Droplet Digital PCR or Real-Time PCR as a Method for Quantifying SARS-CoV-2 RNA in Plasma—Is There a Difference?
Abstract
1. Introduction
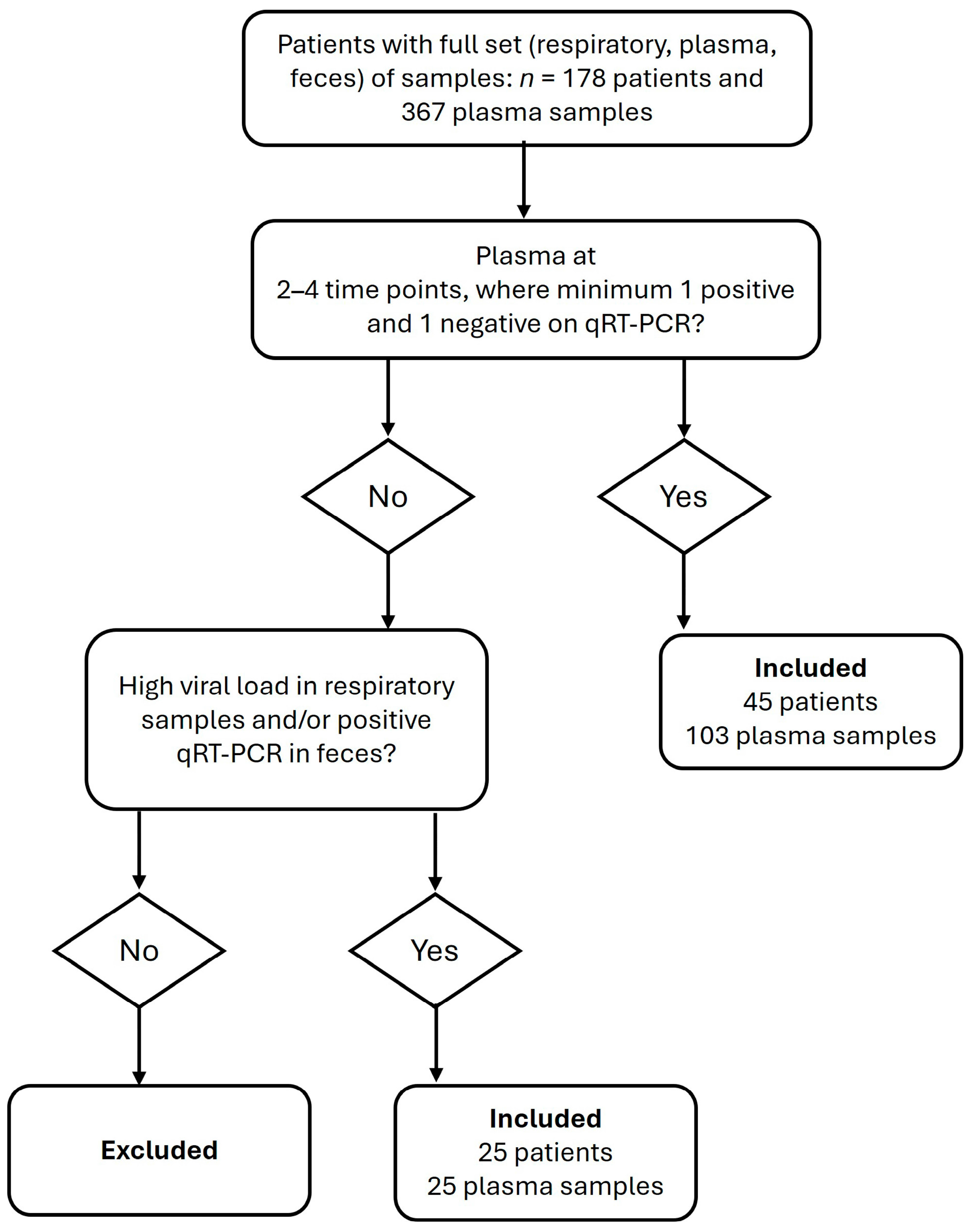
2. Materials and Methods
2.1. Sample Material
2.2. SARS-CoV-2 RNA Standard
2.3. Nucleic Acid Extraction
2.4. Oligos
2.5. qRT-PCR Detection and Quantification
2.6. RT-ddPCR Detection
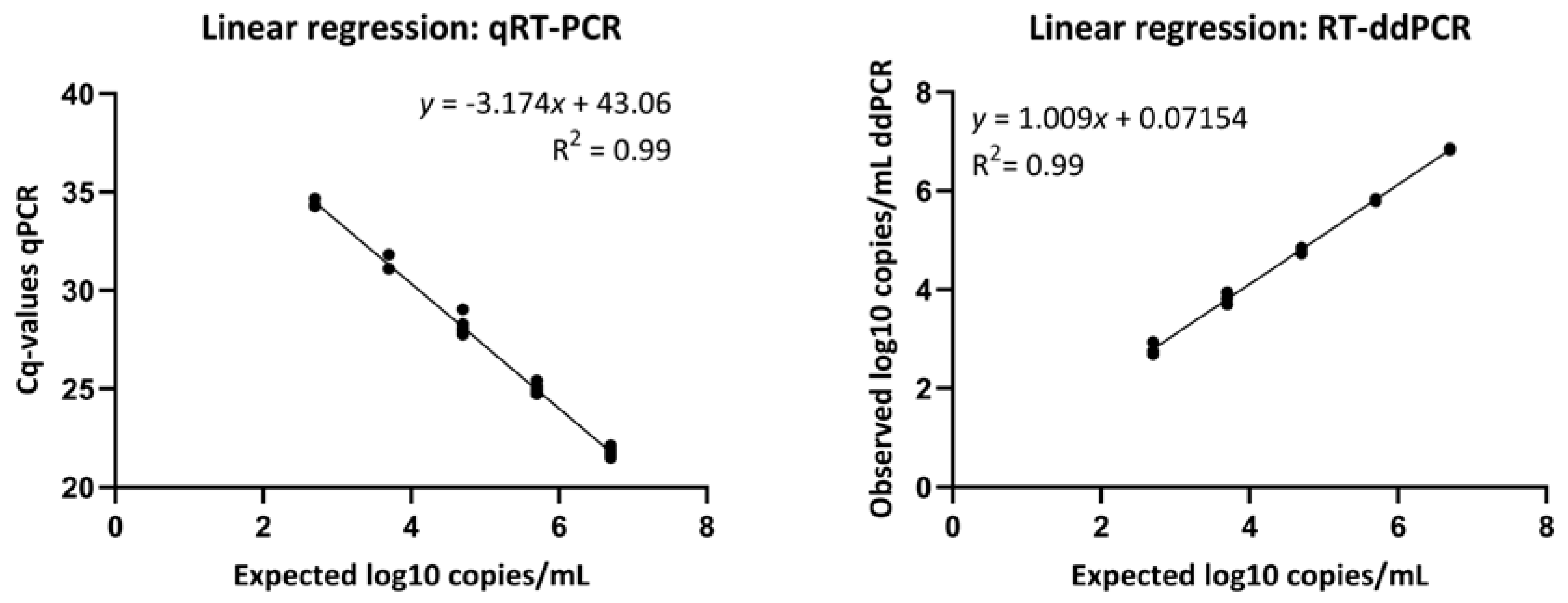
2.7. Verification of Linearity
2.8. Statistical Analysis
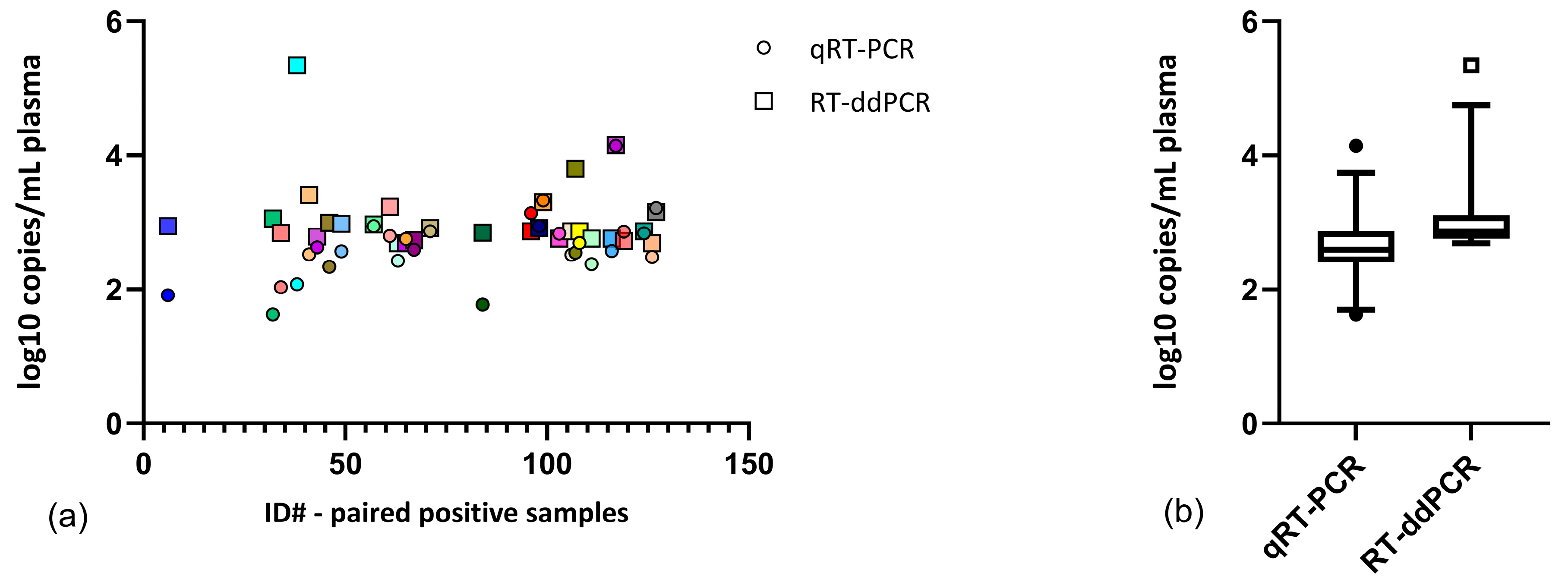
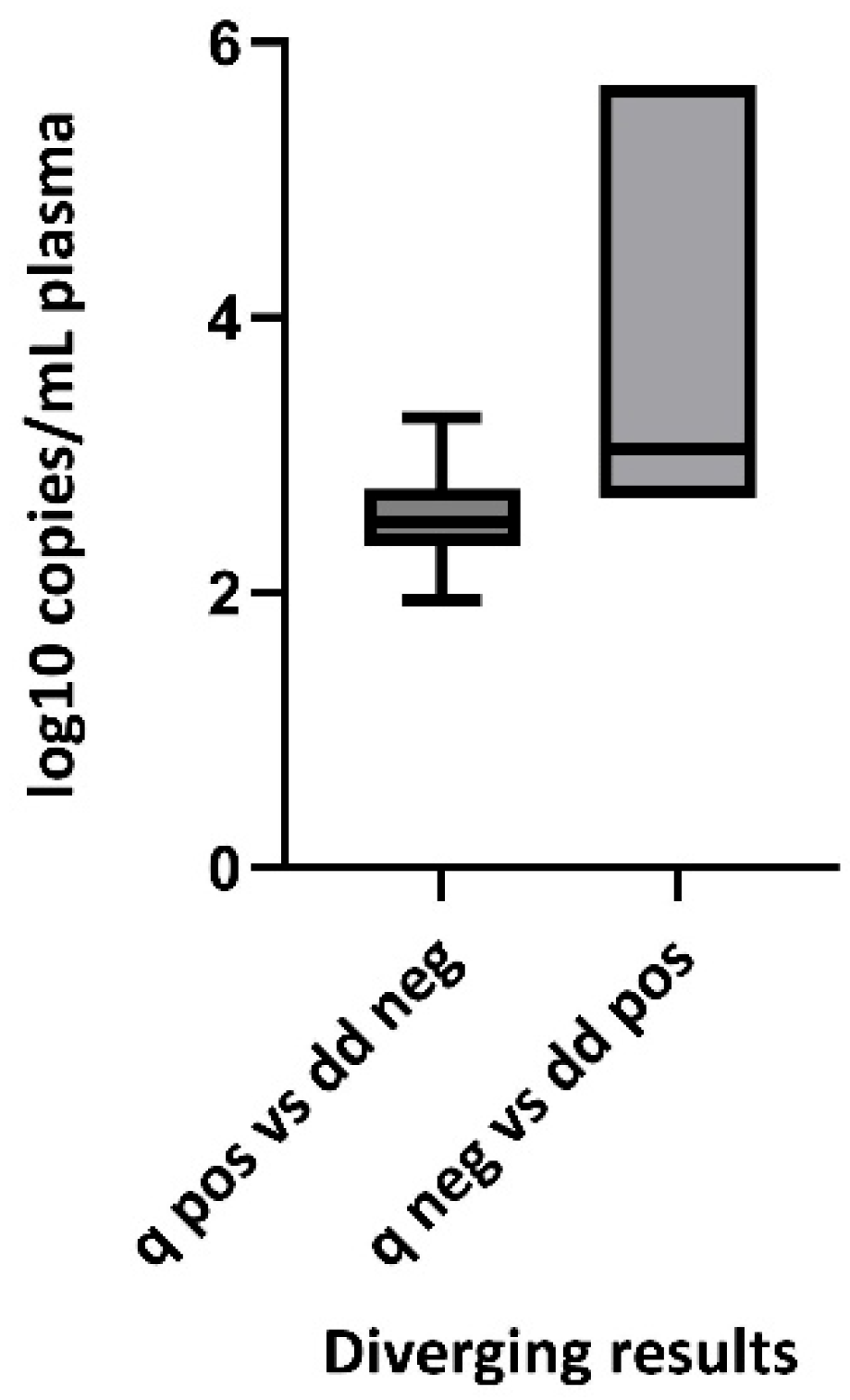
3. Results
| Positive RT-ddPCR | Negative RT-ddPCR | Total | |
|---|---|---|---|
| Positive qRT-PCR | 29 | 16 | 45 |
| Negative qRT-PCR | 23 | 60 | 83 |
| Total | 52 | 76 | 128 |
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Liu, Y.; Wang, J.; Long, T.; Yee Mok, B.W.; Huang, Y.; Peng, Z.; Jia, Q.; Liu, C.; So, P.-K.; et al. Identifications of novel host cell factors that interact with the receptor-binding domain of the SARS-CoV-2 spike protein. J. Biol. Chem. 2024, 300, 107390. [Google Scholar] [CrossRef] [PubMed]
- Puray-Chavez, M.; LaPak, K.M.; Schrank, T.P.; Elliott, J.L.; Bhatt, D.P.; Agajanian, M.J.; Jasuja, R.; Lawson, D.Q.; Keanu Davis, K.; Rothlauf, P.W.; et al. Systematic analysis of SARS-CoV-2 infection of an ACE2-negative human airway cell. Cell Rep. 2021, 36, 109364. [Google Scholar] [CrossRef]
- Arora, P.; Zhang, L.; Nehlmeier, I.; Kempf, A.; Graichen, L.; Kreitz, E.; Sidarovich, A.; Rocha, C.; Gärtner, S.; Winkler, M.; et al. Host cell lectins ASGR1 and DC-SIGN jointly with TMEM106B confer ACE2 independence and imdevimab resistance to SARS-CoV-2 pseudovirus with spike mutation E484D. J. Virol. 2025, 99, e01230-24. [Google Scholar] [CrossRef]
- Nader, D.; Fletcher, N.; Curley, G.F.; Kerrigan, S.W. SARS-CoV-2 uses major endothelial integrin αvβ3 to cause vascular dysregulation in-vitro during COVID-19. PLoS ONE 2021, 16, e0253347. [Google Scholar] [CrossRef]
- Varga, Z.; Flammer, A.J.; Steiger, P.; Haberecker, M.; Andermatt, R.; Zinkernagel, A.S.; Mehra, M.R.; Schuepbach, R.A.; Ruschitzka, F.; Moch, H. Endothelial cell infection and endotheliitis in COVID-19. Lancet 2020, 395, 1417–1418. [Google Scholar] [CrossRef]
- Brunet-Ratnasingham, E.; Anand, S.P.; Gantner, P.; Dyachenko, A.; Moquin-Beaudry, G.; Brassard, N.; Beaudoin-Bussières, G.; Pagliuzza, A.; Gasser, R.; Benlarbi, M.; et al. Integrated immunovirological profiling validates plasma SARS-CoV-2 RNA as an early predictor of COVID-19 mortality. Sci. Adv. 2021, 7, eabj5629. [Google Scholar] [CrossRef]
