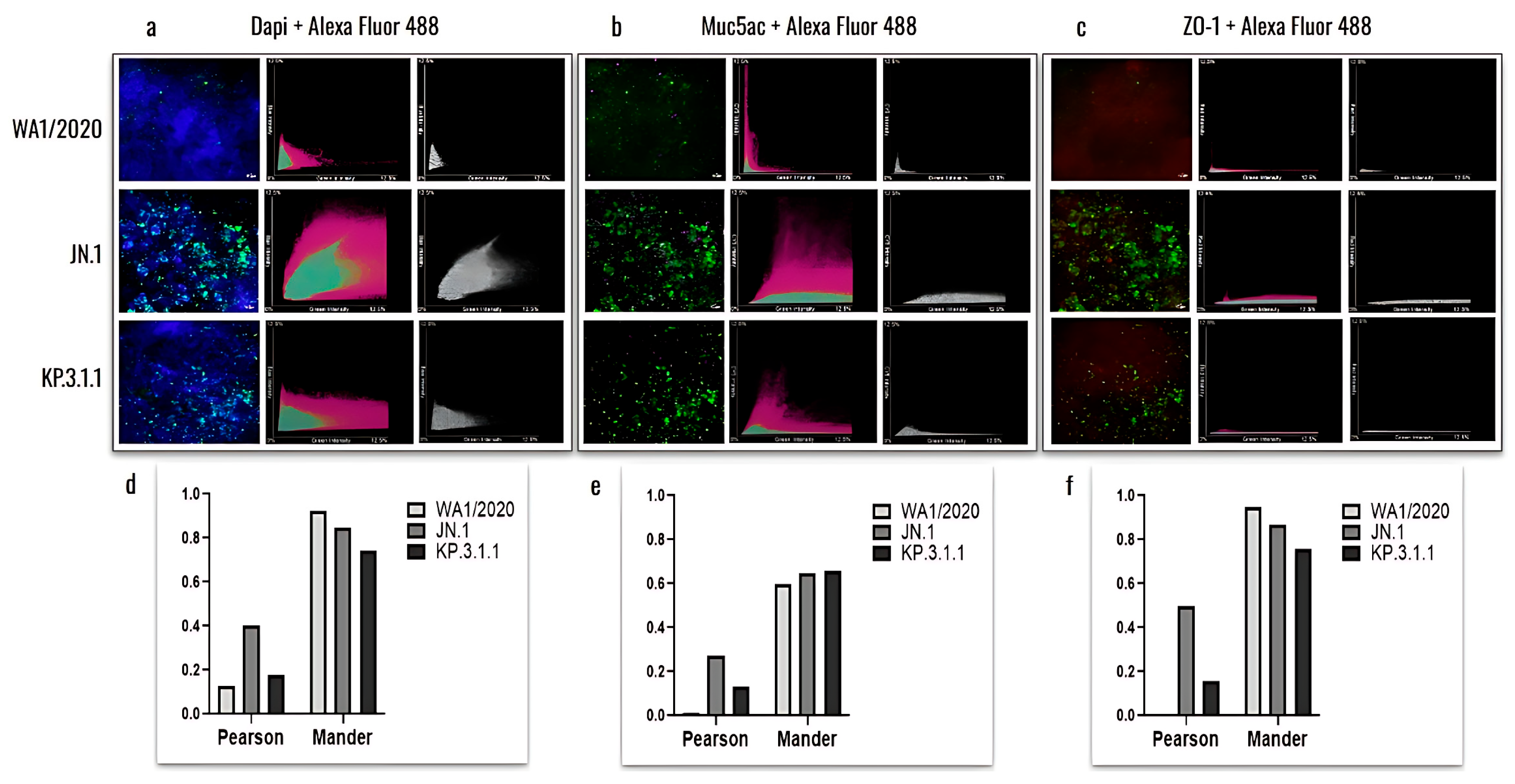
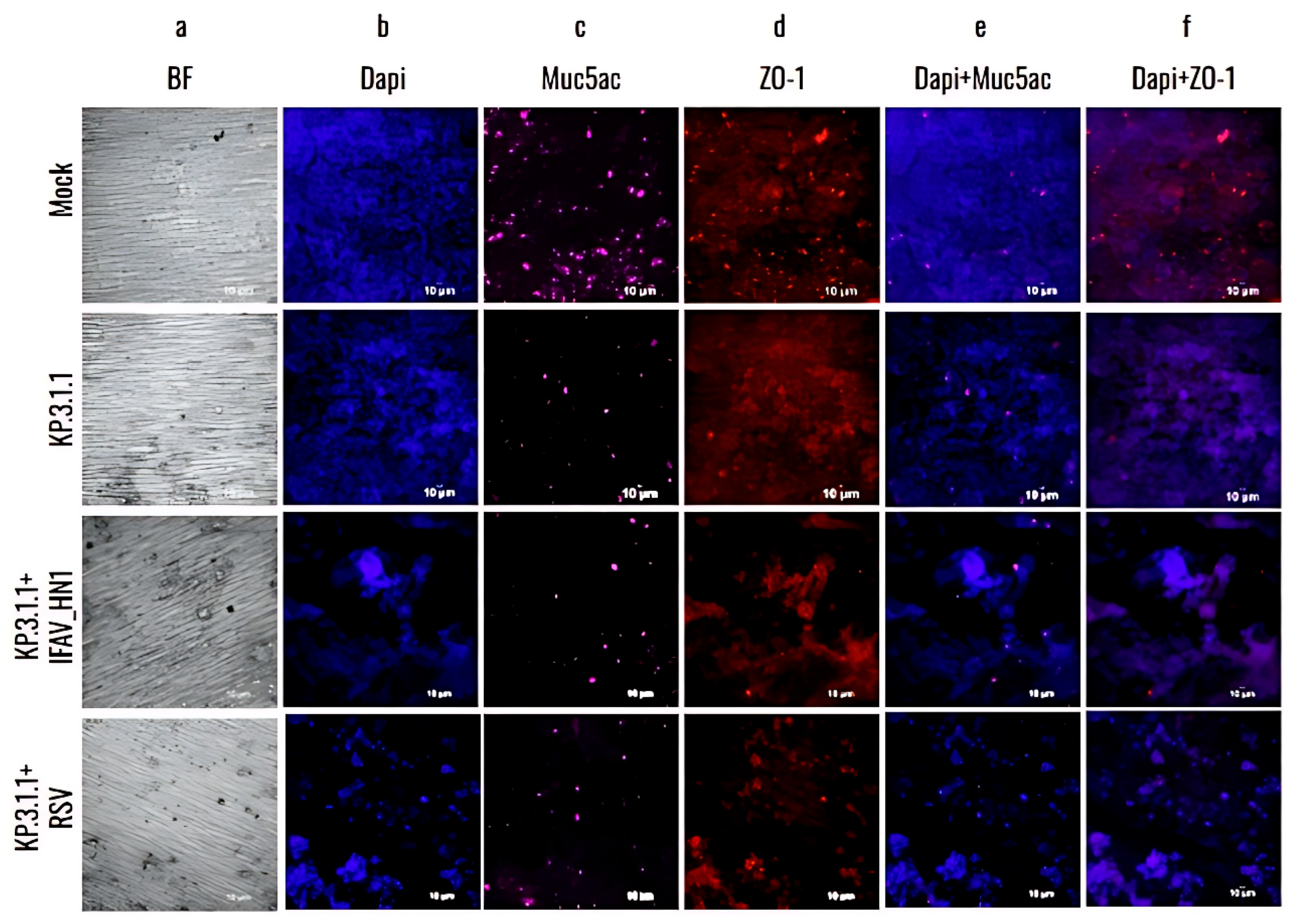
2.7.4. hBAEC Differentiation and Colocalization Evaluated by Immunofluorescence Assay
2.7.5. Profile of Cytokine Expression in hBAEC
2.8. Statistical Analysis
3. Results
3.1. In Vitro Characterization of SARS-CoV-2 Infection Using VeroE6/TMPRSS2
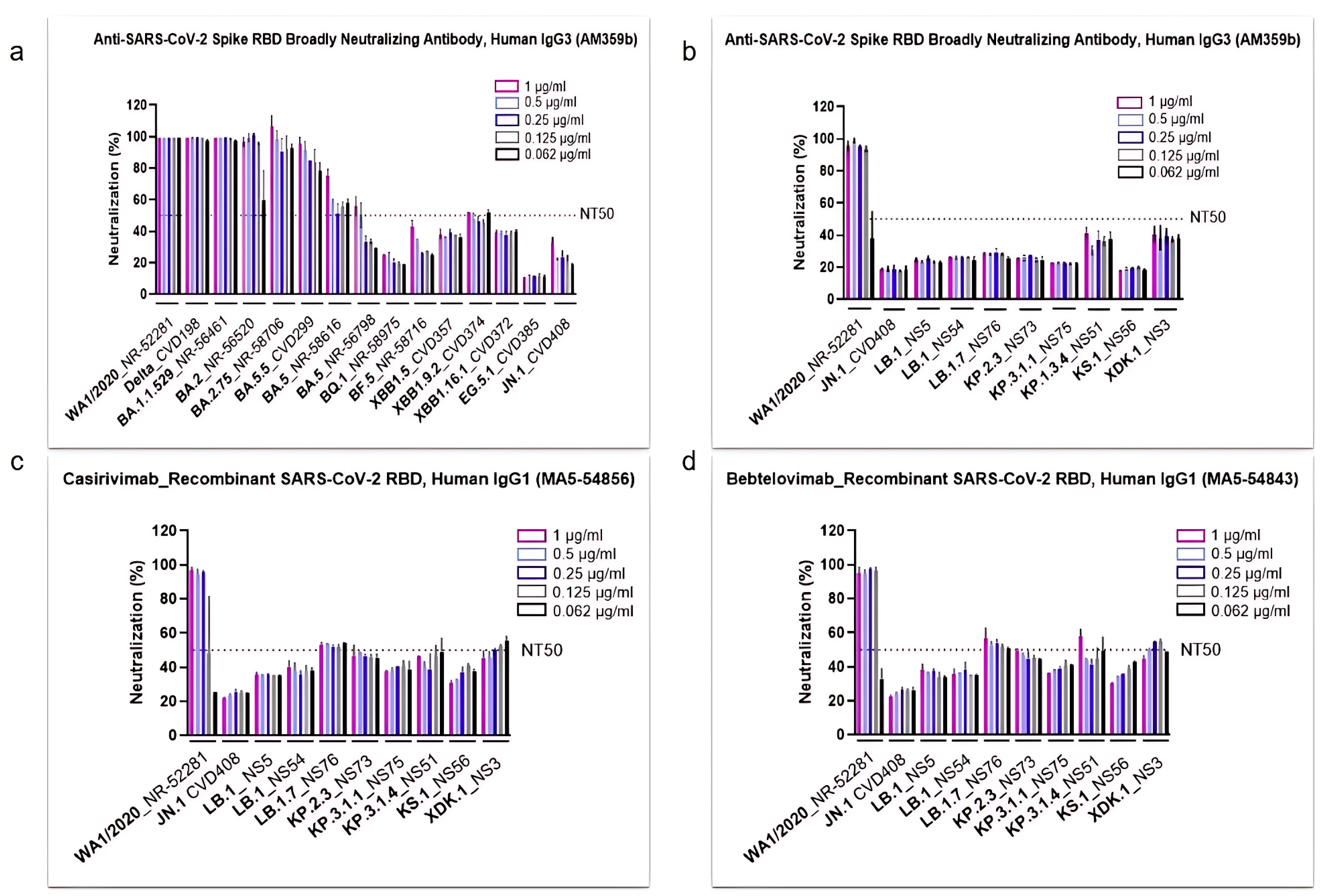
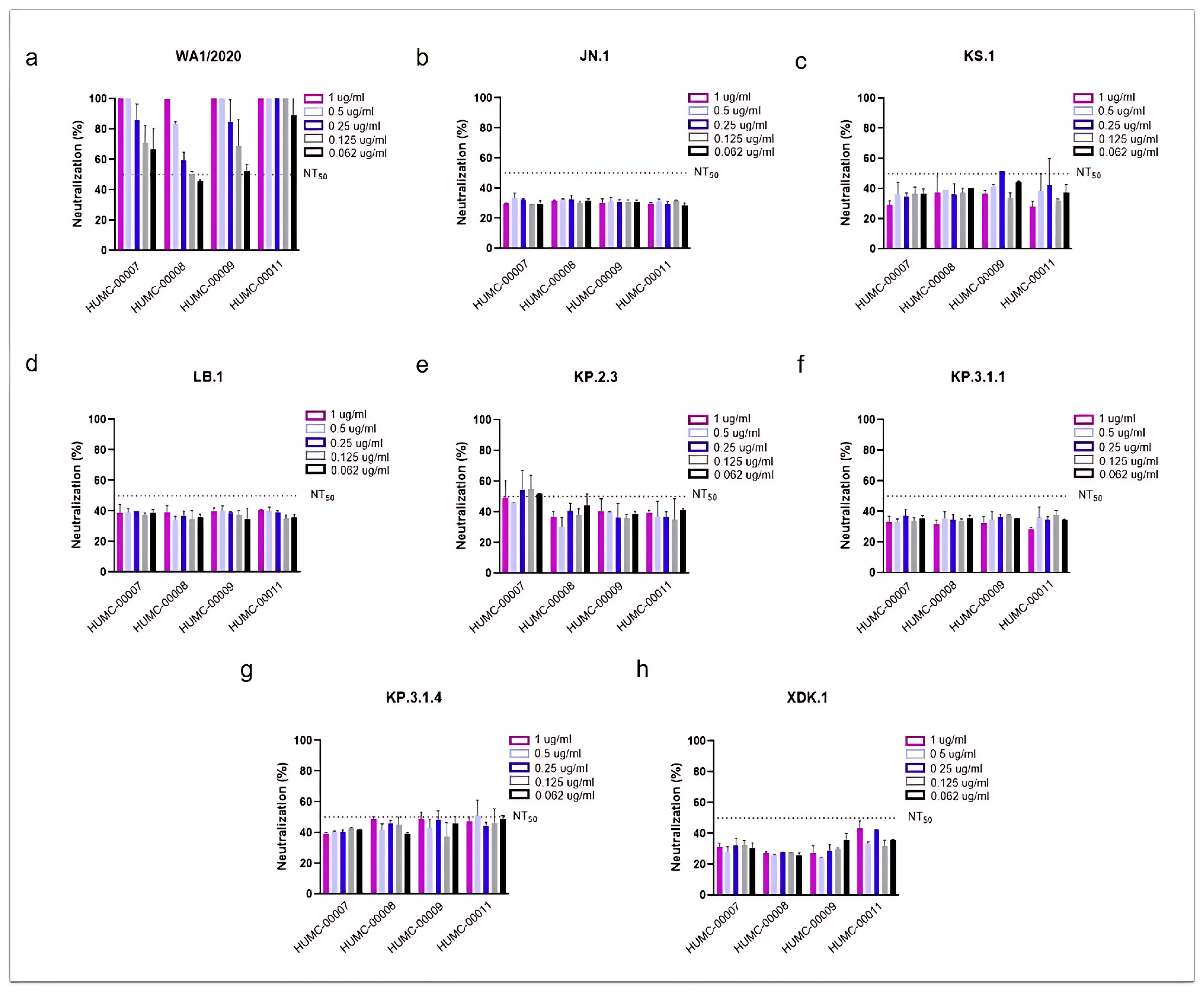
3.2. SARS-CoV-2 Omicron Subvariants Escape the Neutralizing Effects of Antibodies
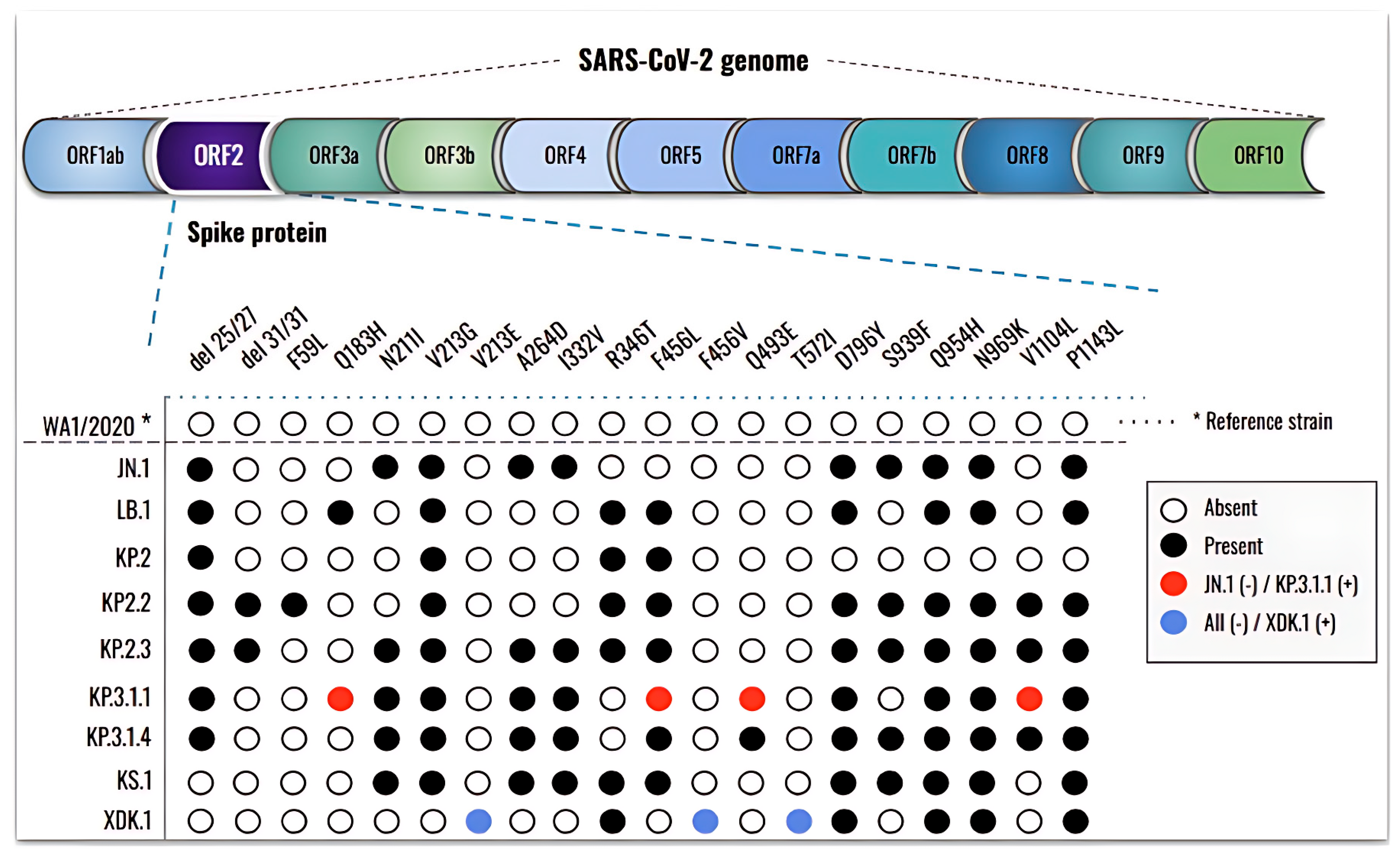
3.3. New Mutations, Rather than an Increase in the Number, Are Present in the Newly Emergent SARS-CoV-2 Subvariants
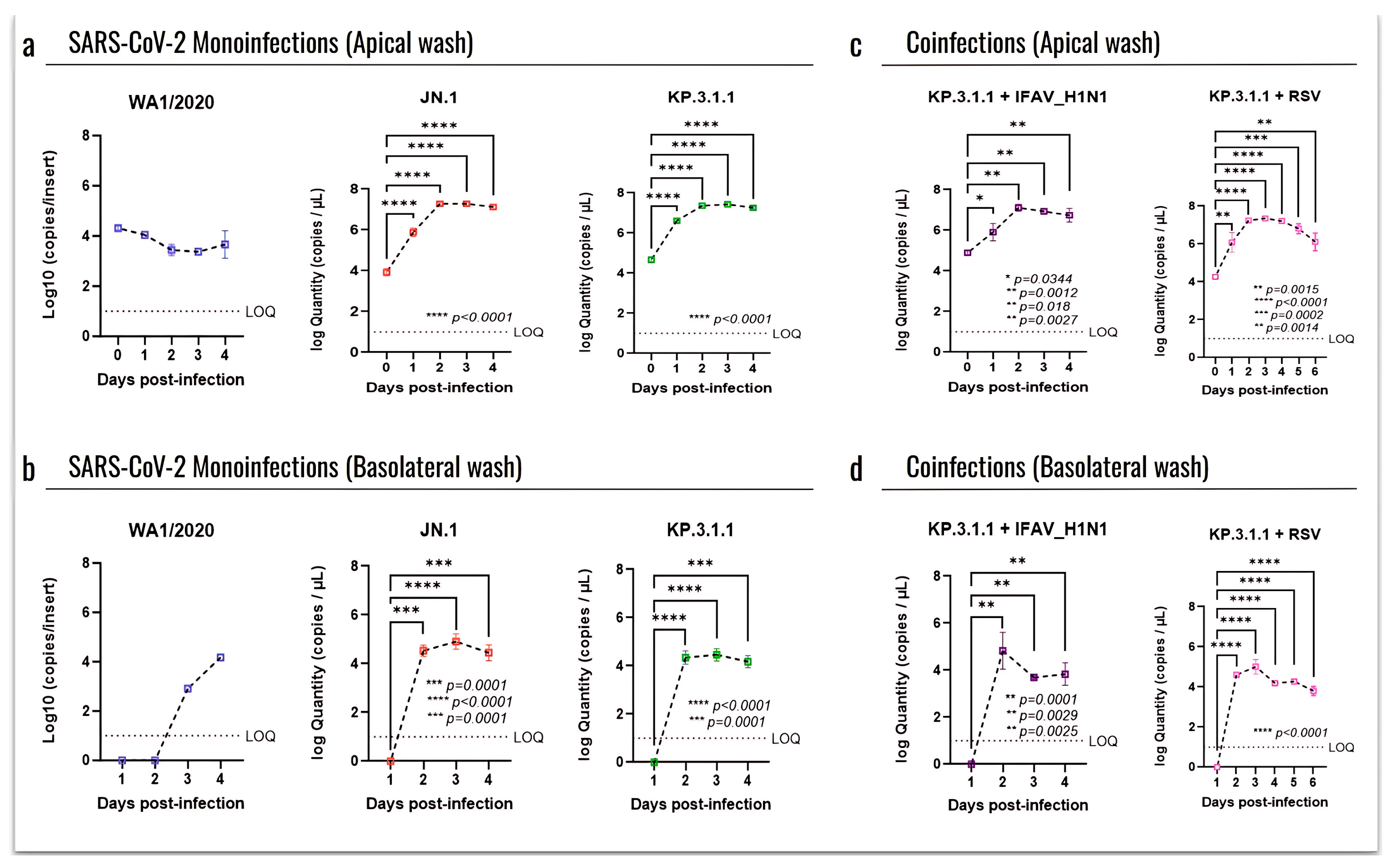
3.4. Omicron SARS-CoV-2 Induces a Strong Cytopathic Effect in hBAEC During Monoinfection or Coinfection with IFAV H1N1 and RSV
3.5. Omicron Subvariants Induce a Higher Level of Infection in hBAEC Compared to the SARS-CoV-2 Parent Strain
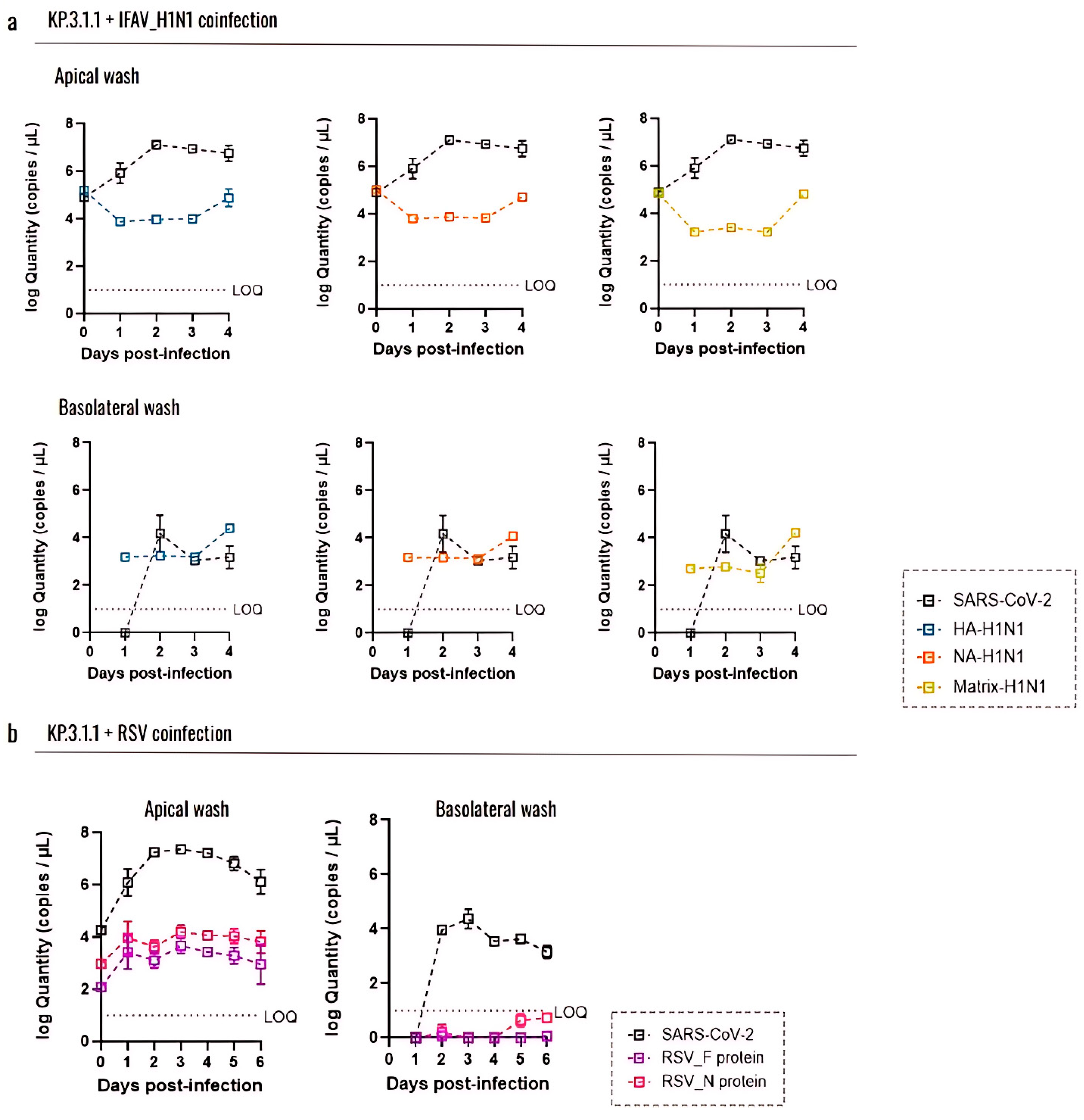
3.6. Coinfection with Other Respiratory Viruses Does Not Reduce the Replication Capacity of SARS-CoV-2 in the Human Bronchial Airway Epithelium
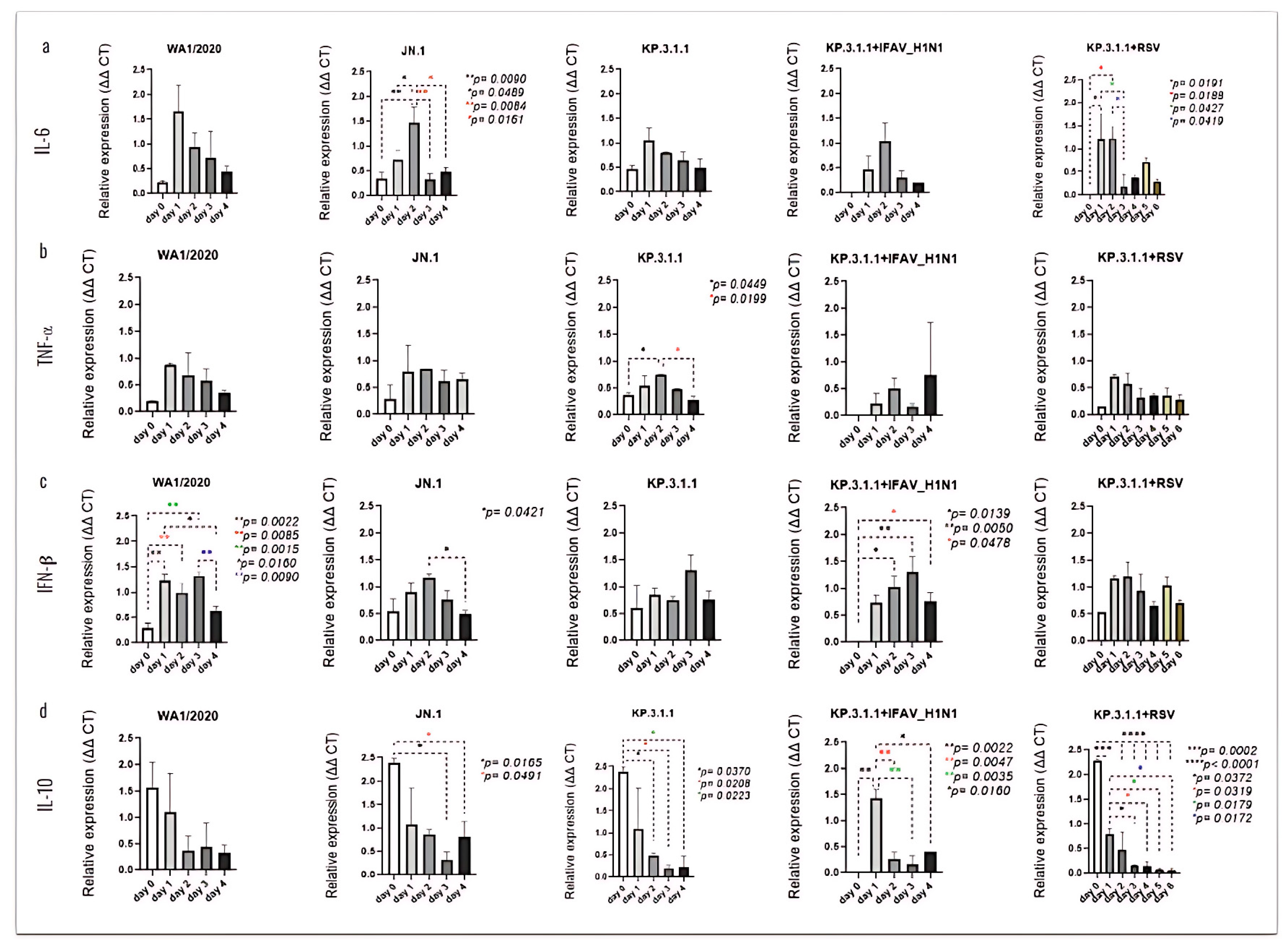
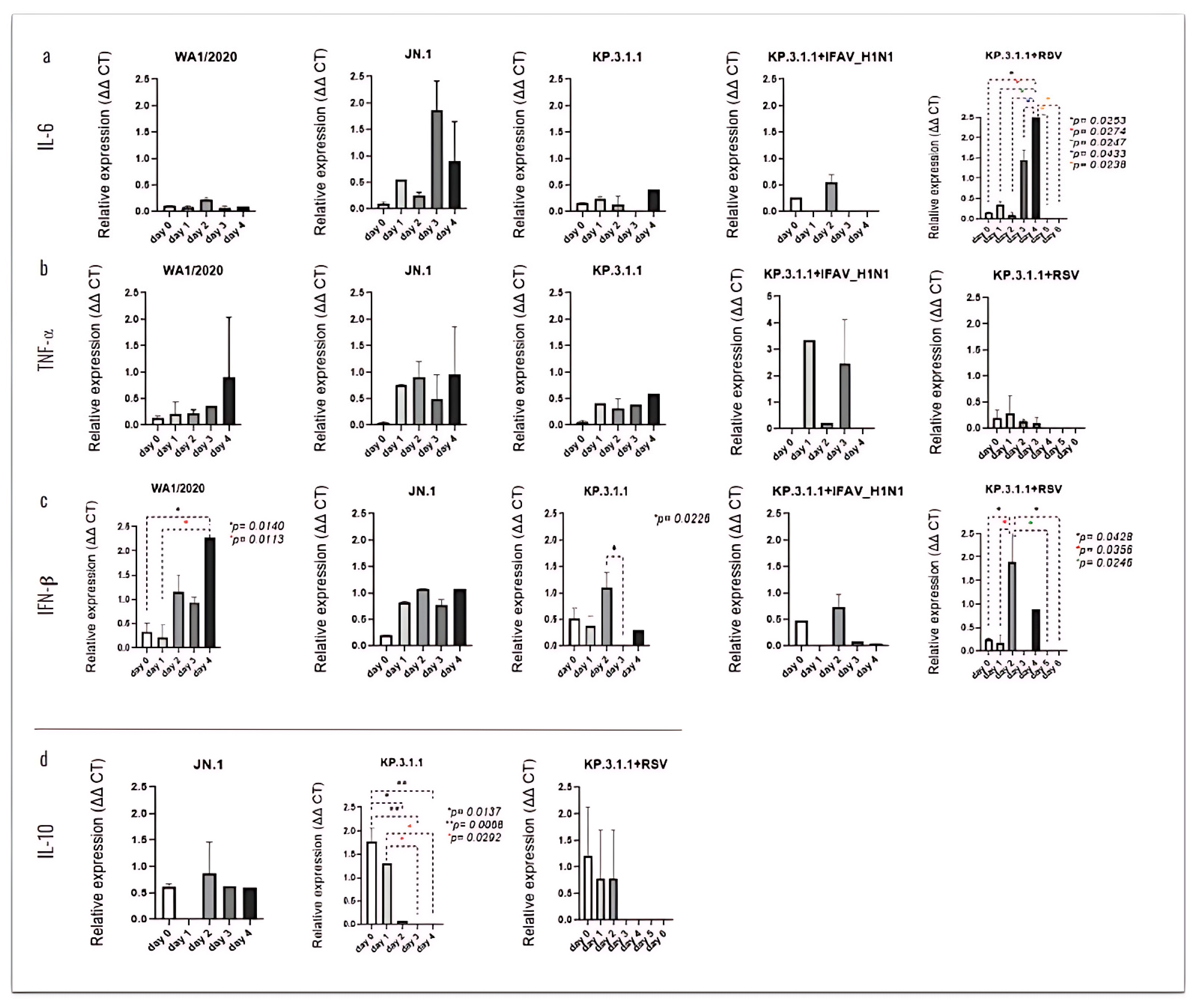
3.7. SARS-CoV-2 Mono- and Coinfection with IFAV_H1N1 and RSV Stimulate Strong Pro-Inflammatory Cytokine Response by hBAEC
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A/A | antibiotic/antimycotic |
| AF | Alexa Fluor |
| ALI | air-liquid interface |
| ANOVA | Analysis of variances |
| Abs | antibodies |
| BEI Resources | Biodefense and Emerging Infections Research Resources |
| BF | brightfield |
| BSA | bovine serum albumin |
| CDC | Centers for Disease Control |
| CDI | Center for Discovery and Innovation |
| COVID-19 | Coronavirus Infectious Disease 2019 |
| CPE | cytopathic effect |
| CV | crystal violet |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DMEM | Dulbecco’s Modified Eagle Medium |
| EMEM | Eagle’s Minimum Essential Medium |
| FBS | fetal bovine serum |
| gDNA | genomic DNA |
| HA | haemagglutinin |
| hAEC | human airway epithelial cells |
