Development of a Triplex Real-Time PCR Method for the Simultaneous Detection of Porcine Circovirus 2, 3, and 4 in China Between 2023 and 2024
Abstract
1. Introduction
2. Materials and Methods
2.1. Primers and Probes
2.2. Viruses and Clinical Samples
2.3. DNA/RNA Extraction and Reverse Transcription
2.4. Construction of Recombinant Plasmids
2.5. Optimization of the Triplex qPCR Assay
2.6. Establishment of Standard Curves
2.7. Specificity, Sensitivity, Repeatability, and Stability
2.8. Comparison of the Detection Results of Different Methods
2.9. Clinical Sample Detection
2.10. PCV2/PCV3/PCV4 Sequence Analyses
3. Results
3.1. Construction of Standard Plasmids
3.2. Optimization of Triplex qPCR Reaction Conditions
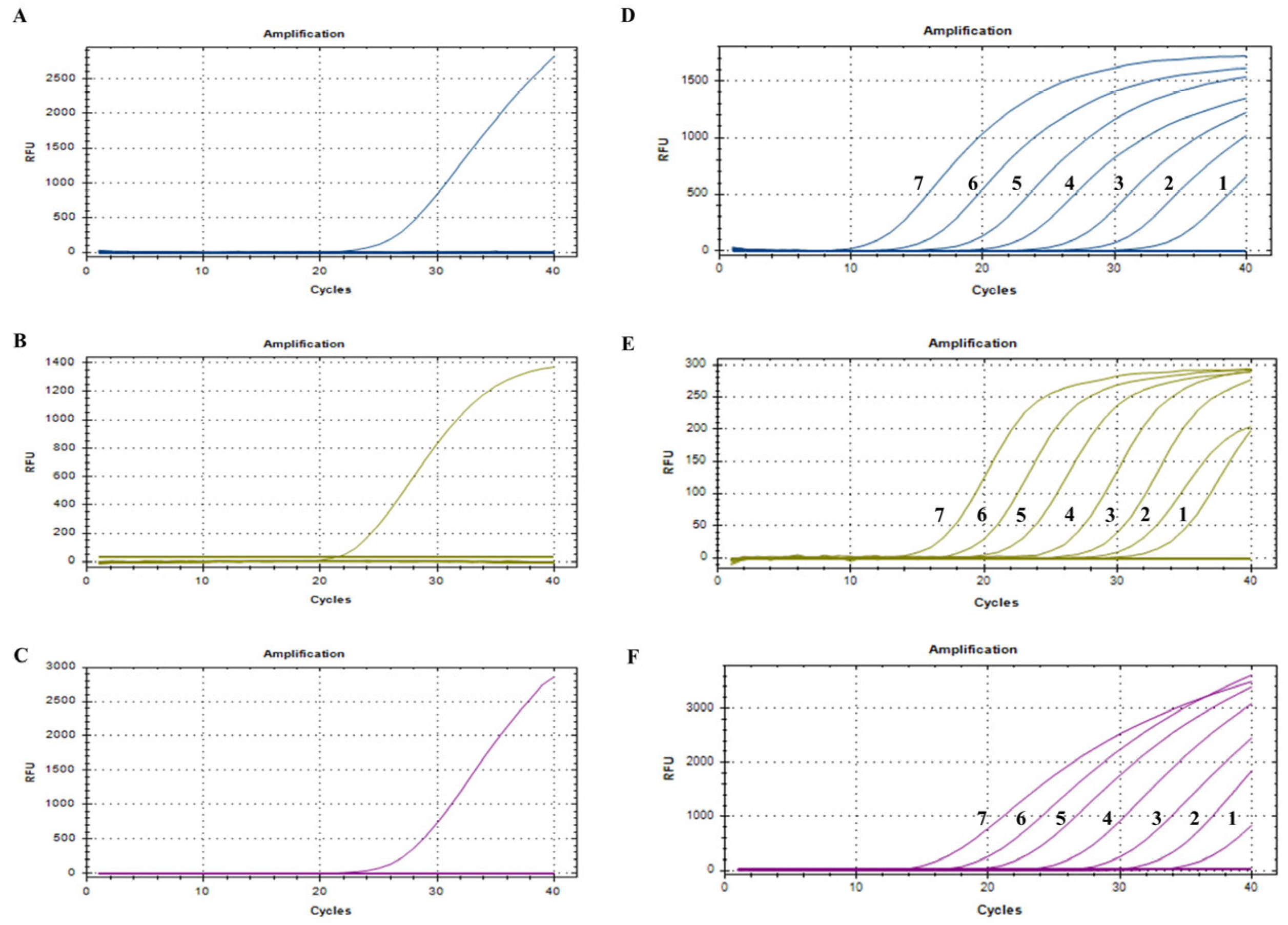
3.3. Establishment of Standard Curves
3.4. Specificity, Sensitivity, Repeatability, and Stability
3.5. Comparison of the Detection Results of Different Methods
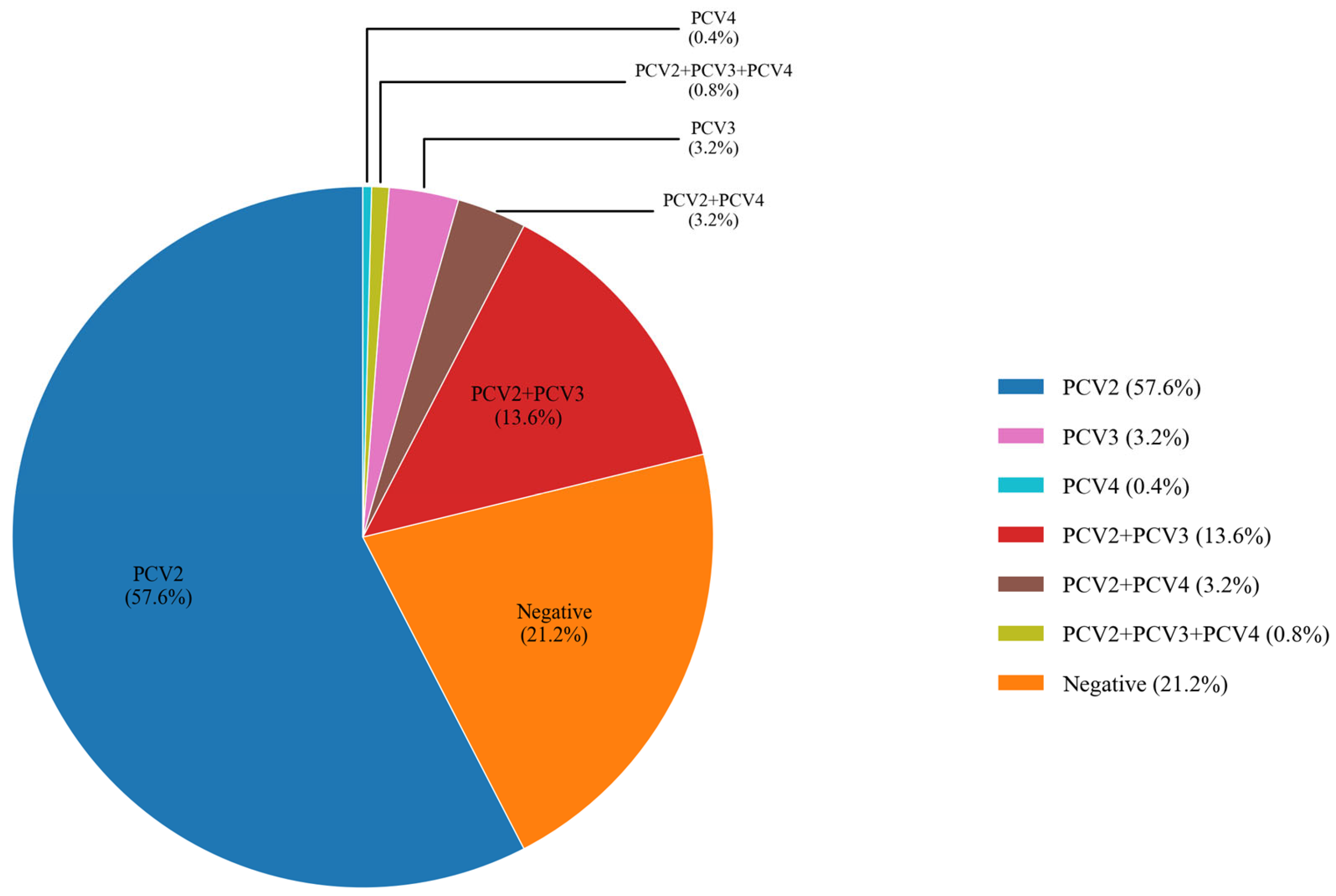
3.6. Clinical Sample Detection
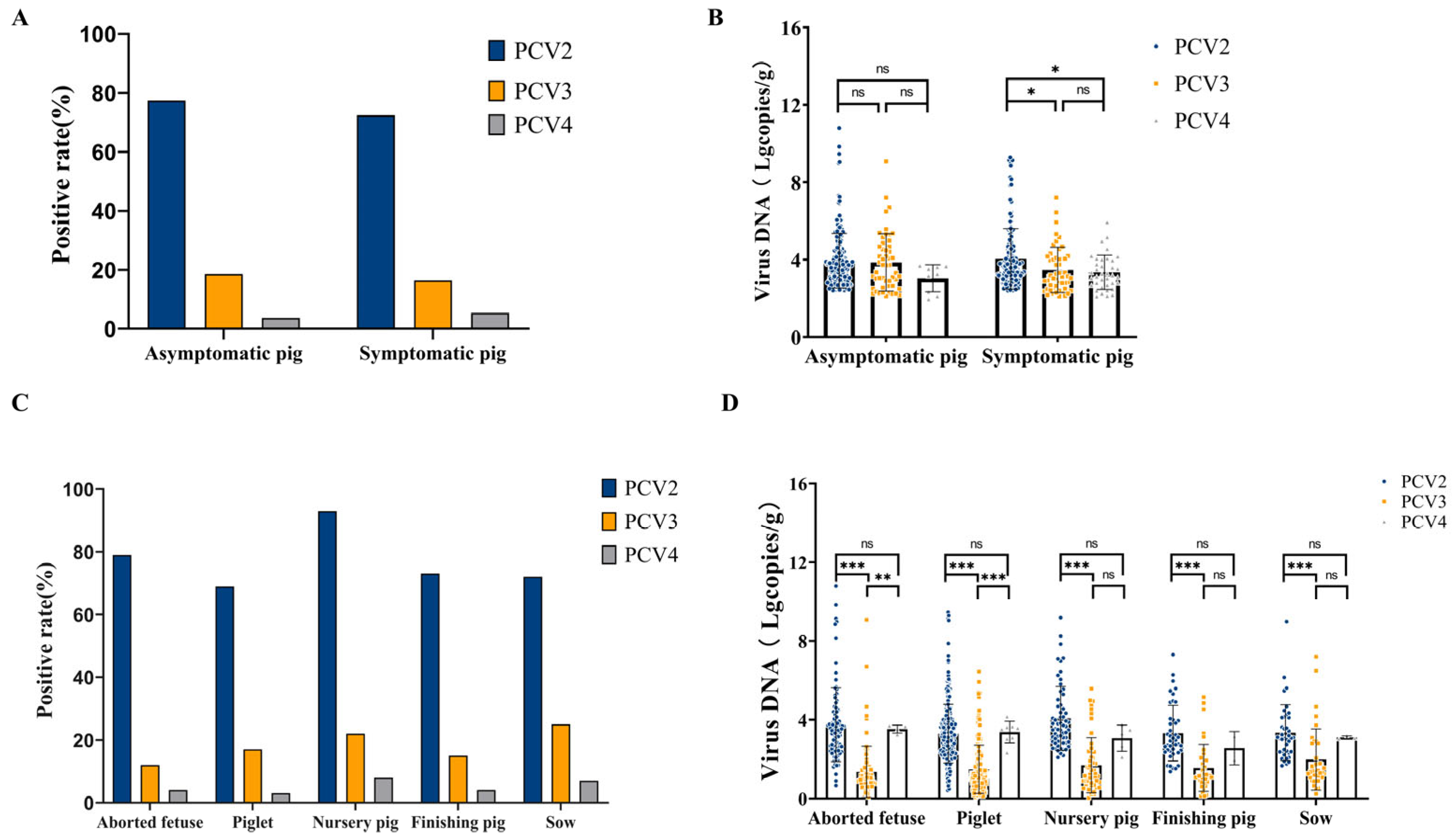
3.7. Detection of PCV2, PCV3, and PCV4 in Different Samples
3.8. PCV2/PCV3/PCV4 Genetic Characteristics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maity, H.K.; Samanta, K.; Deb, R.; Gupta, V.K. Revisiting Porcine Circovirus Infection: Recent Insights and Its Significance in the Piggery Sector. Vaccines 2023, 11, 8. [Google Scholar] [CrossRef]
- Chae, C. A review of porcine circovirus 2-associated syndromes and diseases. Vet. J. 2005, 169, 326–336. [Google Scholar] [CrossRef]
- Rosario, K.; Breitbart, M.; Harrach, B.; Segalés, J.; Delwart, E.; Biagini, P.; Varsani, A. Revisiting the taxonomy of the family Circoviridae: Establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch. Virol. 2017, 162, 1447–1463. [Google Scholar] [CrossRef]
- Klaumann, F.; Correa-Fiz, F.; Franzo, G.; Sibila, M.; Núñez, J.I.; Segalés, J. Current Knowledge on Porcine circovirus 3 (PCV-3): A Novel Virus With a Yet Unknown Impact on the Swine Industry. Front. Vet. Sci. 2018, 5, 315. [Google Scholar] [CrossRef]
