Discovery and Genome Characterization of Three New Rhabdoviruses Infecting Passiflora spp. in Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Double-Stranded RNA Extraction, High-Throughput Sequencing, and Bioinformatic Analysis
2.3. Virus Detection and Sanger Sequence
2.4. Complete Sequence of Rhabdoviruses from Passion Fruit
2.5. 5′ and 3′ End Method for Rapid Amplification of cDNA Ends (RACE)
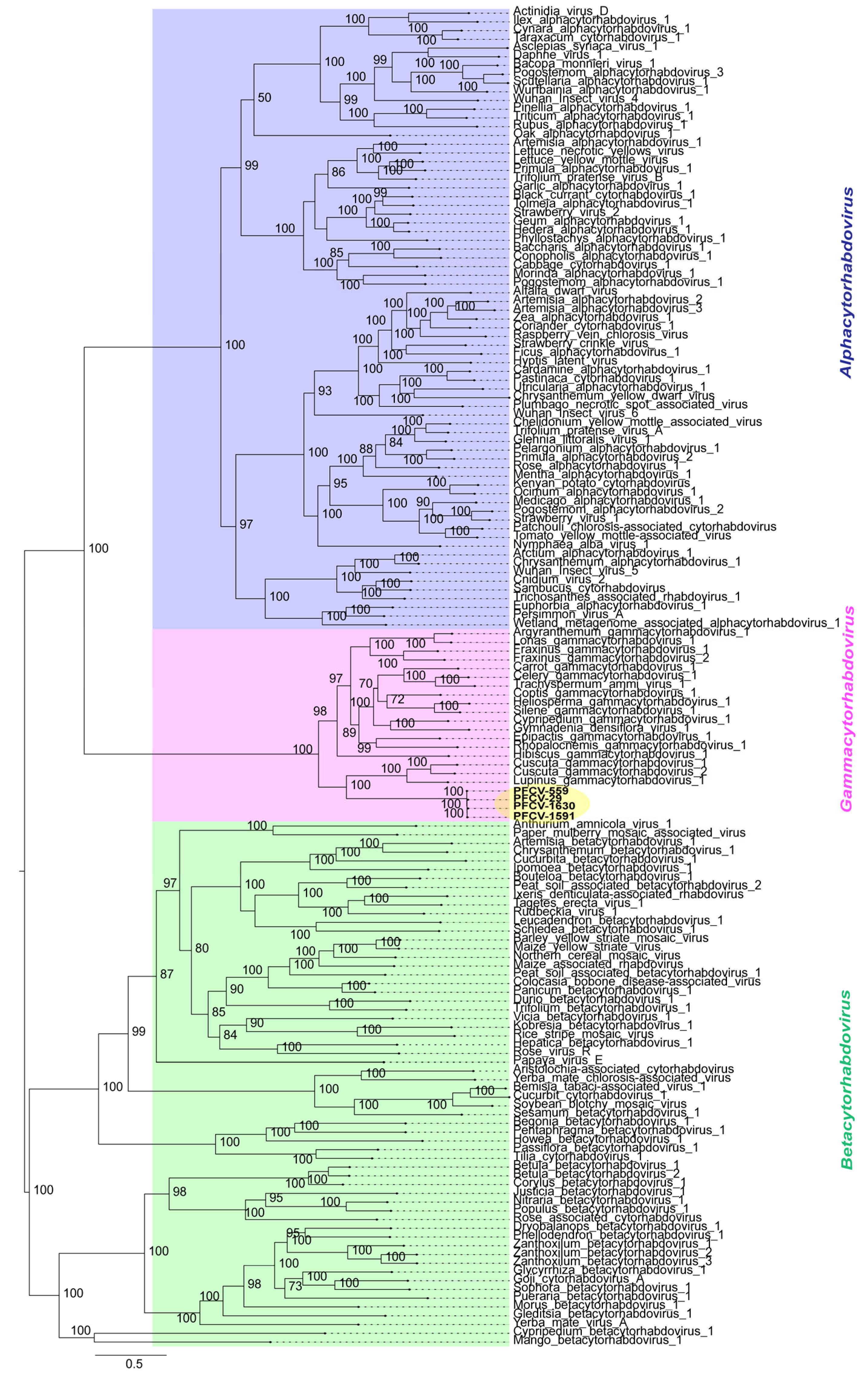
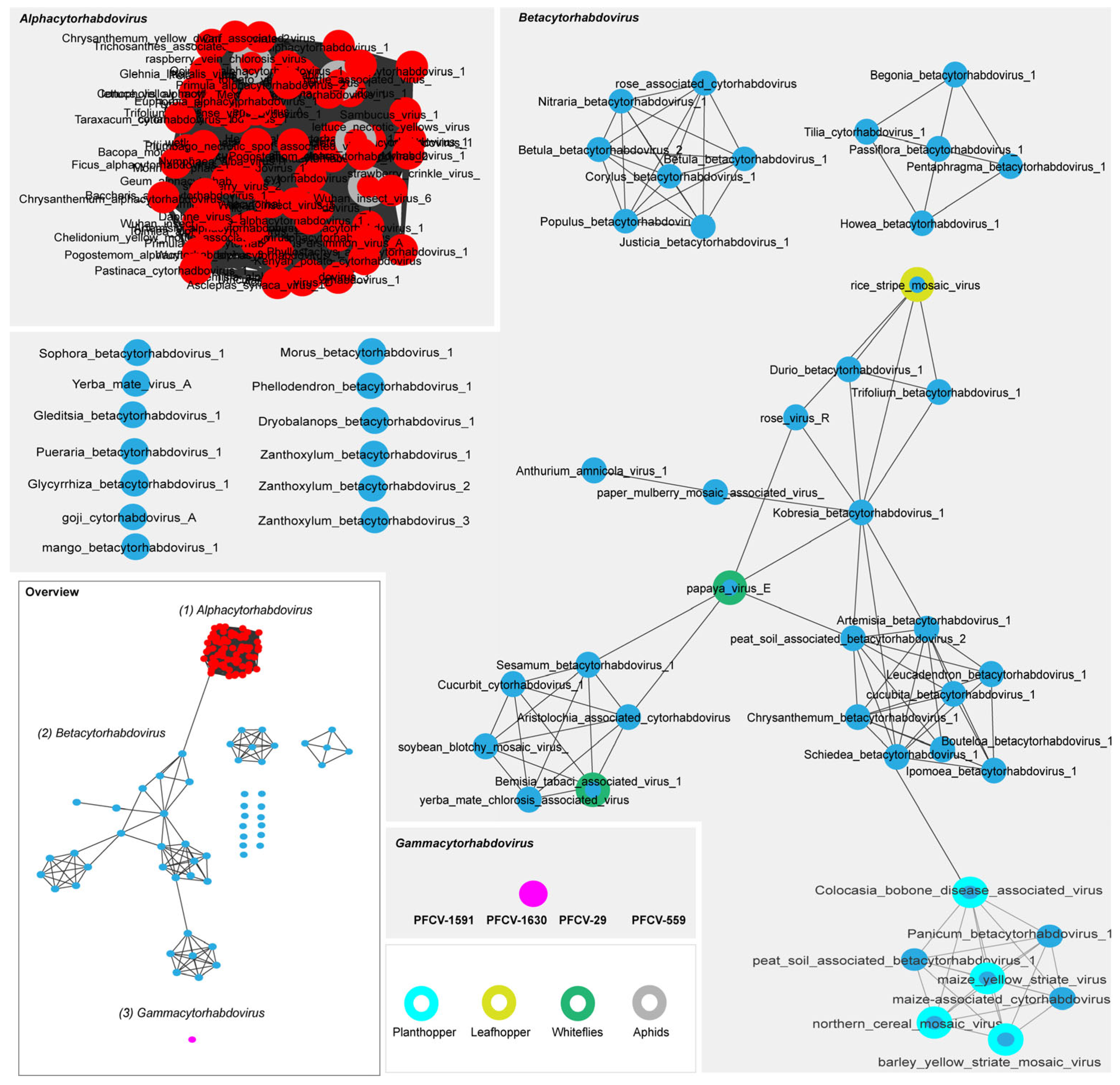
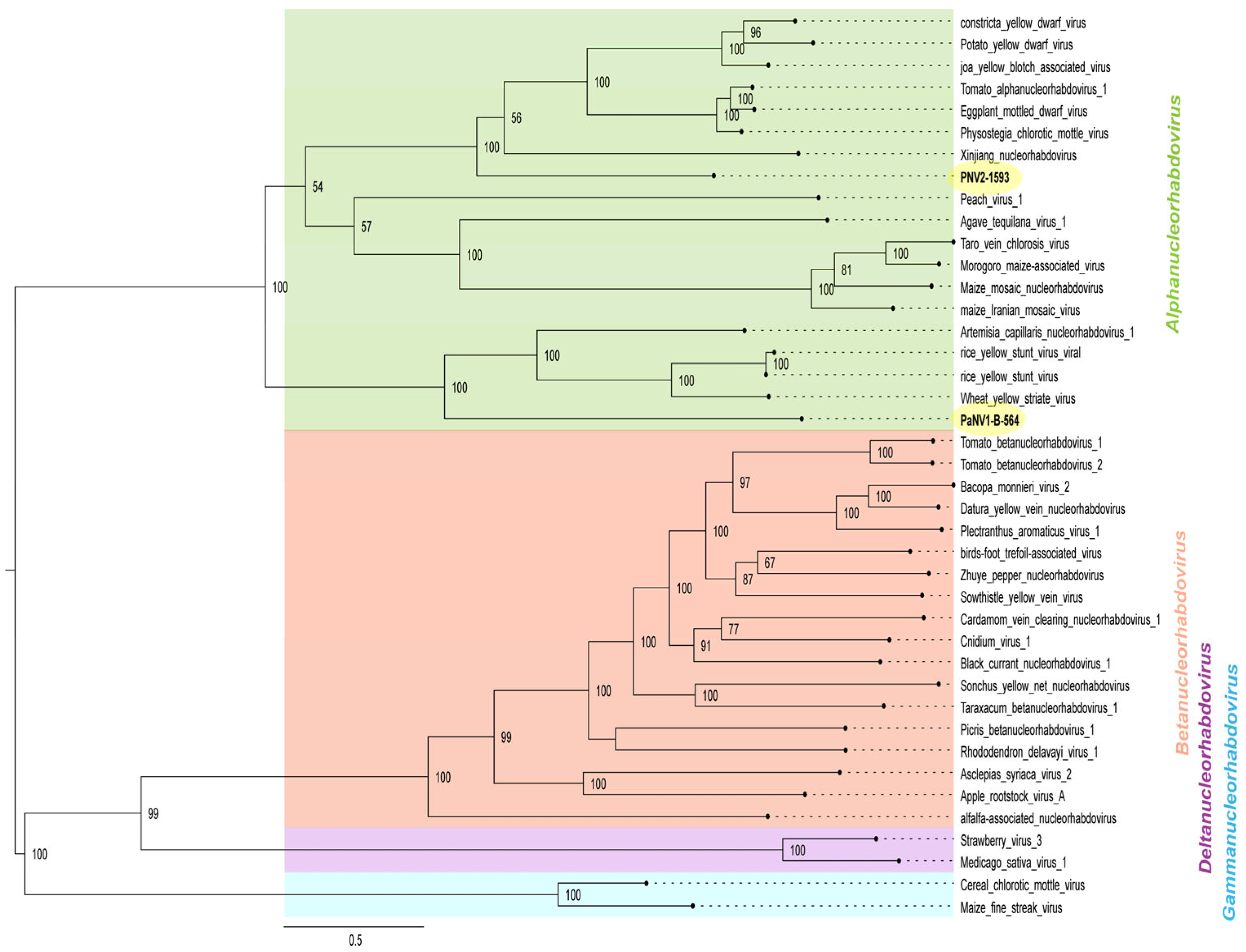
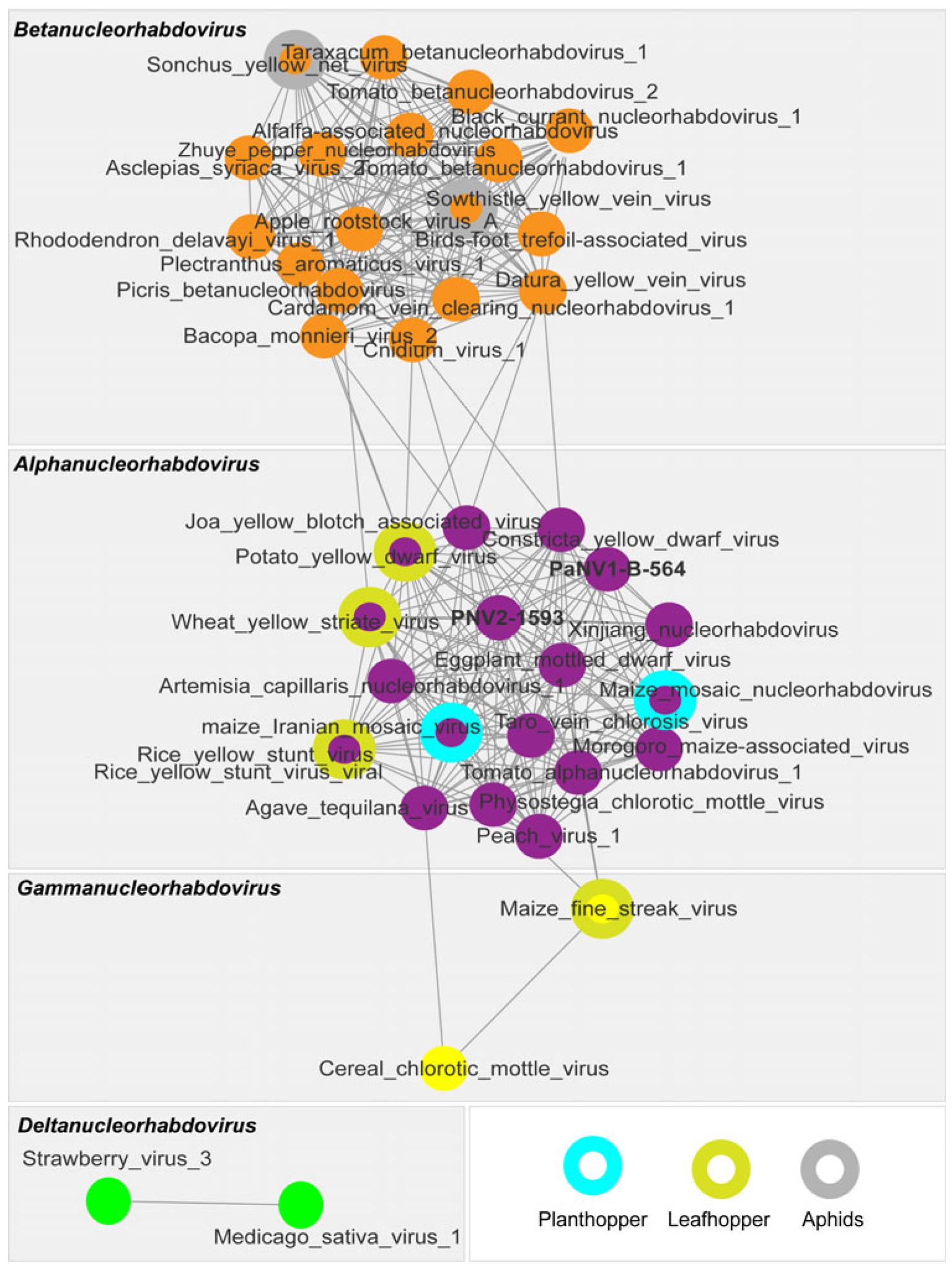
2.6. Phylogenetic Analysis
2.7. Transmission Electron Microscopy
2.8. Distribution of PFCV, PaNV1, and PaNV2 in Passion Fruit Plants in Brazil
3. Results and Discussion
3.1. Preliminary Identification of Rhabdoviruses from Passion Fruit Plants in Brazil
3.2. Prediction of the Genomic Sequence of Bean-Associated Cytorhabdovirus, Cowpea Mild Mottle Virus, and Cowpea Aphid-Borne Mosaic Virus in the BAG2-FP and BAG3-FP Libraries