| hBAEC | human airway epithelial cells derived from bronchiolar tissue |
| HCoP | human convalescent plasma |
| Hep-2 | Human cervix epithelial cells |
| HMH-BioR | Hackensack Meridian Health BioRepository |
| IFAV_H1N1 | Influenza A virus |
| LOQ | limit of quantification |
| M | matrix |
| mAbs | monoclonal antibodies |
| MCC | Mander’s colocalization coefficient |
| MDCK | Madin-Darby Canine Kidney |
| MOI | Multiplicity of infection |
| NA | neuraminidase |
| NBF | neutral buffered formalin |
| NJDOH | New Jersey Department of Health |
| NTD | N-terminal domain |
| NT50 | Neutralization titers at 50% |
| ORF2 | open reading frame 2 |
| PCC | Pearson’s correlation coefficient |
| PET | polyester |
| PFU | Plaque forming unit assay |
| RBD | receptor-binding domain |
| RSV | Respiratory Syncytial Virus |
| RT | room temperature |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| SD | standard deviation |
| SE | standard error |
| TPCK | TPCK-treated trypsin from bovine pancreas |
| WA1/2020 | Washington strain |
| WHO | World Health Organization |
References
- Maison, D.P.; Tasissa, H.; Deitchman, A.; Peluso, M.J.; Deng, Y.; Miller, F.D.; Henrich, T.J.; Gerschenson, M. COVID-19 clinical presentation, management, and epidemiology: A concise compendium. Front. Public Health. 2025, 13, 1498445. [Google Scholar] [CrossRef]
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; COVID-19 Genomics UK Consortium; Peacock, S.J.; Barclay, W.S.; de Silva, T.I.; Towers, G.J.; et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Duchene, S.; Featherstone, L.; Haritopoulou-Sinanidou, M.; Rambaut, A.; Lemey, P.; Baele, G. Temporal signal and the phylodynamic threshold of SARS-CoV-2. Virus Evol. 2020, 6, veaa061. [Google Scholar] [CrossRef]
- Rouzine, I.M. Evolutionary Mechanisms of the Emergence of the Variants of Concern of SARS-CoV-2. Viruses 2025, 17, 197. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, D.; Chen, Y.; Sun, Y.; Jin, C.; Hu, C.; Feng, Y.; Su, J.; Ren, L.; Hao, Y.; et al. Characterization of RBD-specific cross-neutralizing antibodies responses against SARS-CoV-2 variants from COVID-19 convalescents. Front. Immunol. 2023, 14, 1160283. [Google Scholar] [CrossRef]