- Fenaux, M.; Halbur, P.G.; Gill, M.; Toth, T.E.; Meng, X.J. Genetic characterization of type 2 porcine circovirus (PCV-2) from pigs with postweaning multisystemic wasting syndrome in different geographic regions of North America and development of a differential PCR-restriction fragment length polymorphism assay to detect and differentiate between infections with PCV-1 and PCV-2. J. Clin. Microbiol. 2000, 38, 2494–2503. [Google Scholar]
- Fux, R.; Söckler, C.; Link, E.K.; Renken, C.; Krejci, R.; Sutter, G.; Ritzmann, M.; Eddicks, M. Full genome characterization of porcine circovirus type 3 isolates reveals the existence of two distinct groups of virus strains. Virol. J. 2018, 15, 25. [Google Scholar] [CrossRef]
- Zhang, H.H.; Hu, W.Q.; Li, J.Y.; Liu, T.N.; Zhou, J.Y.; Opriessnig, T.; Xiao, C.T. Novel circovirus species identified in farmed pigs designated as Porcine circovirus 4, Hunan province, China. Transbound. Emerg. Dis. 2020, 67, 1057–1061. [Google Scholar] [CrossRef]
- Tischer, I.; Rasch, R.; Tochtermann, G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralblatt Bakteriol. Parasitenkd. Infekt. Hyg. Erste Abt. Originale. Reihe A Med. Mikrobiol. Parasitol. 1974, 226, 153–167. [Google Scholar]
- Tischer, I.; Mields, W.; Wolff, D.; Vagt, M.; Griem, W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch. Virol. 1986, 91, 271–276. [Google Scholar] [CrossRef]
- Allan, G.M.; McNeilly, F.; Kennedy, S.; Daft, B.; Clarke, E.G.; Ellis, J.A.; Haines, D.M.; Meehan, B.M.; Adair, B.M. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagn. Investig. 1998, 10, 3–10. [Google Scholar] [CrossRef]
- Wellenberg, G.J.; Pesch, S.; Berndsen, F.W.; Steverink, P.J.; Hunneman, W.; Van der Vorst, T.J.; Peperkamp, N.H.; Ohlinger, V.F.; Schippers, R.; Van Oirschot, J.T.; et al. Isolation and characterization of porcine circovirus type 2 from pigs showing signs of post-weaning multisystemic wasting syndrome in The Netherlands. Vet. Q. 2000, 22, 167–172. [Google Scholar] [CrossRef]
- Ellis, J.; Hassard, L.; Clark, E.; Harding, J.; Allan, G.; Willson, P.; Strokappe, J.; Martin, K.; McNeilly, F.; Meehan, B.; et al. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can. Vet. J. 1998, 39, 44–51. [Google Scholar]