3.3. Full Genome and Characterization of Rhabdoviruses from Passion Fruit
3.3.1. Passiflora Cytorhabdovirus (PFCV)
3.3.2. Passiflora Nucleorhabdovirus 1 (PaNV1) and 2 (PaNV2)
3.4. Distribution of the Rhabdoviruses PFCV, PaNV1, PaNV2 in the BAG-FP and Producing Regions of Brazil, and Co-Infection with Other Viruses
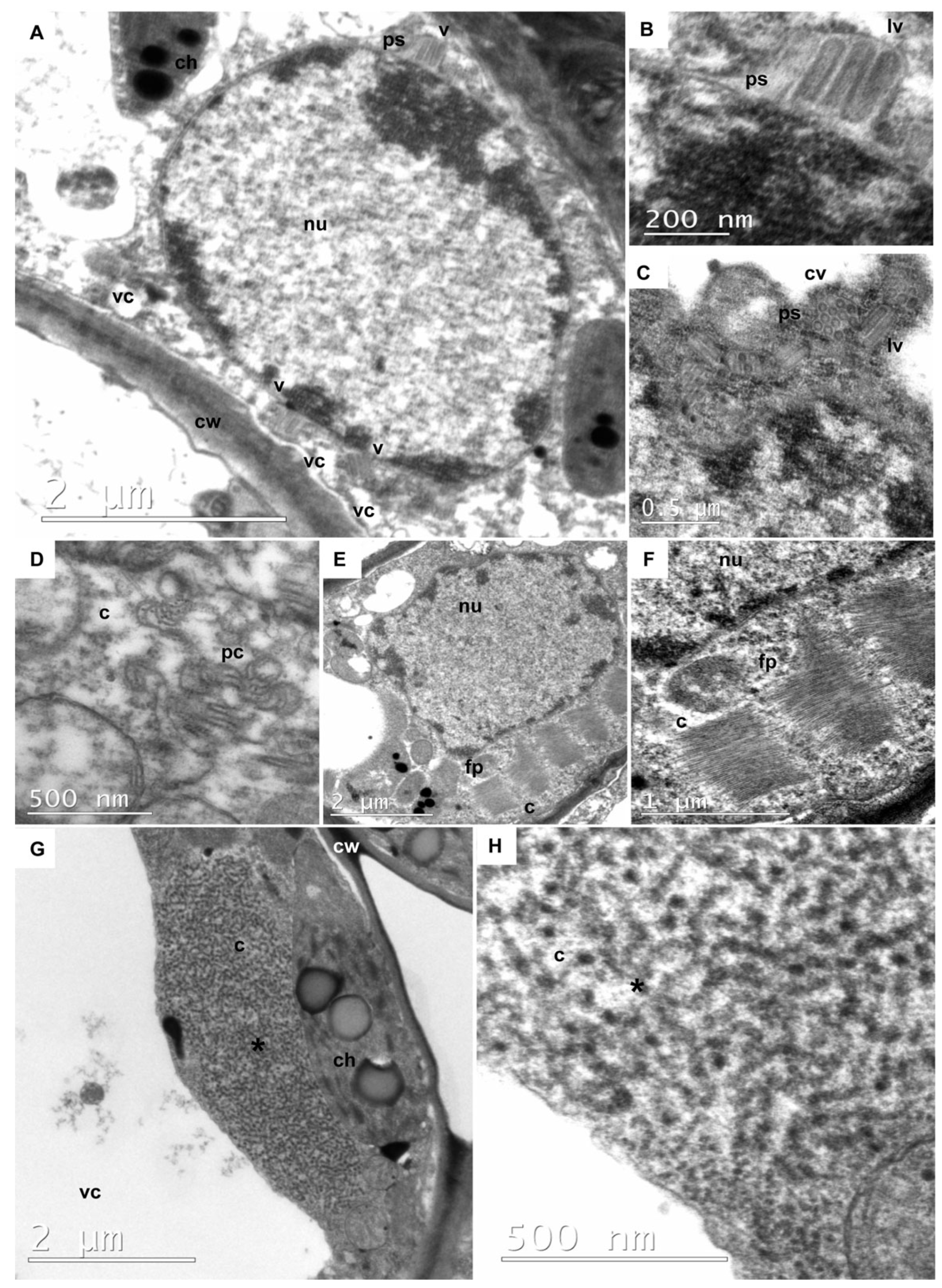
3.5. Rhabdovirus Particles
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Higuita, M.; Sanchez-Yali, J.; Perez, A.; Arias, E.; Gutierrez, P.A. An integrated analysis of the Passifloraceae virome using public-domain data. Phytopathology 2024. [Google Scholar] [CrossRef] [PubMed]
- Bejerman, N.; Dietzgen, R.; Debat, H. Novel tri-segmented rhabdoviruses: A data mining expedition unveils the cryptic diversity of cytorhabdoviruses. Viruses 2023, 15, 2402. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Huang, A.; Zhou, X.; Li, Z.; Dietzgen, R.G.; Zhou, C.; Cao, M. Natural defect of a plant rhabdovirus glycoprotein gene: A case study of virus–plant coevolution. Phytopathology 2021, 111, 227–236. [Google Scholar] [CrossRef]
- Bejerman, N.; Dietzgen, R.G.; Debat, H. Illuminating the plant rhabdovirus landscape through metatranscriptomics Data. Viruses 2021, 13, 1304. [Google Scholar] [CrossRef]
- Bejerman, N.; Debat, H. Alpha- and Betagymnorhavirus: Two new genera of gymnosperm-infecting viruses in the family Rhabdoviridae, subfamily Betarhabdovirinae. Arch. Virol. 2024, 169, 193. [Google Scholar] [CrossRef]
- Ma, Y.; Che, H.; Gao, S.; Lin, Y.; Li, S. Diverse novel viruses coinfecting the tropical ornamental plant Polyscias Balfouriana in China. Viruses 2022, 14, 1120. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.C.; Lopes, Í.S.; Fonseca, P.L.C.; Olmo, R.P.; Bittencourt, F.; Vasconcelos, L.M.; Pirovani, C.P.; Gaiotto, F.A.; Aguiar, E.R.G.R. Expanding the environmental virome: Infection profile in a native rainforest tree species. Front. Microbiol. 2022, 13, 874319. [Google Scholar] [CrossRef]
- Faleiro, F.G.; Oliveira, J.S.; Junqueira, N.T.V.; Santos, R.S. Banco de Germoplasma de Passiflora L.’Flor Da Paixão’no Portal Alelo Recursos Genéticos, 1st ed.; Embrapa Recursos Genéticos e Biotecnologia: Brasília, DF, Brazil, 2019; Volume 1. [Google Scholar]
- IBGE Produção Agrícola Municipal-PAM 2023: Tabela 4—Área Destinada à Colheita, Área Colhida, Quantidade Produzida, Rendimento Médio e Valor da Produção do Brasil, das Grandes Regiões e das Unidades da Federação, Segundo os Produtos das Lavouras Permanentes. Available online: https://www.ibge.gov.br/estatisticas/economicas/agricultura-e-pecuaria/9117-producao-agricola-municipal-culturas-temporarias-e-permanentes.html?=&t=resultados (accessed on 23 February 2025).