- Jacobs, J.L.; Naqvi, A.; Shah, F.A.; Boltz, V.F.; Kearney, M.F.; McVerry, B.J.; Ray, P.; Schaefer, C.; Fitzpatrick, M.; Methé, B.; et al. Plasma SARS-CoV-2 RNA Levels as a Biomarker of Lower Respiratory Tract SARS-CoV-2 Infection in Critically Ill Patients With COVID-19. J. Infect. Dis. 2022, 226, 2089–2094. [Google Scholar] [CrossRef]
- Ynga-Durand, M.; Maaß, H.; Milošević, M.; Krstanović, F.; Pribanić Matešić, M.; Jonjić, S.; Protić, A.; Brizić, I.; Šustić, A.; Čičin-Šain, L. SARS-CoV-2 Viral Load in the Pulmonary Compartment of Critically Ill COVID-19 Patients Correlates with Viral Serum Load and Fatal Outcomes. Viruses 2022, 14, 1292. [Google Scholar] [CrossRef]
- Jacobs, J.L.; Bain, W.; Naqvi, A.; Staines, B.; Castanha, P.M.S.; Yang, H.; Boltz, V.F.; Barratt-Boyes, S.; A Marques, E.T.; Mitchell, S.L.; et al. Severe Acute Respiratory Syndrome Coronavirus 2 Viremia Is Associated With Coronavirus Disease 2019 Severity and Predicts Clinical Outcomes. Clin. Infect. Dis. 2022, 74, 1525–1533. [Google Scholar] [CrossRef]
- Li, M.-Y.; Li, L.; Zhang, Y.; Wang, X.-S. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty 2020, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Pontelli, M.C.; Castro, Í.A.; Martins, R.B.; La Serra, L.; Veras, F.P.; Nascimento, D.C.; Silva, C.M.; Cardoso, R.S.; Rosales, R.; Gomes, R.; et al. SARS-CoV-2 productively infects primary human immune system cells in vitro and in COVID-19 patients. J. Mol. Cell Biol. 2022, 14, mjac021. [Google Scholar] [CrossRef]
- Junqueira, C.; Crespo, Â.; Ranjbar, S.; De Lacerda, L.B.; Lewandrowski, M.; Ingber, J.; Parry, B.; Ravid, S.; Clark, S.; Schrimpf, M.R.; et al. FcγR-mediated SARS-CoV-2 infection of monocytes activates inflammation. Nature 2022, 606, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Delorey, T.M.; Ziegler, C.G.K.; Heimberg, G.; Normand, R.; Yang, Y.; Segerstolpe, Å.; Abbondanza, D.; Fleming, S.J.; Subramanian, A.; Montoro, D.T.; et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 2021, 595, 107–113. [Google Scholar] [CrossRef]
- Shen, X.-R.; Geng, R.; Li, Q.; Chen, Y.; Li, S.-F.; Wang, Q.; Min, J.; Yang, Y.; Li, B.; Jiang, R.-D.; et al. ACE2-independent infection of T lymphocytes by SARS-CoV-2. Signal Transduct. Target. Ther. 2022, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Alexandersen, S.; Chamings, A.; Bhatta, T.R. SARS-CoV-2 genomic and subgenomic RNAs in diagnostic samples are not an indicator of active replication. Nat. Commun. 2020, 11, 6059. [Google Scholar] [CrossRef]
