Antagonism in Orthotospoviruses Is Reflected in Plant Small RNA Profile
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses and Their Maintenance
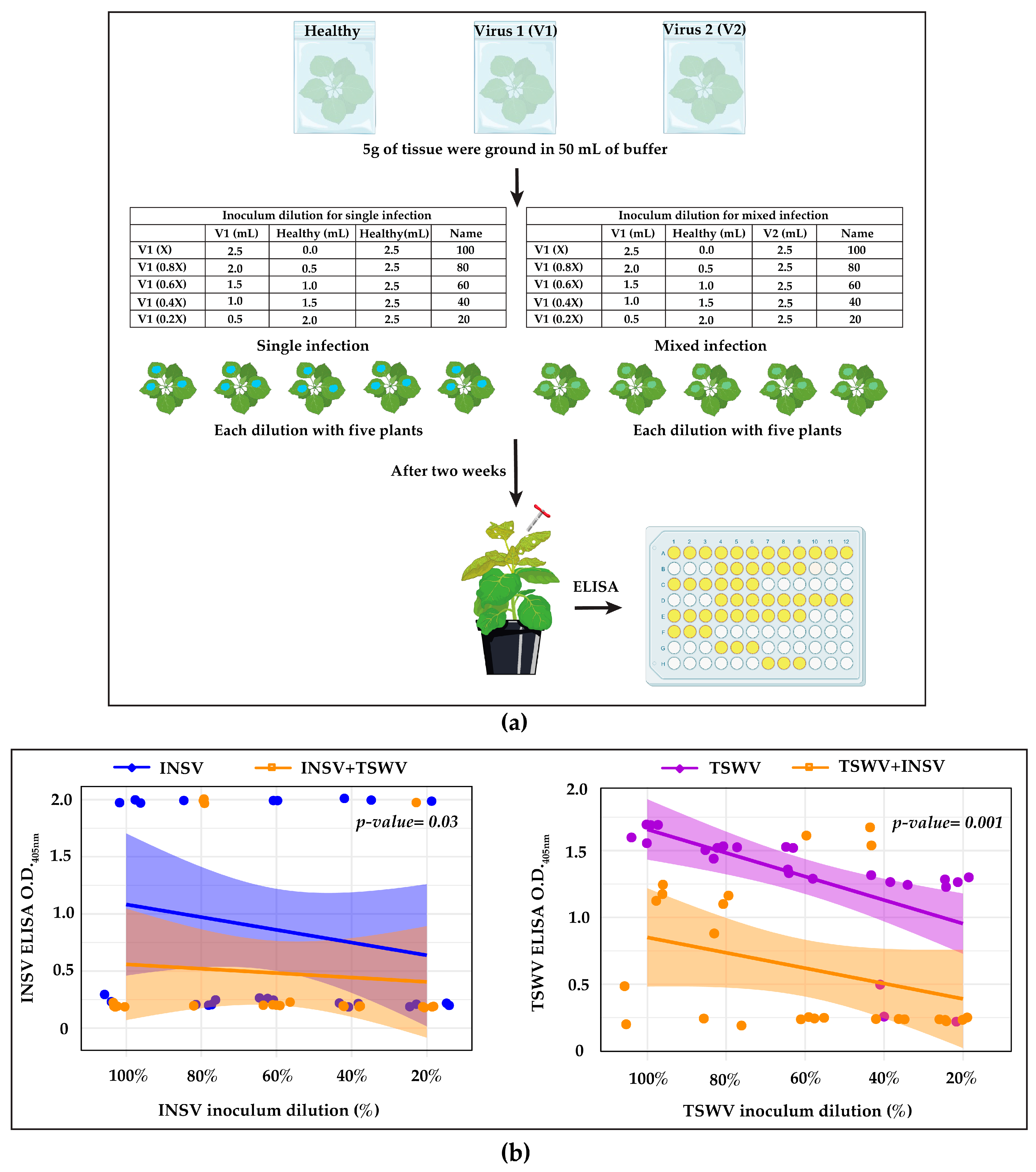
2.2. Virus–Virus Interaction
2.3. Sequential Inoculation for Study of Super Infection Exclusion
2.4. ELISA Testing
2.5. Statistical Analysis
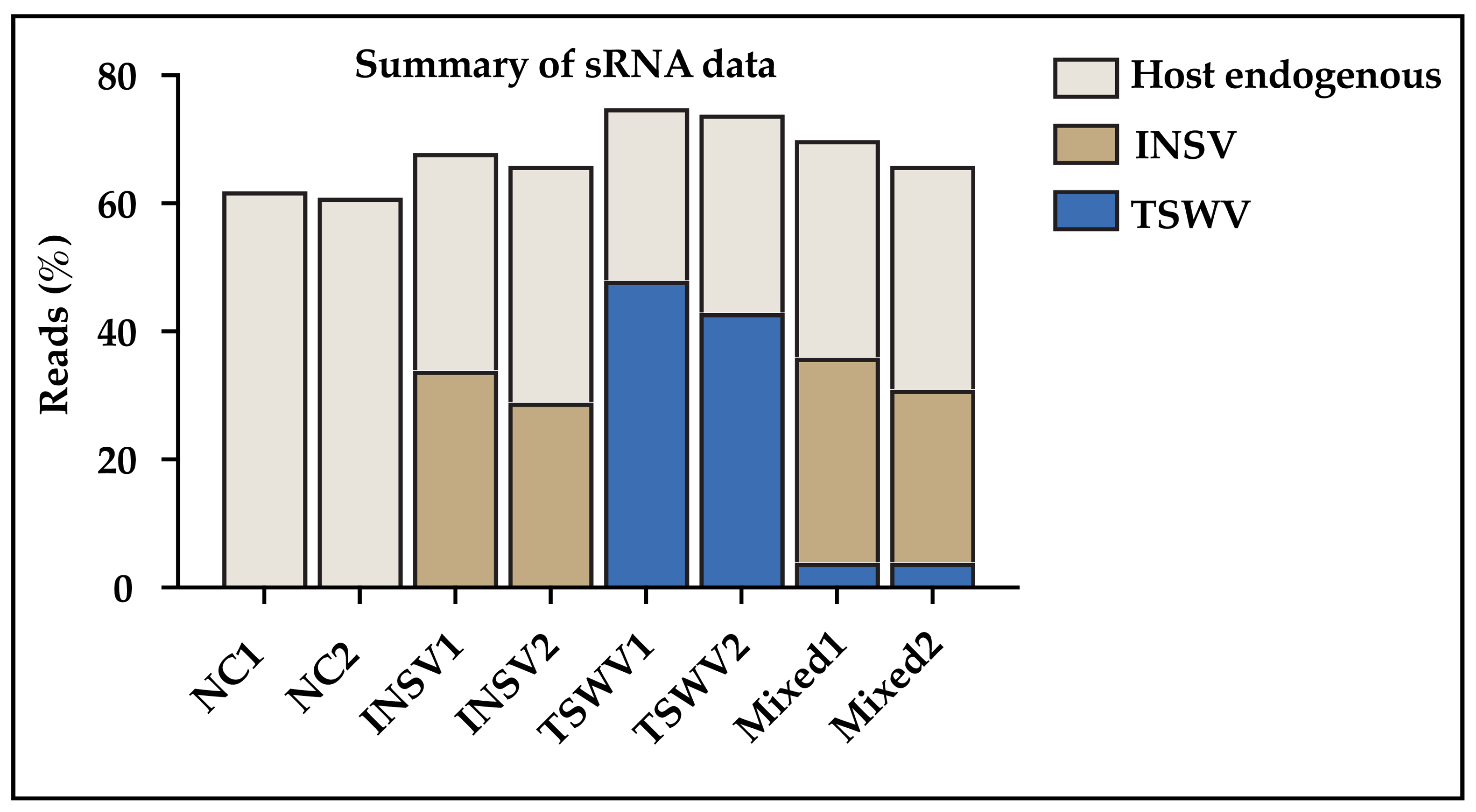
2.6. Virus Inoculations for sRNA Sequencing
2.7. RNA Extraction, sRNA Sequencing, and qPCR
2.8. Bioinformatic Analysis
3. Results and Discussion
3.1. INSV and TSWV Have an Antagonistic Interaction
3.2. Host Plant Appears to Contribute to the Antagonism
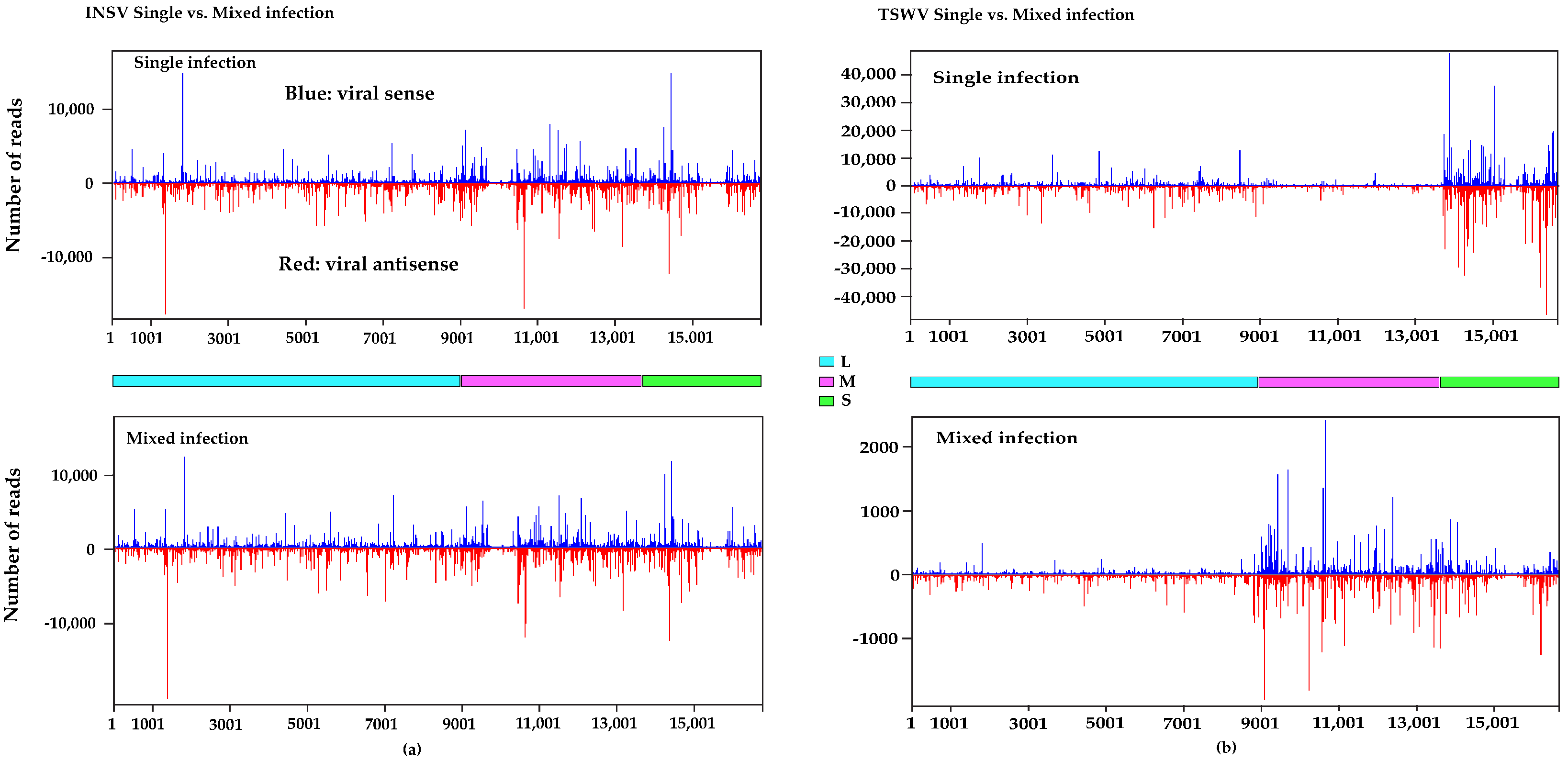
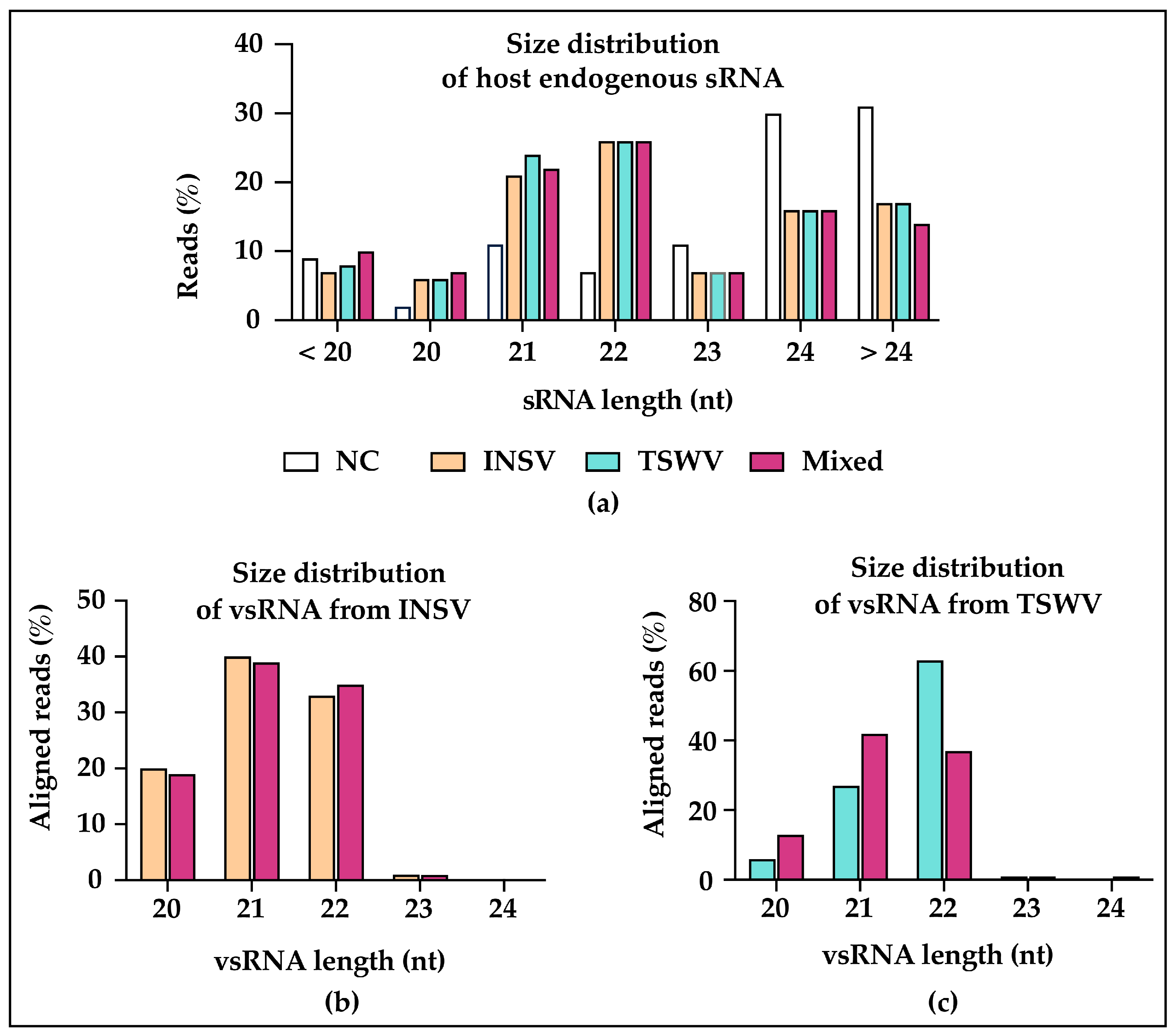
3.3. Small RNA (sRNA) Profiles Reflect Viral Antagonism
3.4. Different DCLs and AGOs Could Be Recruited During Co-Infection
3.5. Virus-Activated Small Interfering RNAs (vasiRNAs) in Virus-Infected Plants
3.6. Conserved Host Response in miRNAs Regulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rochow, W.; Ross, A.F. Virus multiplication in plants doubly infected by potato viruses X and Y. Virology 1955, 1, 10–27. [Google Scholar] [CrossRef]
- bance, V.B. Replication of potato virus X RNA is altered in coinfections with potato virus Y. Virology 1991, 182, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Ross, A. Local lesion formation and virus production following simultaneous inoculation with Potato virus-X and virus-Y. Phytopathology 1950, 40, 24. [Google Scholar]
- Chávez-Calvillo, G.; Contreras-Paredes, C.A.; Mora-Macias, J.; Noa-Carrazana, J.C.; Serrano-Rubio, A.A.; Dinkova, T.D.; Carrillo-Tripp, M.; Silva-Rosales, L. Antagonism or synergism between papaya ringspot virus and papaya mosaic virus in Carica papaya is determined by their order of infection. Virology 2016, 489, 179–191. [Google Scholar] [CrossRef]