- Faleiro, F.G.; Oliveira, J.S.; Walter, B.M.T.; Junqueira, N.T.V. Banco de Germoplasma de Passiflora L.’Flor Da Paixão’: Caracterização Fenotípica, Diversidade Genética, Fotodocumentação e Herborização, 2nd ed.; ProImpress: Brasília, DF, Brazil, 2020; Volume 1. [Google Scholar]
- Pares, R.D.; Martin, A.B.; Morrison, W. Rhabdovirus-like particles in passionfruit. Australas. Plant Pathol. 1983, 12, 51–52. [Google Scholar] [CrossRef]
- Chagas, C.M.; Catroxo, M.H.; Oliveira, J.M.d.; Furtado, E.L. Occurrence of passion fruit vein clearing virus in the state of São Paulo. Fitopatol. Bras. 1987, 12, 275–278. [Google Scholar]
- Kitajima, E.W.; Crestani, O.A. Association of a rhabdovirus with passionfruit vein clearing in Brazil. Fitopatol. Bras. 1985, 3, 681–688. [Google Scholar]
- Kitajima, E.W. An annotated list of plant viruses and viroids described in Brazil (1926–2018). Biota Neotrop. 2020, 20, e20190932. [Google Scholar] [CrossRef]
- Vidal, A.H.; Felix, G.P.; Abreu, E.F.M.; Pinheiro-Lima, B.; Vianna, M.J.X.; Nogueira, I.; Abreu, A.C.R.; Sanches, M.M.; Santos-Jiménez, J.L.; Rosa, R.C.C.; et al. Occurrence of bean-associated cytorhabdovirus and cowpea mild mottle virus infecting cultivated and wild Passiflora spp. in Brazil. Crop Prot. 2023, 168, 106236. [Google Scholar] [CrossRef]
- Walker, P.J.; Freitas-Astúa, J.; Bejerman, N.; Blasdell, K.R.; Breyta, R.; Dietzgen, R.G.; Fooks, A.R.; Kondo, H.; Kurath, G.; Kuzmin, I.V. ICTV virus taxonomy profile: Rhabdoviridae 2022. J. Gen. Virol. 2022, 103, 001689. [Google Scholar] [CrossRef] [PubMed]
- Dietzgen, R.G.; Kuhn, J.H.; Clawson, A.N.; Freitas-Astúa, J.; Goodin, M.M.; Kitajima, E.W.; Kondo, H.; Wetzel, T.; Whitfield, A.E. Dichorhavirus: A Proposed New Genus for Brevipalpus mite-transmitted, nuclear, bacilliform, bipartite, negative-strand RNA plant viruses. Arch. Virol. 2014, 159, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Sasaya, T.; Kusaba, S.; Ishikawa, K.; Koganezawa, H. Nucleotide sequence of RNA2 of lettuce big-vein virus and evidence for a possible transcription termination/initiation strategy similar to that of rhabdoviruses. J. Gen. Virol. 2004, 85, 2709–2717. [Google Scholar] [CrossRef]
- Gao, D.M.; Zhang, Z.J.; Qiao, J.H.; Gao, Q.; Zang, Y.; Xu, W.Y.; Xie, L.; Fang, X.D.; Ding, Z.H.; Yang, Y.Z.; et al. A rhabdovirus accessory protein inhibits jasmonic acid signaling in plants to attract insect vectors. Plant Physiol. 2022, 190, 1349–1364. [Google Scholar] [CrossRef]
- Vidal, A.H.; Felix, G.P.; Abreu, E.F.M.; Nogueira, I.; Alves-Freitas, D.M.T.; Faleiro, F.G.; Fontenele, R.S.; Peixoto, J.R.; Lacorte, C.; Rosa, R.C.C.; et al. Occurrence of lettuce chlorosis virus in Passiflora spp. in Brazil. J. Plant Pathol. 2021, 103, 443–447. [Google Scholar] [CrossRef]
- Vidal, A.H.; Lacorte, C.; Sanches, M.M.; Alves-Freitas, D.M.T.; Abreu, E.F.M.; Pinheiro-Lima, B.; Rosa, R.C.C.; Jesus, O.N.; Campos, M.A.; Felix, G.P.; et al. Characterization of cucurbit aphid-borne yellows virus (CABYV) from passion fruit in Brazil: Evidence of a complex of species within CABYV isolates. Viruses 2023, 15, 410. [Google Scholar] [CrossRef]
- Vidal, A.H.; Sanches, M.M.; Alves-Freitas, D.M.T.; Abreu, E.F.M.; Lacorte, C.; Pinheiro-Lima, B.; Rosa, R.C.C.; Jesus, O.N.; Campos, M.A.; Varsani, A.; et al. First world report of cucurbit aphid-borne yellows virus infecting passionfruit. Plant Dis. 2018, 102, 2665. [Google Scholar] [CrossRef]
- Matvienko, M. CLC Genomics Workbench. Plant and Animal Genome. Sr. Field Application Scientist; CLC Bio: Aarhus, Denmark, 2015. [Google Scholar]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Alves-Freitas, D.M.T.; Pinheiro-Lima, B.; Faria, J.C.; Lacorte, C.; Ribeiro, S.G.; Melo, F.L. Double-stranded RNA high-throughput sequencing reveals a new cytorhabdovirus in a bean golden mosaic virus-resistant common bean transgenic line. Viruses 2019, 11, 90. [Google Scholar] [CrossRef]