- Bermejo-Martin, J.F.; González-Rivera, M.; Almansa, R.; Micheloud, D.; Tedim, A.P.; Domínguez-Gil, M.; Resino, S.; Martín-Fernández, M.; Murua, P.R.; Pérez-García, F.; et al. Viral RNA load in plasma is associated with critical illness and a dysregulated host response in COVID-19. Crit. Care 2020, 24, 691. [Google Scholar] [CrossRef]
- Ram-Mohan, N.; Kim, D.; Zudock, E.J.; Hashemi, M.M.; Tjandra, K.C.; Rogers, A.J.; A Blish, C.; Nadeau, K.C.; A Newberry, J.; Quinn, J.V.; et al. SARS-CoV-2 RNAemia Predicts Clinical Deterioration and Extrapulmonary Complications from COVID-19. Clin. Infect. Dis. 2022, 74, 218–226. [Google Scholar] [CrossRef]
- Andersson, M.; Arancibia-Carcamo, C.; Auckland, K.; Baillie, J.; Barnes, E.; Beneke, T.; Bibi, S.; Brooks, T.; Carroll, M.; Crook, D.; et al. SARS-CoV-2 RNA detected in blood products from patients with COVID-19 is not associated with infectious virus. Wellcome Open Res. 2020, 5, 181. [Google Scholar] [CrossRef]
- Berastegui-Cabrera, J.; Salto-Alejandre, S.; Valerio, M.; Pérez-Palacios, P.; Revillas, F.A.-D.L.; Abelenda-Alonso, G.; Oteo-Revuelta, J.A.; Carretero-Ledesma, M.; Muñoz, P.; Pascual, Á.; et al. SARS-CoV-2 RNAemia is associated with severe chronic underlying diseases but not with nasopharyngeal viral load. J. Infect. 2021, 82, e38–e41. [Google Scholar] [CrossRef] [PubMed]
- Colagrossi, L.; Antonello, M.; Renica, S.; Merli, M.; Matarazzo, E.; Travi, G.; Vecchi, M.; Colombo, J.; Muscatello, A.; Grasselli, G.; et al. SARS-CoV-2 RNA in plasma samples of COVID-19 affected individuals: A cross-sectional proof-of-concept study. BMC Infect. Dis. 2021, 21, 184. [Google Scholar] [CrossRef] [PubMed]
- Eberhardt, K.A.; Meyer-Schwickerath, C.; Heger, E.; Knops, E.; Lehmann, C.; Rybniker, J.; Schommers, P.; Eichenauer, D.A.; Kurth, F.; Ramharter, M.; et al. RNAemia Corresponds to Disease Severity and Antibody Response in Hospitalized COVID-19 Patients. Viruses 2020, 12, 1045. [Google Scholar] [CrossRef] [PubMed]
- Fajnzylber, J.; Regan, J.; Coxen, K.; Corry, H.; Wong, C.; Rosenthal, A.; Worrall, D.; Giguel, F.; Piechocka-Trocha, A.; Atyeo, C.; et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat. Commun. 2020, 11, 5493. [Google Scholar] [CrossRef]