- CDC COVID Data Tracker: Variant Proportions. Available online: https://covid.cdc.gov/covid-data-tracker/#variant-proportions (accessed on 20 May 2025).
- Kim, D.; Quinn, J.; Pinsky, B.; Shah, N.H.; Brown, I. Rates of Co-infection Between SARS-CoV-2 and Other Respiratory Pathogens. J. Am. Med. Assoc. 2020, 323, 2085–2086. [Google Scholar] [CrossRef]
- Giannattasio, A.; Maglione, M.; D’Anna, C.; Muzzica, S.; Angrisani, F.; Acierno, S.; Perrella, A.; Tipo, V. Silent RSV in infants with SARS-CoV-2 infection: A case series. Pediatr. Pulmonol. 2021, 56, 3044–3046. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.L.; Li, Y.Q.; Chen, X.; Lin, H.; Jiang, Z.C.; Gu, D.L.; Chen, X.; Tang, C.X.; Xie, Z.Q. Co-Infection with Common Respiratory Pathogens and SARS-CoV-2 in Patients with COVID-19 Pneumonia and Laboratory Biochemistry Findings: A Retrospective Cross-Sectional Study of 78 Patients from a Single Center in China. Med. Sci. Monit. 2021, 27, e929783. [Google Scholar] [CrossRef]
- Halabi, K.C.; Wang, H.; Leber, A.L.; Sánchez, P.J.; Ramilo, O.; Mejias, A. Respiratory syncytial virus and SARS-CoV-2 coinfections in children. Pediatr. Pulmonol. 2022, 57, 3158–3160. [Google Scholar] [CrossRef]
- Yan, X.; Li, K.; Lei, Z.; Luo, J.; Wang, Q.; Wei, S. Prevalence and associated outcomes of coinfection between SARS-CoV-2 and influenza: A systematic review and meta-analysis. Int. J. Infect. Dis. 2023, 136, 29–36. [Google Scholar] [CrossRef]
- Liang, K.; Barnett, K.C.; Hsu, M.; Chou, W.C.; Bais, S.S.; Riebe, K.; Xie, Y.; Nguyen, T.T.; Ogiun, T.H., III; Vannella, K.M.; et al. Initiator cells death event induced by SARS-CoV-2 in the human airway epithelium. Sci. Immunol. 2024, 9, 97. [Google Scholar] [CrossRef]
- Zhu, N.; Wang, W.; Liu, Z.; Liang, C.; Wang, W.; Ye, F.; Huang, B.; Zhao, L.; Wang, H.; Zhou, W.; et al. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat. Commun. 2020, 11, 3910. [Google Scholar] [CrossRef]