- Crespo, O.; Robles, C.; Ruiz, L.; Janssen, D. Antagonism of Cucumber green mottle mosaic virus against Tomato leaf curl New Delhi virus in zucchini and cucumber. Ann. Appl. Biol. 2020, 176, 147–157. [Google Scholar] [CrossRef]
- Syller, J.; Grupa, A. Antagonistic within-host interactions between plant viruses: Molecular basis and impact on viral and host fitness. Mol. Plant Pathol. 2016, 17, 769–782. [Google Scholar] [CrossRef]
- Rosa, C.; Kuo, Y.W.; Wuriyanghan, H.; Falk, B.W. RNA interference mechanisms and applications in plant pathology. Annu. Rev. Phytopathol. 2018, 56, 581–610. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; Guo, J.; Zhang, X.; Meulia, T.; Paul, P.; Madden, L.V.; Li, D.; Qu, F. Random plant viral variants attain temporal advantages during systemic infections and in turn resist other variants of the same virus. Sci. Rep. 2015, 5, 15346. [Google Scholar] [CrossRef]
- Zhang, X.F.; Sun, R.; Guo, Q.; Zhang, S.; Meulia, T.; Halfmann, R.; Li, D.; Qu, F. A self-perpetuating repressive state of a viral replication protein blocks superinfection by the same virus. PLoS Pathog. 2017, 13, e1006253. [Google Scholar] [CrossRef]
- Goodman, R.M.; Ross, A.F. Enhancement by potato virus Y of potato virus X synthesis in doubly infected tobacco depends on the timing of invasion by the viruses. Virology 1974, 58, 263–271. [Google Scholar] [CrossRef]
- Dong, J.H.; Cheng, X.F.; Yin, Y.Y.; Fang, Q.; Ding, M.; Li, T.T.; Zhang, L.Z.; Su, X.X.; McBeath, J.H.; Zhang, Z.K. Characterization of tomato zonate spot virus, a new tospovirus in China. Arch. Virol. 2008, 153, 855–864. [Google Scholar] [CrossRef] [PubMed]