- Wang, Y.; Noll, L.; Lu, N.; Porter, E.; Stoy, C.; Zheng, W.; Liu, X.; Peddireddi, L.; Niederwerder, M.; Bai, J. Genetic diversity and prevalence of porcine circovirus type 3 (PCV3) and type 2 (PCV2) in the Midwest of the USA during 2016–2018. Transbound. Emerg. Dis. 2020, 67, 1284–1294. [Google Scholar] [CrossRef]
- Franzo, G.; Segalés, J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS ONE 2018, 13, e0208585. [Google Scholar] [CrossRef]
- Phan, T.G.; Giannitti, F.; Rossow, S.; Marthaler, D.; Knutson, T.P.; Li, L.; Deng, X.; Resende, T.; Vannucci, F.; Delwart, E. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol. J. 2016, 13, 184. [Google Scholar] [CrossRef]
- Franzo, G.; Delwart, E.; Fux, R.; Hause, B.; Su, S.; Zhou, J.; Segalés, J. Genotyping Porcine Circovirus 3 (PCV-3) Nowadays: Does It Make Sense? Viruses 2020, 12, 265. [Google Scholar] [CrossRef]
- Opriessnig, T.; Karuppannan, A.K.; Castro, A.; Xiao, C.T. Porcine circoviruses: Current status, knowledge gaps and challenges. Virus Res. 2020, 286, 198044. [Google Scholar] [CrossRef]
- Xu, T.; Hou, C.Y.; Zhang, Y.H.; Li, H.X.; Chen, X.M.; Pan, J.J.; Chen, H.Y. Simultaneous detection and genetic characterization of porcine circovirus 2 and 4 in Henan province of China. Gene 2022, 808, 145991. [Google Scholar] [CrossRef]
- Xu, T.; Zhang, Y.H.; Tian, R.B.; Hou, C.Y.; Li, X.S.; Zheng, L.L.; Wang, L.Q.; Chen, H.Y. Prevalence and genetic analysis of porcine circovirus type 2 (PCV2) and type 3 (PCV3) between 2018 and 2020 in central China. Infect. Genet. Evol. 2021, 94, 105016. [Google Scholar] [CrossRef]
- Ge, M.; Ren, J.; Xie, Y.L.; Zhao, D.; Fan, F.C.; Song, X.Q.; Li, M.X.; Xiao, C.T. Prevalence and Genetic Analysis of Porcine Circovirus 3 in China From 2019 to 2020. Front. Vet. Sci. 2021, 8, 773912. [Google Scholar] [CrossRef]
- Ku, X.; Zhang, C.; Li, P.; Yu, X.; Sun, Q.; Xu, F.; Qian, P.; He, Q. Epidemiological and genetic characteristics of porcine circovirus 3 in 15 provinces and municipalities of China between 2016 and 2020. Virol. J. 2022, 19, 187. [Google Scholar] [CrossRef]