- Giacomelli, A.; Righini, E.; Micheli, V.; Pinoli, P.; Bernasconi, A.; Rizzo, A.; Oreni, L.; Ridolfo, A.L.; Antinori, S.; Ceri, S.; et al. SARS-CoV-2 viremia and COVID-19 mortality: A prospective observational study. PLoS ONE 2023, 18, e0281052. [Google Scholar] [CrossRef]
- Gutmann, C.; Takov, K.; Burnap, S.A.; Singh, B.; Ali, H.; Theofilatos, K.; Reed, E.; Hasman, M.; Nabeebaccus, A.; Fish, M.; et al. SARS-CoV-2 RNAemia and proteomic trajectories inform prognostication in COVID-19 patients admitted to intensive care. Nat. Commun. 2021, 12, 3406. [Google Scholar] [CrossRef]
- Hagman, K.; Hedenstierna, M.; Gille-Johnson, P.; Hammas, B.; Grabbe, M.; Dillner, J.; Ursing, J. Severe Acute Respiratory Syndrome Coronavirus 2 RNA in Serum as Predictor of Severe Outcome in Coronavirus Disease 2019: A Retrospective Cohort Study. Clin. Infect. Dis. 2021, 73, e2995–e3001. [Google Scholar] [CrossRef]
- Hogan, C.A.; Stevens, B.A.; Sahoo, M.K.; Huang, C.; Garamani, N.; Gombar, S.; Yamamoto, F.; Murugesan, K.; Kurzer, J.; Zehnder, J.; et al. High Frequency of SARS-CoV-2 RNAemia and Association With Severe Disease. Clin. Infect. Dis. 2021, 72, e291–e295. [Google Scholar] [CrossRef]
- Kawasuji, H.; Morinaga, Y.; Tani, H.; Yoshida, Y.; Takegoshi, Y.; Kaneda, M.; Murai, Y.; Kimoto, K.; Ueno, A.; Miyajima, Y.; et al. SARS-CoV-2 RNAemia with a higher nasopharyngeal viral load is strongly associated with disease severity and mortality in patients with COVID-19. J. Med. Virol. 2022, 94, 147–153. [Google Scholar] [CrossRef]
- Martin-Vicente, M.; Almansa, R.; Martínez, I.; Tedim, A.P.; Bustamante, E.; Tamayo, L.; Aldecoa, C.; Gómez, J.M.; Renedo, G.; Berezo, J.Á.; et al. Low anti-SARS-CoV-2 S antibody levels predict increased mortality and dissemination of viral components in the blood of critical COVID-19 patients. J. Intern. Med. 2022, 291, 232–240. [Google Scholar] [CrossRef]
- Rodríguez-Serrano, D.A.; Roy-Vallejo, E.; Zurita Cruz, N.D.; Martín Ramírez, A.; Rodríguez-García, S.C.; Arevalillo-Fernández, N.; Galván-Román, J.M.; García-Rodrigo, L.F.; Vega-Piris, L.; Llano, M.C.; et al. Detection of SARS-CoV-2 RNA in serum is associated with increased mortality risk in hospitalized COVID-19 patients. Sci. Rep. 2021, 11, 13134. [Google Scholar] [CrossRef]
- Zheng, S.; Fan, J.; Yu, F.; Feng, B.; Lou, B.; Zou, Q.; Xie, G.; Lin, S.; Wang, R.; Yang, X.; et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January–March 2020: Retrospective cohort study. BMJ 2020, 369, m1443. [Google Scholar] [CrossRef] [PubMed]