- Pappu, H.; Jones, R.; Jain, R. Global status of tospovirus epidemics in diverse cropping systems: Successes achieved and challenges ahead. Virus Res. 2009, 141, 219–236. [Google Scholar] [CrossRef]
- Margaria, P.; Rosa, C. First complete genome sequence of a tomato spotted wilt virus isolate from the United States and its relationship to other TSWV isolates of different geographic origin. Arch. Virol. 2015, 160, 2915–2920. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Margaria, P.; Rosa, C. Characterization of the first complete genome sequence of an Impatiens necrotic spot orthotospovirus isolate from the United States and worldwide phylogenetic analyses of INSV isolates. BMC Res. Notes 2018, 11, 288. [Google Scholar] [CrossRef]
- Sialer, M.F.; Gallitelli, D. The occurrence of Impatiens necrotic spot virus and Tomato spotted wilt virus in mixed infection in tomato. J. Plant Pathol. 2000, 82, 244. [Google Scholar]
- Martinez-Ochoa, N.; Csinos, A.; Whitty, E.; Johnson, A.; Parrish, M. First Report on the Incidence of Mixed Infections of Impatiens necrotic spot virus (INSV) and Tomato spotted wilt virus (TSWV) in Tobacco Grown in Georgia, South Carolina, and Virginia. Plant Health Prog. 2003, 4, 40. [Google Scholar] [CrossRef]
- Martínez-Ochoa, N.; Csinos, A.; Webster, T.; Bertrand, P. Occurrence of mixed infections of Tomato spotted wilt virus (TSWV) and Impatiens necrotic spot virus (INSV) in weeds around tobacco fields in Georgia. In Proceedings of the American Phytopathological Society Abstracts, Charlotte, NC, USA,, 9–13 August 2003; Volume 93, p. S58. [Google Scholar]
- Zhao, K.; Rosa, C. Thrips as the transmission bottleneck for mixed infection of two orthotospoviruses. Plants 2020, 9, 509. [Google Scholar] [CrossRef] [PubMed]
- Csorba, T.; Kontra, L.; Burgyán, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479, 85–103. [Google Scholar] [CrossRef]