- Wu, H.; Hou, C.; Wang, Z.; Meng, P.; Chen, H.; Cao, H. First complete genomic sequence analysis of porcine circovirus type 4 (PCV4) in wild boars. Vet. Microbiol. 2022, 273, 109547. [Google Scholar] [CrossRef]
- Opriessnig, T.; Meng, X.J.; Halbur, P.G. Porcine circovirus type 2 associated disease: Update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J. Vet. Diagn. Investig. 2007, 19, 591–615. [Google Scholar] [CrossRef]
- Segalés, J. Porcine circovirus type 2 (PCV2) infections: Clinical signs, pathology and laboratory diagnosis. Virus Res. 2012, 164, 10–19. [Google Scholar] [CrossRef]
- Opriessnig, T.; Langohr, I. Current state of knowledge on porcine circovirus type 2-associated lesions. Vet. Pathol. 2013, 50, 23–38. [Google Scholar] [CrossRef]
- Arruda, B.; Piñeyro, P.; Derscheid, R.; Hause, B.; Byers, E.; Dion, K.; Long, D.; Sievers, C.; Tangen, J.; Williams, T.; et al. PCV3-associated disease in the United States swine herd. Emerg. Microbes Infect. 2019, 8, 684–698. [Google Scholar] [CrossRef]
- Kim, S.C.; Nazki, S.; Kwon, S.; Juhng, J.H.; Mun, K.H.; Jeon, D.Y.; Jeong, C.G.; Khatun, A.; Kang, S.J.; Kim, W.I. The prevalence and genetic characteristics of porcine circovirus type 2 and 3 in Korea. BMC Vet. Res. 2018, 14, 294. [Google Scholar] [CrossRef]
- Xu, T.; You, D.; Wu, F.; Zhu, L.; Sun, X.G.; Lai, S.Y.; Ai, Y.R.; Zhou, Y.C.; Xu, Z.W. First molecular detection and genetic analysis of porcine circovirus 4 in the Southwest of China during 2021–2022. Front. Microbiol. 2022, 13, 1052533. [Google Scholar] [CrossRef]
- Saporiti, V.; Huerta, E.; Correa-Fiz, F.; Grosse Liesner, B.; Duran, O.; Segalés, J.; Sibila, M. Detection and genotyping of Porcine circovirus 2 (PCV-2) and detection of Porcine circovirus 3 (PCV-3) in sera from fattening pigs of different European countries. Transbound. Emerg. Dis. 2020, 67, 2521–2531. [Google Scholar] [CrossRef]
- Yao, L.; Li, C.; Wang, J.; Cheng, Y.; Ghonaim, A.H.; Sun, Q.; Yu, X.; Niu, W.; Fan, S.; He, Q. Development of an indirect immunofluorescence assay for PCV3 antibody detection based on capsid protein. Anim. Dis. 2021, 1, 11. [Google Scholar] [CrossRef]