- Prebensen, C.; Myhre, P.L.; Jonassen, C.; Rangberg, A.; Blomfeldt, A.; Svensson, M.; Omland, T.; Berdal, J.-E. Severe Acute Respiratory Syndrome Coronavirus 2 RNA in Plasma Is Associated With Intensive Care Unit Admission and Mortality in Patients Hospitalized With Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 73, e799–e802. [Google Scholar] [CrossRef]
- Rovito, R.; Bono, V.; Augello, M.; Tincati, C.; Mainoldi, F.; Beaudoin-Bussières, G.; Tauzin, A.; Bianchi, S.; Hadla, M.; Yellenki, V.; et al. Association between SARS-CoV-2 RNAemia and dysregulated immune response in acutely ill hospitalized COVID-19 patients. Sci. Rep. 2022, 12, 19658. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Thierbach, A.; Di Cristanziano, V.; Eberhardt, K.A.; Pirkl, M.; Steger, G.; Heger, E.; Kaiser, R.; Koch, M.; Klein, F.; Rauschning, D.; et al. High rate of RNAemia and impaired immunity in patients with immunodeficiency in the vaccination era. J. Clin. Virol. 2025, 177, 105774. [Google Scholar] [CrossRef] [PubMed]
- Menezes, S.M.; Jamoulle, M.; Carletto, M.P.; Moens, L.; Meyts, I.; Maes, P.; Van Weyenbergh, J. Blood transcriptomic analyses reveal persistent SARS-CoV-2 RNA and candidate biomarkers in post-COVID-19 condition. Lancet Microbe 2024, 5, 100849. [Google Scholar] [CrossRef]
- Quan, P.-L.; Sauzade, M.; Brouzes, E. dPCR: A Technology Review. Sensors 2018, 18, 1271. [Google Scholar] [CrossRef]
- Martin, A.; Storto, A.; Le Hingrat, Q.; Collin, G.; André, B.; Mallory, A.; Dangla, R.; Descamps, D.; Visseaux, B.; Gossner, O. High-sensitivity SARS-CoV-2 group testing by digital PCR among symptomatic patients in hospital settings. J. Clin. Virol. 2021, 141, 104895. [Google Scholar] [CrossRef]
- Sun, Y.; Ding, C.; Chen, Q.; Xie, J.; Yu, J.; Shi, Y.; Jiang, C.; Zhang, Z.; He, H.; Ge, Y.; et al. Digital PCR assay for the effective detection of COVID-19 patients with SARS-CoV-2 low viral load. J. Virol. Methods 2021, 295, 114185. [Google Scholar] [CrossRef]
- Deiana, M.; Mori, A.; Piubelli, C.; Scarso, S.; Favarato, M.; Pomari, E. Assessment of the direct quantitation of SARS-CoV-2 by droplet digital PCR. Sci. Rep. 2020, 10, 18764. [Google Scholar] [CrossRef]
- Liu, X.; Feng, J.; Zhang, Q.; Guo, D.; Zhang, L.; Suo, T.; Hu, W.; Guo, M.; Wang, X.; Huang, Z.; et al. Analytical comparisons of SARS-CoV-2 detection by qRT-PCR and ddPCR with multiple primer/probe sets. Emerg. Microbes Infect. 2020, 9, 1175–1179. [Google Scholar] [CrossRef] [PubMed]