- Nicolini, C.; Pio-Ribeiro, G.; Andrade, G.P.; Melo, F.L.; Oliveira, V.C.; Guimarães, F.C.; Resende, R.O.; Kitajima, E.W.; Rezende, J.A.M.; Nagata, T. A distinct tymovirus infecting Cassia hoffmannseggii in Brazil. Virus Genes 2012, 45, 190–194. [Google Scholar] [CrossRef]
- Schuster, D.M.; Buchman, G.W.; Rashtchian, A. A simple and efficient method for amplification of cDNA ends using 5′ RACE. Focus 1992, 14, 46–52. [Google Scholar]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Gerlt, J.A.; Bouvier, J.T.; Davidson, D.B.; Imker, H.J.; Sadkhin, B.; Slater, D.R.; Whalen, K.L. Enzyme function initiative-enzyme similarity tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2015, 1854, 1019–1037. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Pinheiro-Lima, B.; Pereira-Carvalho, R.C.; Alves-Freitas, D.M.T.; Kitajima, E.W.; Vidal, A.H.; Lacorte, C.; Godinho, M.T.; Fontenele, R.S.; Faria, J.C.; Abreu, E.F.M.; et al. Transmission of the bean-associated cytorhabdovirus by the whitefly Bemisia Tabaci MEAM1. Viruses 2020, 12, 1028. [Google Scholar] [CrossRef]
- Kassem, M.A.; Juarez, M.; Gómez, P.; Mengual, C.M.; Sempere, R.N.; Plaza, M.; Elena, S.F.; Moreno, A.; Fereres, A.; Aranda, M.A. Genetic diversity and potential vectors and reservoirs of cucurbit aphid-borne yellows virus in southeastern Spain. Phytopathology 2013, 103, 1188–1197. [Google Scholar] [CrossRef]
- Abreu, A.C.R.; Vidal, A.H.; Santos-Jiménez, J.L.; Rosa, R.C.C.; Vaslin, M.F.; Ribeiro, S. Mixed infection cucurbit aphid-borne yellows virus and cowpea aphid-borne mosaic virus on passion fruit in Rio de Janeiro State, Brazil. In Proceedings of the XXXII Congresso Brasileiro de Virologia, Virtual, 19–23 October 2021. [Google Scholar]
- Zanardo, L.G.; Carvalho, C.M. Cowpea Mild Mottle Virus (Carlavirus, Betaflexiviridae): A Review. Trop. Plant Pathol. 2017, 42, 417–430. [Google Scholar] [CrossRef]
- Mlotshwa, S.; Verver, J.; Sithole-Niang, I.; Van Kampen, T.; Van Kammen, A.; Wellink, J. The genomic sequence of cowpea aphid-borne mosaic virus and its similarities with other potyviruses. Arch. Virol. 2002, 147, 1043–1052. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Liu, Y.; Zhou, X.; Yin, C.; Zhang, P.; Yang, Q.; Mao, L.; Shentu, X.; Yu, X. Nanopore sequencing technology and its application in plant virus diagnostics. Front. Microbiol. 2022, 13, 939666. [Google Scholar] [CrossRef]
- Liefting, L.W.; Waite, D.W.; Thompson, J.R. Application of Oxford Nanopore technology to plant virus detection. Viruses 2021, 13, 1424. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Kim, H.J.; Cho, I.S.; Jeong, R.D. Identification of viruses infecting phalaenopsis orchids using nanopore sequencing and development of an RT-RPA-CRISPR/Cas12a for rapid visual detection of nerine latent virus. Int. J. Mol. Sci. 2024, 25, 2666. [Google Scholar] [CrossRef]
- Maragathavalli, P.; Jana, S. The hybrid feature incorporated dual deep learning architecture for the automatic jasmine plant disease detection and classification. Curr. Sci. 2024, 127, 1108. [Google Scholar] [CrossRef]
- Kim, H.J.; Cho, I.S.; Choi, S.R.; Jeong, R.D. Identification of an isolate of citrus tristeza virus by Nanopore sequencing in Korea and development of a CRISPR/Cas12a-based assay for rapid visual detection of the virus. Phytopathology 2024, 114, 1421–1428. [Google Scholar] [CrossRef] [PubMed]
- Walker, P.J.; Firth, C.; Widen, S.G.; Blasdell, K.R.; Guzman, H.; Wood, T.G.; Paradkar, P.N.; Holmes, E.C.; Tesh, R.B.; Vasilakis, N. Evolution of genome size and complexity in the Rhabdoviridae. PLoS Pathog. 2015, 11, e1004664. [Google Scholar] [CrossRef]