- Xia, D.; Huang, L.; Xie, Y.; Zhang, X.; Wei, Y.; Liu, D.; Zhu, H.; Bian, H.; Feng, L.; Liu, C. The prevalence and genetic diversity of porcine circovirus types 2 and 3 in Northeast China from 2015 to 2018. Arch. Virol. 2019, 164, 2435–2449. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Liu, H.; Chen, J.; Zhang, J.; Li, X.; Long, Y.; Jiang, Y.; Li, W.; Zhou, B. Development of a TaqMan-Probe-Based Multiplex Real-Time PCR for the Simultaneous Detection of Porcine Circovirus 2, 3, and 4 in East China from 2020 to 2022. Vet. Sci. 2022, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.G.; Do, H.Q.; Huynh, T.M.; Park, Y.H.; Park, B.K.; Chung, H.C. Molecular-based detection, genetic characterization and phylogenetic analysis of porcine circovirus 4 from Korean domestic swine farms. Transbound. Emerg. Dis. 2022, 69, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Bai, C.; Ge, K.; Li, Y.; Gao, W.; Jiang, S.; Wang, Y. Establishment of an SYBR Green-based real-time PCR assay for porcine circovirus type 4 detection. J. Virol. Methods 2020, 285, 113963. [Google Scholar] [CrossRef]
- Segalés, J.; Kekarainen, T.; Cortey, M. The natural history of porcine circovirus type 2: From an inoffensive virus to a devastating swine disease? Vet. Microbiol. 2013, 165, 13–20. [Google Scholar] [CrossRef]
- Fort, M.; Sibila, M.; Nofrarías, M.; Pérez-Martín, E.; Olvera, A.; Mateu, E.; Segalés, J. Porcine circovirus type 2 (PCV2) Cap and Rep proteins are involved in the development of cell-mediated immunity upon PCV2 infection. Vet. Immunol. Immunopathol. 2010, 137, 226–234. [Google Scholar] [CrossRef]
- Olvera, A.; Cortey, M.; Segalés, J. Molecular evolution of porcine circovirus type 2 genomes: Phylogeny and clonality. Virology 2007, 357, 175–185. [Google Scholar] [CrossRef]
- Yang, S.; Yin, S.; Shang, Y.; Liu, B.; Yuan, L.; Zafar Khan, M.U.; Liu, X.; Cai, J. Phylogenetic and genetic variation analyses of porcine circovirus type 2 isolated from China. Transbound. Emerg. Dis. 2018, 65, e383–e392. [Google Scholar] [CrossRef]