- Suo, T.; Liu, X.; Feng, J.; Guo, M.; Hu, W.; Guo, D.; Ullah, H.; Yang, Y.; Zhang, Q.; Wang, X.; et al. ddPCR: A more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes Infect. 2020, 9, 1259–1268. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shi, Q.; Peng, M.; Lu, R.; Li, H.; Cai, Y.; Chen, J.; Xu, J.; Shen, B. Evaluation of droplet digital PCR for quantification of SARS-CoV-2 Virus in discharged COVID-19 patients. Aging 2020, 12, 20997–21003. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Lee, J.; Hassan, Z.U.; Ku, K.B.; Kim, S.-J.; Kim, H.G.; Park, E.C.; Park, G.-S.; Park, D.; Baek, S.-H.; et al. Comparison of Digital PCR and Quantitative PCR with Various SARS-CoV-2 Primer-Probe Sets. J. Microbiol. Biotechnol. 2021, 31, 358–367. [Google Scholar] [CrossRef] [PubMed]
- Poggio, P.; Songia, P.; Vavassori, C.; Ricci, V.; Banfi, C.; Barbieri, S.S.; Garoffolo, G.; Myasoedova, V.A.; Piacentini, L.; Raucci, A.; et al. Digital PCR for high sensitivity viral detection in false-negative SARS-CoV-2 patients. Sci. Rep. 2021, 11, 4310. [Google Scholar] [CrossRef]
- Vasudevan, H.N.; Xu, P.; Servellita, V.; Miller, S.; Liu, L.; Gopez, A.; Chiu, C.Y.; Abate, A.R. Digital droplet PCR accurately quantifies SARS-CoV-2 viral load from crude lysate without nucleic acid purification. Sci. Rep. 2021, 11, 780. [Google Scholar] [CrossRef]
- Dong, L.; Zhou, J.; Niu, C.; Wang, Q.; Pan, Y.; Sheng, S.; Wang, X.; Zhang, Y.; Yang, J.; Liu, M.; et al. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. Talanta 2021, 224, 121726. [Google Scholar] [CrossRef]
- Alteri, C.; Cento, V.; Antonello, M.; Colagrossi, L.; Merli, M.; Ughi, N.; Renica, S.; Matarazzo, E.; Di Ruscio, F.; Tartaglione, L.; et al. Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients. PLoS ONE 2020, 15, e0236311. [Google Scholar] [CrossRef]
- Kinloch, N.N.; Ritchie, G.; Dong, W.; Cobarrubias, K.D.; Sudderuddin, H.; Lawson, T.; Matic, N.; Montaner, J.S.; Leung, V.; Romney, M.G.; et al. SARS-CoV-2 RNAs Quantification Using Droplet Digital RT-PCR. J. Mol. Diagn. 2021, 23, 907–919. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.W.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed]
- NIBSC Code: 20/146; WHO International Standard First WHO International Standard for SARS-CoV-2 RNA. The National Institute for Biological Standards and Control: Hertfordshire, UK, 2021.