- Bejerman, N.; Acevedo, R.M.; de Breuil, S.; Ruiz, O.A.; Sansberro, P.; Dietzgen, R.G.; Nome, C.; Debat, H. Molecular characterization of a novel cytorhabdovirus with a unique genomic organization infecting yerba mate (Ilex paraguariensis) in Argentina. Arch. Virol. 2020, 165, 1475–1479. [Google Scholar] [CrossRef]
- Prabowo, E.Y.; Valmonte-Cortes, G.R.; Darling, T.L.; Buckley, E.; Duxbury, M.; Seale, B.; Higgins, C.M. Is the Glycoprotein responsible for the differences in dispersal rates between lettuce necrotic yellows virus subgroups? Viruses 2022, 14, 1574. [Google Scholar] [CrossRef]
- Bejerman, M.; Debat, H.; Dietzgen, R.; Freitas-Astua, J.; Kondo, H.; Ramos-Gonzalez, P.; Whitfield, A.; Walker, P. 2024.015P.A.v1.Rhabdoviridae_Cytorhabdovirus_splitgen: Abolish one genus and create three new genera to Include 98 New Species in the Subfamily Betarhabdovirinae (Mononegavirales: Rhabdoviridae). Available online: https://www.google.com/url?sa=t&source=web&rct=j&opi=89978449&url=https://2cms.ictv.global/system/files/proposals/pending/All_Proposals_%2528Compressed_.zip_files%2529/Taxonomy-Proposal-Summaries-for-Ratification-Vote%252C-2025.pdf&ved=2ahUKEwiM-Obtx9KMAxUkpZUCHYimD-MQFnoECBkQAQ&usg=AOvVaw1k_-ImRhdiTVwrZd8UZQiW (accessed on 15 January 2025).
- Wang, Y.; Wang, Y.; Li, Z.; Zeng, Q.; Xu, Q.; Wang, Z.; Zhou, X. Genomic characterization of a novel cytorhabdovirus infecting Ixeris denticulata in China. Arch. Virol. 2024, 169, 46. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.S.; Kim, J.; Jun, M.; Shin, S.; Lee, S.J.; Lim, S. Complete genome sequence of a putative novel cytorhabdovirus isolated from Rudbeckia sp. Arch. Virol. 2022, 167, 2381–2385. [Google Scholar] [CrossRef] [PubMed]
- Kauffmann, C.M.; Jesus, B.; Silva, J.M.F.; Blawid, R.; Nagata, T. Complete genome sequence of patchouli chlorosis-associated cytorhabdovirus, a new cytorhabdovirus infecting patchouli plants in Brazil. Arch. Virol. 2022, 167, 2817–2820. [Google Scholar] [CrossRef]
- Ramos-González, P.L.; Alexandre, M.A.V.; Potsclam-Barro, M.; Duarte, L.M.L.; Michea Gonzalez, G.L.; Chabi-Jesus, C.; Ramos, A.F.; Harakava, R.; Lorenzi, H.; Freitas-Astúa, J.; et al. Two novel betarhabdovirins infecting ornamental plants and the peculiar intracellular behavior of the cytorhabdovirus in the liana Aristolochia gibertii. Viruses 2024, 16, 322. [Google Scholar] [CrossRef] [PubMed]
- Lefeuvre, P.; Martin, D.P.; Elena, S.F.; Shepherd, D.N.; Roumagnac, P.; Varsani, A. Evolution and Ecology of Plant Viruses. Nat. Rev. Microbiol. 2019, 17, 632–644. [Google Scholar] [CrossRef]
- Fontenele, R.S.; Salywon, A.M.; Majure, L.C.; Cobb, I.N.; Bhaskara, A.; Avalos-Calleros, J.A.; Argüello-Astorga, G.R.; Schmidlin, K.; Khalifeh, A.; Smith, K.; et al. New World cactaceae plants harbor diverse geminiviruses. Viruses 2021, 13, 694. [Google Scholar] [CrossRef]
- Alexandre, R.S.; Wagner Júnior, A.; Negreiros, J.R.d.S.; Parizzotto, A.; Bruckner, C.H. Germinação de sementes de genótipos de maracujazeiro. Pesqui. Agropecu. Bras. 2004, 39, 1239–1245. [Google Scholar] [CrossRef]
- Fránová, J.; Přibylová, J.; Koloniuk, I. Molecular and biological characterization of a new strawberry cytorhabdovirus. Viruses 2019, 11, 982. [Google Scholar] [CrossRef]
- Sylvester, E.S.; Richardson, J.; Frazier, N.W. Serial passage of strawberry crinkle virus in the aphid Chaetosiphon Jacobi. Virology 1974, 59, 301–306. [Google Scholar] [CrossRef]
- Stubbs, L.L.; Grogan, R.G. Necrotic yellows: A newly recognized virus disease of lettuce. Aust. J. Agric. Res. 1963, 14, 439–459. [Google Scholar] [CrossRef]
- Dumón, A.D.; Mattio, M.F.; Argüello Caro, E.B.; Alemandri, V.; Puyané, N.; del Vas, M.; López Lambertini, P.; Truol, G. Occurrence of a closely-related isolate to maize yellow striate virus in wheat plants. Agriscientia 2015, 32, 107–112. [Google Scholar] [CrossRef]