- Xiao, C.T.; Halbur, P.G.; Opriessnig, T. Global molecular genetic analysis of porcine circovirus type 2 (PCV2) sequences confirms the presence of four main PCV2 genotypes and reveals a rapid increase of PCV2d. J. Gen. Virol. 2015, 96, 1830–1841. [Google Scholar] [CrossRef]
- Yang, Y.; Xu, T.; Wen, J.; Yang, L.; Lai, S.; Sun, X.; Xu, Z.; Zhu, L. Prevalence and phylogenetic analysis of porcine circovirus type 2 (PCV2) and type 3 (PCV3) in the Southwest of China during 2020–2022. Front. Vet. Sci. 2022, 9, 1042792. [Google Scholar] [CrossRef]
- Li, X.; Chen, S.; Niu, G.; Zhang, X.; Ji, W.; Ren, Y.; Zhang, L.; Ren, L. Porcine Circovirus Type 4 Strains Circulating in China Are Relatively Stable and Have Higher Homology with Mink Circovirus than Other Porcine Circovirus Types. Int. J. Mol. Sci. 2022, 23, 3288. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, T.; Niu, G.; Liu, X.; Zhang, X.; Zhang, Y.; Ren, L. Recent progress on porcine circovirus type 3. Infect. Genet. Evol. 2019, 73, 227–233. [Google Scholar] [CrossRef]
- Ouyang, T.; Zhang, X.; Liu, X.; Ren, L. Co-Infection of Swine with Porcine Circovirus Type 2 and Other Swine Viruses. Viruses 2019, 11, 185. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Xu, F.; Lin, Y.; Wang, Y.; Zhang, Y.; Su, K.; Li, T.; Li, H.; Song, Q. Detection of Porcine Circovirus Type 2a and Pasteurella multocida Capsular Serotype D in Growing Pigs Suffering from Respiratory Disease. Vet. Sci. 2022, 9, 528. [Google Scholar] [CrossRef]
- Zheng, S.; Wu, X.; Zhang, L.; Xin, C.; Liu, Y.; Shi, J.; Peng, Z.; Xu, S.; Fu, F.; Yu, J.; et al. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound. Emerg. Dis. 2017, 64, 1337–1341. [Google Scholar] [CrossRef]
- Liu, S.; Li, W.; Wang, Y.; Gu, C.; Liu, X.; Charreyre, C.; Fan, S.; He, Q. Coinfection with Haemophilus parasuis serovar 4 increases the virulence of porcine circovirus type 2 in piglets. Virol. J. 2017, 14, 227. [Google Scholar] [CrossRef]


| Pathogen | Primer | Sequence (5′-3′) | Length (bp) | Gene |
|---|---|---|---|---|
| PCV2 | PCV2-F | GGAGTCTGGTGACCGTTGC | 109 bp | ORF1 |
| PCV2-R | TTCCAATCCCGCTTCTGCATT | |||