- Yeung-Luk, B.H.; Narayanan, G.A.; Ghosh, B.; Wally, A.; Lee, E.; Mokaya, M.; Wankhade, E.; Zhang, R.; Lee, B.; Park, B.; et al. SARS-CoV-2 infection alters mitochondrial and cytoskeletal function in human respiratory epithelial cells mediated by expression of spike protein. mBio 2023, 14, e0082023. [Google Scholar] [CrossRef] [PubMed]
- Stölting, H.; Baillon, L.; Frise, R.; Bonner, K.; Hewitt, R.J.; Molyneaux, P.L.; Gore, M.L.; Breathing Together Consortium; Barclay, W.S.; Saglani, S.; et al. Distinct airway epithelial immune responses after infection with SARS-CoV-2 compared to H1N1. Mucosal Immunol. 2022, 15, 952–963. [Google Scholar] [CrossRef] [PubMed]
- Cheemarla, N.R.; Mihaylova, V.T.; Watkins, T.A.; Foxman, E.F. Counterintuitive effect of antiviral therapy on influenza A-SARS-CoV-2 coinfection due to viral interference. bioRxiv 2023. [Google Scholar] [CrossRef]
- Mendoza, E.J.; Manguiat, K.; Wood, H.; Drebot, M. Two Detailed Plaque Assay Protocols for the Quantification of Infectious SARS-CoV-2. Curr. Protoc. Microbiol. 2020, 57, ecpmc105. [Google Scholar] [CrossRef]
- Li, Y.; Wan, Y.; Liu, P.; Zhao, J.; Lu, G.; Qi, J.; Wang, Q.; Lu, X.; Wu, Y.; Liu, W.; et al. A humanized neutralizing antibody against MERS-CoV targeting the receptor-binding domain of the spike protein. Cell Res. 2015, 25, 1237–1249. [Google Scholar] [CrossRef]
- Corman, V.M.; Müller, M.A.; Costabel, U.; Timm, J.; Binger, T.; Meyer, B.; Kreher, P.; Lattwein, E.; Eschbach-Bludau, M.; Nitsche, A.; et al. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Eurosurveillance 2012, 17, 20334. [Google Scholar] [CrossRef]
- World Health Organization. WHO Information for the Molecular Detection of Influenza Viruses, 7th ed.; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Ohol, Y.M.; Wang, Z.; Kemble, G.; Duke, G. Direct Inhibition of Cellular Fatty Acid Synthase Impairs Replication of Respiratory Syncytial Virus and Other Respiratory Viruses. PLoS ONE 2015, 10, e0144648. [Google Scholar] [CrossRef]