- Kontra, L.; Csorba, T.; Tavazza, M.; Lucioli, A.; Tavazza, R.; Moxon, S.; Tisza, V.; Medzihradszky, A.; Turina, M.; Burgyán, J. Distinct effects of p19 RNA silencing suppressor on small RNA mediated pathways in plants. PLoS Pathog. 2016, 12, e1005935. [Google Scholar] [CrossRef]
- Jin, Y.; Zhao, J.H.; Guo, H.S. Recent advances in understanding plant antiviral RNAi and viral suppressors of RNAi. Curr. Opin. Virol. 2021, 46, 65–72. [Google Scholar] [CrossRef]
- Ding, S.W. Transgene silencing, RNA interference, and the antiviral defense mechanism directed by small interfering RNAs. Phytopathology 2023, 113, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature 2004, 431, 343–349. [Google Scholar] [CrossRef] [PubMed]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 447–459. [Google Scholar] [CrossRef]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.O.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef]
- Pérez-Quintero, Á.L.; Neme, R.; Zapata, A.; López, C. Plant microRNAs and their role in defense against viruses: A bioinformatics approach. BMC Plant Biol. 2010, 10, 138. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Bashir, N.H.; Li, Z.; Liu, C.; Shi, Y.; Chu, H. Plant microRNAs regulate the defense response against pathogens. Front. Microbiol. 2024, 15, 1434798. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, L.; Yang, Y.; Schmid, M.; Wang, Y. miRNA mediated regulation and interaction between plants and pathogens. Int. J. Mol. Sci. 2021, 22, 2913. [Google Scholar] [CrossRef]
- Sánchez-Tovar, M.R.; Rivera-Bustamante, R.F.; Saavedra-Trejo, D.L.; Guevara-González, R.G.; Torres-Pacheco, I. Mixed Plant Viral Infections: Complementation, Interference and Their Effects, a Review. Agronomy 2025, 15, 620. [Google Scholar] [CrossRef]