| PCV2-P | FAM-CCGCTCACTTTCAAAAGTTCAGCCA-BHQ1 | |||
| PCV3 | PCV3-F | AGTGCTCCCCATTGAACG | 130 bp | ORF2 |
| PCV3-R | GCCGTTACTTCACCCCCAA | |||
| PCV3-P | HEX-AGAGGCTTTGTCCTGGGTGAGC- BHQ1 | |||
| PCV4 | PCV4-F | AAGCGCAGCGACCTTAAA | 105 bp | ORF1 |
| PCV4-R | GCCACGCCCATACCTTAT | |||
| PCV4-P | VIC-GCCCGTGAGTTCCCGTCTGT-BHQ1 |
| Reagent | Volume (μL) |
|---|---|
| PCV2-F (10 μM) | 0.6 μL |
| PCV2-R (10 μM) | 0.6 μL |
| PCV2-P (10 μM) | 0.4 μL |
| PCV3-F (10 μM) | 0.5 μL |
| PCV3-R (10 μM) | 0.5 μL |
| PCV3-P (10 μM) | 0.4 μL |
| PCV4-F (10 μM) | 0.6 μL |
| PCV4-R (10 μM) | 0.6 μL |
| PCV4-P (10 μM) | 0.4 μL |
| Nucleic acids | 3 μL |
| RNase-free distilled water | 2.4 μL |
| AceQ® Universal U+ Probe Master Mix | 10 μL |
| Total | 20 μL |
| Plasmid Construct | Concentration (Copies/µL) | Intra-Assay Ct Values | Inter-Assay Ct Values | ||
|---|---|---|---|---|---|
| Mean ± SD | CV (%) | Mean ± SD | CV (%) | ||
| pMD18T-PCV2 | 5.23 × 107 | 12.68 ± 0.08 | 0.63 | 12.73 ± 0.12 | 0.94 |
| 5.23 × 105 | 19.58 ± 0.11 | 0.56 | 19.62 ± 0.15 | 0.76 | |
| 5.23 × 103 | 26.44 ± 0.13 | 0.49 | 26.48 ± 0.17 | 0.64 | |
| pMD18T-PCV3 | 1.20 × 107 | 14.35 ± 0.07 | 0.49 | 14.38 ± 0.10 | 0.70 |
| 1.20 × 105 | 21.03 ± 0.10 | 0.48 | 21.07 ± 0.13 | 0.62 | |
| 1.20 × 103 | 27.68 ± 0.15 | 0.54 | 27.74 ± 0.20 | 0.72 | |
| pMD18T-PCV4 | 1.38 × 107 | 13.64 ± 0.06 | 0.44 | 13.66 ± 0.09 | 0.66 |
| 1.38 × 105 | 20.69 ± 0.09 | 0.43 | 20.73 ± 0.14 | 0.68 | |
| 1.38 × 103 | 27.72 ± 0.14 | 0.50 | 27.77 ± 0.19 | 0.68 | |
| Kappa Test = 1 | Triple qPCR Method | Total | ||
| + | − | |||
| PCV2 Reference Method | + | 48 | 0 | 48 |
| − | 0 | 12 | 12 | |
| Total | 48 | 12 | 60 | |
| Kappa Test = 1 | Triple qPCR Method | Total | ||
| + | − | |||
| PCV3 Reference Method | + | 42 | 0 | 42 |
| − | 0 | 18 | 18 | |
| Total | 42 | 18 | 60 | |
| Kappa Test = 0.9667 | Triple qPCR Method | Total | ||
| + | − | |||
| PCV4 Reference Method | + | 29 | 0 | 29 |
| − | 1 | 30 | 31 | |
| Total | 30 | 30 | 60 | |
| PCV2 | PCV3 | PCV4 | |||||
|---|---|---|---|---|---|---|---|