- Sankuntaw, N.; Punyadee, N.; Chantratita, W.; Lulitanond, V. Coinfection with respiratory syncytial virus and rhinovirus increases IFN-λ1 and CXCL10 expression in human primary bronchial epithelial cells. New Microbiol. 2024, 47, 60–67. [Google Scholar]
- Dunn, K.W.; Kamocka, M.M.; McDonald, J.H. A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 2011, 300, C723–C742. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−∆∆CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Mediavilla, J.R.; Lozy, T.; Lee, A.; Kim, J.; Kan, V.W.; Titova, E.; Amin, A.; Zody, M.C.; Corvelo, A.; Oschwald, D.M.; et al. Molecular and Clinical Epidemiology of SARS-CoV-2 Infection among Vaccinated and Unvaccinated Individuals in a Large Healthcare Organization from New Jersey. Viruses 2023, 15, 1699. [Google Scholar] [CrossRef] [PubMed]
- Qudus, M.S.; Tian, M.; Sirajuddin, S.; Liu, S.; Afaq, U.; Wali, M.; Liu, J.; Pan, P.; Luo, Z.; Zhang, Q.; et al. The roles of critical pro-inflammatory cytokines in the drive of cytokine storm during SARS-CoV-2 infection. J. Med. Virol. 2023, 95, e28751. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, H.F.L.; Domachowske, J.B. Inflammatory responses to respiratory syncytial virus (RSV) infection and the development of immunomodulatory pharmacotherapeutics. Curr. Med. Chem. 2012, 19, 1424–1431. [Google Scholar] [CrossRef] [PubMed]
- Khalil, B.A.L.; Elemam, N.M.; Maghazachi, A.A. Chemokines and chemokine receptors during COVID-19 infection. Comput. Struct. Biotechnol. J. 2021, 19, 976–988. [Google Scholar] [CrossRef]
- Gu, Y.; Zuo, X.; Zhang, S.; Ouyang, Z.; Jiang, S.; Wang, F.; Wang, G. The Mechanism behind Influenza Virus Cytokine Storm. Viruses 2021, 13, 1362. [Google Scholar] [CrossRef]
- Somersan-Karakaya, S.; Mylonakis, E.; Menon, V.P.; Wells, J.C.; Ali, S.; Sivapalasingam, S.; Sun, Y.; Bhore, R.; Mei, J.; Miller, J.; et al. Casirivimab and Imdevimab for the Treatment of Hospitalized Patients with COVID-19. J. Infect. Dis. 2022, 227, 23–34. [Google Scholar] [CrossRef]