- Singhal, P.; Nabi, S.U.; Yadav, M.K.; Dubey, A. Mixed infection of plant viruses: Diagnostics, interactions and impact on host. J. Plant Dis. Prot. 2021, 128, 353–368. [Google Scholar] [CrossRef]
- Schnettler, E.; Hemmes, H.; Huismann, R.; Goldbach, R.; Prins, M.; Kormelink, R. Diverging affinity of tospovirus RNA silencing suppressor proteins, NSs, for various RNA duplex molecules. J. Virol. 2010, 84, 11542–11554. [Google Scholar] [CrossRef]
- de Ronde, D.; Pasquier, A.; Ying, S.; Butterbach, P.; Lohuis, D.; Kormelink, R. Analysis of T omato spotted wilt virus NSs protein indicates the importance of the N-terminal domain for avirulence and RNA silencing suppression. Mol. Plant Pathol. 2014, 15, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Bag, S.; Mitter, N.; Turina, M.; Pappu, H.R. Mutational analysis of two highly conserved motifs in the silencing suppressor encoded by tomato spotted wilt virus (genus Tospovirus, family Bunyaviridae). Arch. Virol. 2014, 159, 1499–1504. [Google Scholar] [CrossRef]
- Murray, G.G.; Kosakovsky Pond, S.L.; Obbard, D.J. Suppressors of RNAi from plant viruses are subject to episodic positive selection. Proc. R. Soc. B Biol. Sci. 2013, 280, 20130965. [Google Scholar] [CrossRef]
- Takeshita, M.; Koizumi, E.; Noguchi, M.; Sueda, K.; Shimura, H.; Ishikawa, N.; Matsuura, H.; Ohshima, K.; Natsuaki, T.; Kuwata, S.; et al. Infection dynamics in viral spread and interference under the synergism between Cucumber mosaic virus and Turnip mosaic virus. Mol. Plant-Microbe Interact. 2012, 25, 18–27. [Google Scholar] [CrossRef]
- Bombarely, A.; Rosli, H.G.; Vrebalov, J.; Moffett, P.; Mueller, L.A.; Martin, G.B. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant-Microbe Interact. 2012, 25, 1523–1530. [Google Scholar] [CrossRef]