| Region | Number of Samples | Positive Samples | Positivity Rate | Positive Samples | Positivity Rate | Positive Samples | Positivity Rate |
| North China | 117 | 89 | 76.07% (89/117) | 22 | 18.80% (22/117) | 5 | 4.27% (5/117) |
| Northeast China | 21 | 16 | 76.19% (16/21) | 5 | 23.81% (5/21) | 2 | 9.52% (2/21) |
| Eastern China | 91 | 76 | 83.52% (76/91) | 17 | 18.68% (17/91) | 3 | 3.30% (3/91) |
| Central China | 77 | 49 | 63.64% (49/77) | 11 | 14.29% (11/77) | 4 | 5.19% (4/77) |
| South China | 79 | 63 | 79.75% (63/79) | 10 | 12.66% (10/79) | 4 | 5.06% (4/79) |
| Southwest China | 81 | 53 | 65.43% (53/81) | 17 | 20.99% (17/81) | 3 | 3.70% (3/81) |
| Northwest China | 34 | 30 | 88.24% (30/34) | 6 | 17.65% (6/34) | 1 | 2.94% (1/34) |
| Total | 500 | 376 | 75.20% (376/500) | 88 | 17.60% (88/500) | 22 | 4.40% (22/500) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Lu, Y.; Li, D.; Dong, L.; Zeng, Y.; Mei, Z.; Ghonaim, A.H.; USAMA; Yu, Z.; Zhang, S.; et al. Development of a Triplex Real-Time PCR Method for the Simultaneous Detection of Porcine Circovirus 2, 3, and 4 in China Between 2023 and 2024. Viruses 2025, 17, 777. https://doi.org/10.3390/v17060777
Chen Y, Lu Y, Li D, Dong L, Zeng Y, Mei Z, Ghonaim AH, USAMA, Yu Z, Zhang S, et al. Development of a Triplex Real-Time PCR Method for the Simultaneous Detection of Porcine Circovirus 2, 3, and 4 in China Between 2023 and 2024. Viruses. 2025; 17(6):777. https://doi.org/10.3390/v17060777
Chicago/Turabian StyleChen, Yanhong, Yi Lu, Dongfan Li, Ling Dong, Yang Zeng, Zhijing Mei, Ahmed H. Ghonaim, USAMA, Zhixian Yu, Shuo Zhang, and et al. 2025. "Development of a Triplex Real-Time PCR Method for the Simultaneous Detection of Porcine Circovirus 2, 3, and 4 in China Between 2023 and 2024" Viruses 17, no. 6: 777. https://doi.org/10.3390/v17060777
APA StyleChen, Y., Lu, Y., Li, D., Dong, L., Zeng, Y., Mei, Z., Ghonaim, A. H., USAMA, Yu, Z., Zhang, S., Bai, P., Li, W., Yu, X., & He, Q. (2025). Development of a Triplex Real-Time PCR Method for the Simultaneous Detection of Porcine Circovirus 2, 3, and 4 in China Between 2023 and 2024. Viruses, 17(6), 777. https://doi.org/10.3390/v17060777