- Regeneron Pharmaceuticals Inc. Regeneron’s Next Generation Monoclonal Antibodies are Active Against All Known Variants of Concern, Including Both Omicron and Delta. Available online: https://investor.regeneron.com/static-files/4aed42a1-3d26-48af-bd01-3f0c92938c11 (accessed on 2 April 2025).
- Sherchan, R.; Cannady, P., Jr. Casirivimab. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2025. [Google Scholar] [PubMed]
- FDA Updates on Bebtelovimab|FDA. Available online: https://www.fda.gov/drugs/drug-safety-and-availability/fda-updates-bebtelovimab (accessed on 2 April 2025).
- Shrestha, L.B.; Foster, C.; Rawlinson, W.; Tedla, N.; Bull, R.A. Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission. Rev. Med. Virol. 2022, 32, e2381. [Google Scholar] [CrossRef]
- Parsons, R.J.; Acharya, P. Evolution of the SARS-CoV-2 Omicron spike. Cell Rep. 2023, 42, 113444. [Google Scholar] [CrossRef]
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.; et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. Nature 2022, 603, 679–686. [Google Scholar] [CrossRef] [PubMed]
- Cele, S.; Jackson, L.; Khoury, D.S.; Khan, K.; Moyo-Gwete, T.; Tegally, H.; San, J.E.; Cromer, D.; Scheepers, C.; Amoako, D.; et al. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. Nature 2022, 602, 654–656. [Google Scholar] [CrossRef] [PubMed]
- Dejnirattisai, W.; Shaw, R.H.; Supasa, P.; Liu, C.; Stuart, A.S.; Pollard, A.J.; Liu, X.; Lambe, T.; Crook, D.; Stuart, D.I.; et al. Reduced neutralization of SARS-CoV-2 omicron B.1.1.529 variant by post-immunization serum. Lancet 2022, 399, 234–236. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.O.; Jette, C.A.; Abernathy, M.E.; Dam, K.A.; Esswein, S.R.; Gristick, H.B.; Malyutin, A.G.; Sharaf, N.G.; Huey-Tubman, K.E.; Lee, Y.E.; et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature 2020, 588, 682–687. [Google Scholar] [CrossRef]
- Planas, D.; Staropoli, I.; Michel, V.; Lemoine, F.; Donati, F.; Prot, M.; Porrot, F.; Guivel-Benhassine, F.; Jeyarajah, B.; Brisebarre, A.; et al. Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion. Nat. Commun. 2024, 15, 2254. [Google Scholar] [CrossRef]