- Johnson, N.R.; Yeoh, J.M.; Coruh, C.; Axtell, M.J. Improved placement of multi-mapping small RNAs. G3 Genes Genomes Genet. 2016, 6, 2103–2111. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Seguin, J.; Otten, P.; Baerlocher, L.; Farinelli, L.; Pooggin, M.M. MISIS: A bioinformatics tool to view and analyze maps of small RNAs derived from viruses and genomic loci generating multiple small RNAs. J. Virol. Methods 2014, 195, 120–122. [Google Scholar] [CrossRef] [PubMed]
- Blevins, T.; Rajeswaran, R.; Shivaprasad, P.V.; Beknazariants, D.; Si-Ammour, A.; Park, H.S.; Vazquez, F.; Robertson, D.; Meins, F., Jr.; Hohn, T.; et al. Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 2006, 34, 6233–6246. [Google Scholar] [CrossRef]
- Donaire, L.; Wang, Y.; Gonzalez-Ibeas, D.; Mayer, K.F.; Aranda, M.A.; Llave, C. Deep-sequencing of plant viral small RNAs reveals effective and widespread targeting of viral genomes. Virology 2009, 392, 203–214. [Google Scholar] [CrossRef]
- Mitter, N.; Koundal, V.; Williams, S.; Pappu, H. Differential expression of tomato spotted wilt virus-derived viral small RNAs in infected commercial and experimental host plants. PLoS ONE 2013, 8, e76276. [Google Scholar] [CrossRef] [PubMed]
- Margaria, P.; Miozzi, L.; Rosa, C.; Axtell, M.J.; Pappu, H.R.; Turina, M. Small RNA profiles of wild-type and silencing suppressor-deficient tomato spotted wilt virus infected Nicotiana benthamiana. Virus Res. 2015, 208, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Du, P.; Wang, X.; Yu, Y.Q.; Qiu, Y.H.; Li, W.; Gal-On, A.; Zhou, C.; Li, Y.; Ding, S.W. Virus infection triggers widespread silencing of host genes by a distinct class of endogenous siRNAs in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14613–14618. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Elvira-González, L.; Medina, V.; Rubio, L.; Galipienso, L. The persistent southern tomato virus modifies miRNA expression without inducing symptoms and cell ultra-structural changes. Eur. J. Plant Pathol. 2020, 156, 615–622. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, C.; Shi, C.; Yang, Z.; Wang, Y.; Zhou, T.; Sun, F.; Wang, H.; Zhao, S.; Qin, Q.; et al. Rice stripe virus NS3 protein regulates primary miRNA processing through association with the miRNA biogenesis factor OsDRB1 and facilitates virus infection in rice. PLoS Pathog. 2017, 13, e1006662. [Google Scholar] [CrossRef]
- Li, J.; Song, Q.; Zuo, Z.F.; Liu, L. MicroRNA398: A master regulator of plant development and stress responses. Int. J. Mol. Sci. 2022, 23, 10803. [Google Scholar] [CrossRef]
- Lin, K.Y.; Wu, S.Y.; Hsu, Y.H.; Lin, N.S. MiR398-regulated antioxidants contribute to Bamboo mosaic virus accumulation and symptom manifestation. Plant Physiol. 2022, 188, 593–607. [Google Scholar] [CrossRef]
- Gao, X.; Du, Z.; Hao, K.; Zhang, S.; Li, J.; Guo, J.; Wang, Z.; Zhao, S.; Sang, L.; An, M.; et al. ZmmiR398b negatively regulates maize resistance to sugarcane mosaic virus infection by targeting ZmCSD2/4/9. Mol. Plant Pathol. 2024, 25, e13462. [Google Scholar] [CrossRef]
- Naqvi, A.R.; Haq, Q.M.; Mukherjee, S.K. MicroRNA profiling of tomato leaf curl new delhi virus (tolcndv) infected tomato leaves indicates that deregulation of mir159/319 and mir172 might be linked with leaf curl disease. Virol. J. 2010, 7, 281. [Google Scholar] [CrossRef]
- Wu, C.; Li, X.; Guo, S.; Wong, S.M. Analyses of RNA-Seq and sRNA-Seq data reveal a complex network of anti-viral defense in TCV-infected Arabidopsis thaliana. Sci. Rep. 2016, 6, 36007. [Google Scholar] [CrossRef] [PubMed]