- Dreier, J.; Störmer, M.; Kleesiek, K. Use of bacteriophage MS2 as an internal control in viral reverse transcription-PCR assays. J. Clin. Microbiol. 2005, 43, 4551–4557. [Google Scholar] [CrossRef]
- Python Software Foundation. The Python Language Reference, version 3.10; Python Software Foundation: Wilmington, DE, USA, 2024.
- Cook, L.; Starr, K.; Boonyaratanakornkit, J.; Hayden, R.; Sam, S.S.; Caliendo, A.M. Does Size Matter? Comparison of Extraction Yields for Different-Sized DNA Fragments by Seven Different Routine and Four New Circulating Cell-Free Extraction Methods. J. Clin. Microbiol. 2018, 56, e01061-18. [Google Scholar] [CrossRef]
- Falzone, L.; Musso, N.; Gattuso, G.; Bongiorno, D.; Palermo, C.; Scalia, G.; Libra, M.; Stefani, S. Sensitivity assessment of droplet digital PCR for SARS-CoV-2 detection. Int. J. Mol. Med. 2020, 46, 957–964. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Liu, N.; Tan, C.; Feng, Y.; Yuan, X.; Fan, D.; Peng, Y.; Jin, R.; Guo, Y.; Lou, J. Comparison of qualitative and quantitative analyses of COVID-19 clinical samples. Clin. Chim. Acta 2020, 510, 613–616. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.B.; Choi, H.; Lee, G.D.; Lee, J.; Lee, S.; Kim, Y.; Cho, S.-Y.; Lee, D.-G.; Kim, M. Analytical and Clinical Performance of Droplet Digital PCR in the Detection and Quantification of SARS-CoV-2. Mol. Diagn. Ther. 2021, 25, 617–628. [Google Scholar] [CrossRef]
- Tedim, A.P.; Almansa, R.; Domínguez-Gil, M.; González-Rivera, M.; Micheloud, D.; Ryan, P.; Méndez, R.; Blanca-López, N.; Pérez-García, F.; Bustamante, E.; et al. Comparison of real-time and droplet digital PCR to detect and quantify SARS-CoV-2 RNA in plasma. Eur. J. Clin. Investig. 2021, 51, e13501. [Google Scholar] [CrossRef]
- Vogels, C.B.F.; Brito, A.F.; Wyllie, A.L.; Fauver, J.R.; Ott, I.M.; Kalinich, C.C.; Petrone, M.E.; Casanovas-Massana, A.; Muenker, M.C.; Moore, A.J.; et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020, 5, 1299–1305. [Google Scholar] [CrossRef]
- Etievant, S.; Bal, A.; Escuret, V.; Brengel-Pesce, K.; Bouscambert, M.; Cheynet, V.; Generenaz, L.; Oriol, G.; Destras, G.; Billaud, G.; et al. Performance Assessment of SARS-CoV-2 PCR Assays Developed by WHO Referral Laboratories. J. Clin. Med. 2020, 9, 1871. [Google Scholar] [CrossRef]
- Au, W.Y.; Cheung, P.P.H. Diagnostic performances of common nucleic acid tests for SARS-CoV-2 in hospitals and clinics: A systematic review and meta-analysis. Lancet Microbe 2021, 2, e704–e714. [Google Scholar] [CrossRef]
- De Kock, R.; Baselmans, M.; Scharnhorst, V.; Deiman, B. Sensitive detection and quantification of SARS-CoV-2 by multiplex droplet digital RT-PCR. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 807–813. [Google Scholar] [CrossRef]
- Nalla, A.K.; Casto, A.M.; Huang, M.-L.W.; Perchetti, G.A.; Sampoleo, R.; Shrestha, L.; Wei, Y.; Zhu, H.; Jerome, K.R.; Greninger, A.L. Comparative Performance of SARS-CoV-2 Detection Assays Using Seven Different Primer-Probe Sets and One Assay Kit. J. Clin. Microbiol. 2020, 58, e00557-20. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Yue, Y.; Qiu, G.; Ji, Z.; Spillman, M.; Gai, Z.; Chen, Q.; Bielecki, M.; Huber, M.; Trkola, A.; et al. Comparison of analytical sensitivity and efficiency for SARS-CoV-2 primer sets by TaqMan-based and SYBR Green-based RT-qPCR. Appl. Microbiol. Biotechnol. 2022, 106, 2207–2218. [Google Scholar] [CrossRef]
- van Kasteren, P.B.; van der Veer, B.; van den Brink, S.; Wijsman, L.; de Jonge, J.; van den Brandt, A.; Molenkamp, R.; Reusken, C.B.E.M.; Meijer, A. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. Comparison of seven commercial RT-PCR diagnostic kits for COVID-19. J. Clin. Virol. 2020, 128, 104412. [Google Scholar] [CrossRef]