- Wang, Q.; Iketani, S.; Li, Z.; Liu, L.; Guo, Y.; Huang, Y.; Bowen, A.D.; Liu, M.; Wang, M.; Yu, J.; et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell 2023, 186, 279–286.e8. [Google Scholar] [CrossRef]
- Li, P.; Faraone, J.N.; Hsu, C.C.; Chamblee, M.; Zheng, Y.M.; Carlin, C.; Bednash, J.S.; Horowitz, J.C.; Mallampalli, R.K.; Saif, L.J.; et al. Neutralization escape, infectivity, and membrane fusion of JN.1-derived SARS-CoV-2 SLip, FLiRT, and KP.2 variants. Cell Rep. 2024, 43, 114520. [Google Scholar] [CrossRef]
- Wang, Q.; Mellis, I.A.; Ho, J.; Bowen, A.; Kowalski-Dobson, T.; Valdez, R.; Katsamba, P.S.; Wu, M.; Lee, C.; Shapiro, L.; et al. Recurrent SARS-CoV-2 spike mutations confer growth advantages to select JN.1 sublineages. Emerg. Microbes Infect. 2024, 13, 2402880. [Google Scholar] [CrossRef]
- Li, P.; Faraone, J.N.; Hsu, C.C.; Chamblee, M.; Liu, Y.; Zheng, Y.; Xu, Y.; Carlin, C.; Horowitz, J.C.; Mallampalli, R.K.; et al. Neutralization and spike stability of JN.1-derived LB.1, KP.2.3, KP.3, and KP.3.1.1 subvariants. mBio 2025, 16, e0046425. [Google Scholar] [CrossRef]
- Escalera, A.; Laporte, M.; Turner, S.; Karakus, U.; Gonzalez-Reiche, A.S.; van de Guchte, A.; Farrugia, K.; Khalil, Z.; van Bakel, H.; Smith, D.; et al. The impact of S2 mutations on Omicron SARS-CoV-2 cell surface expression and fusogenicity. Emerg. Microbes Infect. 2024, 13, 2297553. [Google Scholar] [CrossRef]
- Pastorio, C.; Zech, F.; Noettger, S.; Jung, C.; Jacob, T.; Sanderson, T.; Sparrer, K.M.J.; Kirchhoff, F. Determinants of Spike infectivity, processing, and neutralization in SARS-CoV-2 Omicron subvariants BA.1 and BA.2. Cell Host Microbe 2022, 30, 1255–1268.e5. [Google Scholar] [CrossRef] [PubMed]
- Maltezou, H.C.; Papanikolopoulou, A.; Vassiliu, S.; Theodoridou, K.; Nikolopoulou, G.; Sipsas, N.V. COVID-19 and Respiratory Virus Co-Infections: A Systematic Review of the Literature. Viruses 2023, 15, 865. [Google Scholar] [CrossRef]
- About Respiratory Illnesses|Respiratory Illnesses|CDC. Available online: https://www.cdc.gov/respiratory-viruses/about/index.html (accessed on 4 April 2025).