| INSV | Disease | Disease Incidence | |||
|---|---|---|---|---|---|
| Single | Incidence | INSV Mixed | INSV Infected | TSWV Infected | Mixed-Infected |
| INSV (X) | 60% | INSV (X) + TSWV (X) | 0% | 100% | 0% |
| INSV (0.8X) | 20% | INSV (0.8X) + TSWV (X) | 60% | 100% | 60% |
| INSV (0.6X) | 40% | INSV (0.6X) + TSWV (X) | 0% | 100% | 0% |
| INSV (0.4X) | 40% | INSV (0.4X) + TSWV (X) | 0% | 100% | 0% |
| INSV (0.2X) | 20% | INSV (0.2X) + TSWV (X) | 20% | 100% | 20% |
| TSWV | Disease | Disease Incidence | |||
| Single | Incidence | TSWV Mixed | TSWV Infected | INSV Infected | Mixed-Infected |
| TSWV (X) | 100% | TSWV (X) + INSV (X) | 60% | 100% | 60% |
| TSWV (0.8X) | 100% | TSWV (0.8X) + INSV (X) | 60% | 100% | 60% |
| TSWV (0.6X) | 100% | TSWV (0.6X) + INSV (X) | 20% | 100% | 20% |
| TSWV (0.4X) | 60% | TSWV (0.4X) + INSV (X) | 40% | 100% | 40% |
| TSWV (0.2X) | 80% | TSWV (0.2X) + INSV (X) | 0% | 100% | 0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, M.T.; Zhao, K.; Johnson, N.; Axtell, M.; Rosa, C. Antagonism in Orthotospoviruses Is Reflected in Plant Small RNA Profile. Viruses 2025, 17, 789. https://doi.org/10.3390/v17060789
Islam MT, Zhao K, Johnson N, Axtell M, Rosa C. Antagonism in Orthotospoviruses Is Reflected in Plant Small RNA Profile. Viruses. 2025; 17(6):789. https://doi.org/10.3390/v17060789
Chicago/Turabian StyleIslam, Md Tariqul, Kaixi Zhao, Nathan Johnson, Michael Axtell, and Cristina Rosa. 2025. "Antagonism in Orthotospoviruses Is Reflected in Plant Small RNA Profile" Viruses 17, no. 6: 789. https://doi.org/10.3390/v17060789
APA StyleIslam, M. T., Zhao, K., Johnson, N., Axtell, M., & Rosa, C. (2025). Antagonism in Orthotospoviruses Is Reflected in Plant Small RNA Profile. Viruses, 17(6), 789. https://doi.org/10.3390/v17060789