- Han, M.S.; Byun, J.-H.; Cho, Y.; Rim, J.H. RT-PCR for SARS-CoV-2: Quantitative versus qualitative. Lancet Infect. Dis. 2021, 21, 165. [Google Scholar] [CrossRef]
- Fan, G.; Jin, Y.; Wang, Q.; Yue, Y. Assessing the comparability of cycle threshold values derived from five external quality assessment rounds for omicron nucleic acid testing. Virol. J. 2023, 20, 119. [Google Scholar] [CrossRef]
- The dMIQE Group; Huggett, J.F. The Digital MIQE Guidelines Update: Minimum Information for Publication of Quantitative Digital PCR Experiments for 2020. Clin. Chem. 2020, 66, 1012–1029. [Google Scholar] [CrossRef] [PubMed]
- Kuypers, J.; Jerome, K.R. Applications of Digital PCR for Clinical Microbiology. J. Clin. Microbiol. 2017, 55, 1621–1628. [Google Scholar] [CrossRef] [PubMed]
- Emslie, K.R.; McLaughlin, J.L.H.; Griffiths, K.; Forbes-Smith, M.; Pinheiro, L.B.; Burke, D.G. Droplet Volume Variability and Impact on Digital PCR Copy Number Concentration Measurements. Anal. Chem. 2019, 91, 4124–4131. [Google Scholar] [CrossRef]
- Niu, C.; Wang, X.; Zhang, Y.; Lu, L.; Wang, D.; Gao, Y.; Wang, S.; Luo, J.; Jiang, Y.; Wang, N.; et al. Interlaboratory assessment of quantification of SARS-CoV-2 RNA by reverse transcription digital PCR. Anal. Bioanal. Chem. 2021, 413, 7195–7204. [Google Scholar] [CrossRef]
| Target | Primer/Probe | Sequence (5′-3′) | Coordinates * |
|---|---|---|---|
| E-gene (SARS-CoV-2) | Forward | ACAGGTACGTTAATAGTTAATAGCGT | 26,269–26,294 |
| Reverse | ATATTGCAGCAGTACGCACACA | 26,381–26,360 | |
| Probe | FAM-ACACTAG/ZEN/CCATCCTTACTGCGCTTCG-IBFQ | 26,332–26,357 | |
| Replicase gene (MS2) | Forward | TGCTCGCGGATACCCG | 3169–3184 |
| Reverse | AACTTGCGTTCTCGAGCGAT | 3229–3210 | |
| Probe | HEX-ACCTCGGGTTTCCGTCTTGCTCGT-BBQ | 3186–3209 |
| Dilution Factor | Expected * log10 copies/mL | Observed log10 copies/mL (Mean ± SD) | Cq Value (Mean ± SD) |
|---|---|---|---|
| 10−1 | 6.7 | 6.85 ± 0.02 | 21.77 ± 0.20 |
| 10−2 | 5.7 | 5.82 ± 0.03 | 24.98 ± 0.17 |
| 10−3 | 4.7 | 4.79 ± 0.06 | 28.14 ± 0.30 |
| 10−4 | 3.7 | 3.82 ± 0.12 | 31.35 ± 0.42 |
| 10−5 | 2.7 | 2.8 ± 0.13 | 34.44 ± 0.22 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Granerud, B.K.; Kaarbø, M.; Al-Baldawi, H.; The Norwegian SARS-CoV-2 Study Group Investigators; Otterdal, K.; Halvorsen, B.; Lind, A.; Rayner, S.; Holter, J.C.; Dudman, S. Droplet Digital PCR or Real-Time PCR as a Method for Quantifying SARS-CoV-2 RNA in Plasma—Is There a Difference? Viruses 2025, 17, 772. https://doi.org/10.3390/v17060772
Granerud BK, Kaarbø M, Al-Baldawi H, The Norwegian SARS-CoV-2 Study Group Investigators, Otterdal K, Halvorsen B, Lind A, Rayner S, Holter JC, Dudman S. Droplet Digital PCR or Real-Time PCR as a Method for Quantifying SARS-CoV-2 RNA in Plasma—Is There a Difference? Viruses. 2025; 17(6):772. https://doi.org/10.3390/v17060772
Chicago/Turabian StyleGranerud, Beathe Kiland, Mari Kaarbø, Huda Al-Baldawi, The Norwegian SARS-CoV-2 Study Group Investigators, Kari Otterdal, Bente Halvorsen, Andreas Lind, Simon Rayner, Jan Cato Holter, and Susanne Dudman. 2025. "Droplet Digital PCR or Real-Time PCR as a Method for Quantifying SARS-CoV-2 RNA in Plasma—Is There a Difference?" Viruses 17, no. 6: 772. https://doi.org/10.3390/v17060772
APA StyleGranerud, B. K., Kaarbø, M., Al-Baldawi, H., The Norwegian SARS-CoV-2 Study Group Investigators, Otterdal, K., Halvorsen, B., Lind, A., Rayner, S., Holter, J. C., & Dudman, S. (2025). Droplet Digital PCR or Real-Time PCR as a Method for Quantifying SARS-CoV-2 RNA in Plasma—Is There a Difference? Viruses, 17(6), 772. https://doi.org/10.3390/v17060772