- Takashita, E.; Ichikawa, M.; Fujisaki, S.; Morita, H.; Nagata, S.; Miura, H.; Watanabe, S.; Hasegawa, H.; Kawaoka, Y. Antiviral susceptibility of SARS-CoV-2 and influenza viruses from 3 co-infected pediatric patients. Int. J. Infect. Dis. 2024, 146, 107134. [Google Scholar] [CrossRef] [PubMed]
- Oppenlander, K.E.; Chung, A.A.; Clabaugh, D. Respiratory Syncytial Virus Bronchiolitis: Rapid Evidence Review. Am. Fam. Physician 2023, 108, 52–57. [Google Scholar] [PubMed]
- Mandelia, Y.; Procop, G.W.; Richter, S.S.; Worley, S.; Liu, W.; Esper, F. Dynamics and predisposition of respiratory viral co-infections in children and adults. Clin. Microbiol. Infect. 2021, 27, 631.e1–631.e6. [Google Scholar] [CrossRef]
- Paul, L.A.; Daneman, N.; Schwartz, K.L.; Science, M.; Brown, K.A.; Whelan, M.; Chan, E.; Buchan, S.A. Association of Age and Pediatric Household Transmission of SARS-CoV-2 Infection. JAMA Pediatr. 2021, 175, 1151–1158. [Google Scholar] [CrossRef] [PubMed]
- Do, T.N.D.; Donckers, K.; Vangeel, L.; Chatterjee, A.K.; Gallay, P.A.; Bobardt, M.D.; Bilello, J.P.; Cihlar, T.; De Jonghe, S.; Neyts, J.; et al. A robust SARS-CoV-2 replication model in primary human epithelial cells at the air liquid interface to assess antiviral agents. Antivir. Res. 2021, 192, 105122. [Google Scholar] [CrossRef]
- Sano, E.; Suzuki, T.; Hashimoto, R.; Itoh, Y.; Sakamoto, A.; Sakai, Y.; Saito, A.; Okuzaki, D.; Motooka, D.; Muramoto, Y.; et al. Cell response analysis in SARS-CoV-2 infected bronchial organoids. Commun. Biol. 2022, 5, 516. [Google Scholar] [CrossRef]
- Tanneti, N.S.; Patel, A.K.; Tan, L.H.; Marques, A.D.; Perera, R.A.P.M.; Sherrill-Mix, S.; Kelly, B.J.; Renner, D.M.; Collman, R.G.; Rodino, K.; et al. Comparison of SARS-CoV-2 variants of concern in primary human nasal cultures demonstrates Delta as most cytopathic and Omicron as fastest replicating. mBio 2024, 15, e0312923. [Google Scholar] [CrossRef]
- Rahman, M.; Irmler, M.; Keshavan, S.; Introna, M.; Beckers, J.; Palmberg, L.; Johanson, G.; Ganguly, K.; Upadhyay, S. Differential Effect of SARS-CoV-2 Spike Glycoprotein 1 on Human Bronchial and Alveolar Lung Mucosa Models: Implications for Pathogenicity. Viruses 2021, 13, 2537. [Google Scholar] [CrossRef]
- Dichtl, S.; Zaderer, V.; Kozubowski, V.; Abd, E.L.; Halim, H.; Lafon, E.; Lanser, L.; Weiss, G.; Lass-Flörl, C.; Wilflingseder, D.; et al. Cilgavimab/Tixagevimab as alternative therapeutic approach for BA.2 infections. Front. Med. 2022, 9, 1005589. [Google Scholar] [CrossRef] [PubMed]
- Zarkoob, H.; Allué-Guardia, A.; Chen, Y.C.; Garcia-Vilanova, A.; Jung, O.; Coon, S.; Song, M.J.; Park, J.G.; Oladunni, F.; Miller, J.; et al. Modeling SARS-CoV-2 and influenza infections and antiviral treatments in human lung epithelial tissue equivalents. Commun. Biol. 2022, 5, 810. [Google Scholar] [CrossRef]
- Liu, D.; Leung, K.Y.; Lam, H.Y.; Zhang, R.; Fan, Y.; Xie, X.; Chan, K.H.; Hung, I.F. Interaction and antiviral treatment of coinfection between SARS-CoV-2 and influenza in vitro. Virus Res. 2024, 345, 199371. [Google Scholar] [CrossRef] [PubMed]
- Sigal, A. Milder disease with Omicron: Is it the virus or the pre-existing immunity? Nat. Rev. Immunol. 2022, 22, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Abdullahi, A.; Ferreira, I.A.T.M.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarez, N.; Gonzalez-Jimenez, I.; Rasheed, R.; Goldgirsh, K.; Park, S.; Perlin, D.S. Genetic and Immunological Profiling of Recent SARS-CoV-2 Omicron Subvariants: Insights into Immune Evasion and Infectivity in Monoinfections and Coinfections. Viruses 2025, 17, 918. https://doi.org/10.3390/v17070918
Alvarez N, Gonzalez-Jimenez I, Rasheed R, Goldgirsh K, Park S, Perlin DS. Genetic and Immunological Profiling of Recent SARS-CoV-2 Omicron Subvariants: Insights into Immune Evasion and Infectivity in Monoinfections and Coinfections. Viruses. 2025; 17(7):918. https://doi.org/10.3390/v17070918
Chicago/Turabian StyleAlvarez, Nadine, Irene Gonzalez-Jimenez, Risha Rasheed, Kira Goldgirsh, Steven Park, and David S. Perlin. 2025. "Genetic and Immunological Profiling of Recent SARS-CoV-2 Omicron Subvariants: Insights into Immune Evasion and Infectivity in Monoinfections and Coinfections" Viruses 17, no. 7: 918. https://doi.org/10.3390/v17070918
APA StyleAlvarez, N., Gonzalez-Jimenez, I., Rasheed, R., Goldgirsh, K., Park, S., & Perlin, D. S. (2025). Genetic and Immunological Profiling of Recent SARS-CoV-2 Omicron Subvariants: Insights into Immune Evasion and Infectivity in Monoinfections and Coinfections. Viruses, 17(7), 918. https://doi.org/10.3390/v17070918