- Cao, Q.; Xu, W.Y.; Gao, Q.; Jiang, Z.H.; Liu, S.Y.; Fang, X.D.; Gao, D.M.; Wang, Y.; Wang, X.B. Transmission characteristics of barley yellow striate mosaic virus in its planthopper vector Laodelphax Striatellus. Front. Microbiol. 2018, 9, 1419. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Huang, J.; Liu, C.; Chen, B.; Zhang, T.; Zhou, G. Rice stripe mosaic virus, a novel cytorhabdovirus infecting rice via leafhopper transmission. Front. Microbiol. 2017, 7, 2140. [Google Scholar] [CrossRef] [PubMed]
- Cornejo-Franco, J.F.; Reyes-Proaño, E.; Mollov, D.; Mowery, J.; Quito-Avila, D.F. Transmission and pathogenicity of papaya virus E: Insights from an experimental papaya orchard. Plant Dis. 2022, 106, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Pappi, P.G.; Maliogka, V.I.; Amoutzias, G.D.; Katis, N.I. Genetic variation of Eggplant mottled dwarf virus from annual and perennial plant hosts. Arch. Virol. 2016, 161, 631–639. [Google Scholar] [CrossRef]
- Medberry, A.N.; Srivastava, A.; Diaz-Lara, A.; Rwahnih, M.A.; Villamor, D.E.V.; Tzanetakis, I.E. A novel, divergent member of the Rhabdoviridae family infects strawberry. Plant Dis. 2023, 107, 620–623. [Google Scholar] [CrossRef]
- Wang, H.; Wang, J.; Xie, Y.; Fu, Z.; Wei, T.; Zhang, X.F. Development of leafhopper cell culture to trace the early infection process of a nucleorhabdovirus, rice yellow stunt virus, in insect vector cells. Virol. J. 2018, 15, 72. [Google Scholar] [CrossRef]
- Liu, Y.; Du, Z.; Wang, H.; Zhang, S.; Cao, M.; Wang, X. Identification and characterization of wheat yellow striate virus, a novel leafhopper-transmitted nucleorhabdovirus infecting wheat. Front. Microbiol. 2018, 9, 468. [Google Scholar] [CrossRef]
- Babaie, G.H.; Izadpanah, K. Vector transmission of Eggplant mottled dwarf virus in Iran. J. Phytopathol. 2003, 151, 679–682. [Google Scholar] [CrossRef]
- Ammar, E.D.; Hogenhout, S.A. A neurotropic route for maize mosaic virus (Rhabdoviridae) in its planthopper vector Peregrinus Maidis. Virus Res. 2008, 131, 77–85. [Google Scholar] [CrossRef]
- Hortamani, M.; Massah, A.; Izadpanah, K. Maize Iranian mosaic virus shows a descending transcript accumulation order in plant and insect hosts. Arch. Virol. 2018, 163, 887–893. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.O.; Francki, R.I.B.; Zuidema, D. Biology, structure, and replication of plant rhabdoviruses. In The Rhabdoviruses; Springer: Boston, MA, USA, 1987; pp. 427–508. [Google Scholar] [CrossRef]
- Duffus, J.E. Possible multiplication in the aphid vector of Sowthistle yellow vein virus, a virus with an extremely long insect latent period. Virology 1963, 21, 194–202. [Google Scholar] [CrossRef]
- Li, J.; Shang, Q.; Liu, Y.; Dai, W.; Li, X.; Wei, S.; Hu, G.; McNeill, M.R.; Ban, L. Occurrence, distribution, and transmission of alfalfa viruses in China. Viruses 2022, 14, 1519. [Google Scholar] [CrossRef] [PubMed]
- Redinbaugh, M.G.; Seifers, D.L.; Meulia, T.; Abt, J.J.; Anderson, R.J.; Styer, W.E.; Ackerman, J.; Salomon, R.; Houghton, W.; Creamer, R. Maize fine streak virus, a new leafhopper-transmitted rhabdovirus. Phytopathology 2002, 92, 1167–1174. [Google Scholar] [CrossRef]
- Machado, C.F.; Faleiro, F.G.; Santos Filho, H.P.; Fancelli, M.; Carvalho, R.S.; Ritzinger, C.H.S.P.; Araujo, F.P.; Junqueira, N.T.V.; Jesus, O.N.; Novaes, Q.S. Guia de Identificação e Controle de Pragas Na Cultura Do Maracujazeiro; Embrapa: Brasília, DF, Brazil, 2017; Volume 1. [Google Scholar]
- Jayakar, H.R.; Jeetendra, E.; Whitt, M.A. Rhabdovirus assembly and budding. Virus Res. 2004, 106, 117–132. [Google Scholar] [CrossRef]
- Jackson, A.O.; Dietzgen, R.G.; Goodin, M.M.; Bragg, J.N.; Deng, M. Biology of plant rhabdoviruses. Annu. Rev. Phytopathol. 2005, 43, 623–660. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vidal, A.H.; Abreu, A.C.R.; Dantas-Filho, J.F.S.; Vianna, M.J.X.; Lacorte, C.; Abreu, E.F.M.; Felix, G.P.; Alves-Freitas, D.M.T.; Pinheiro-Lima, B.; Nogueira, I.; et al. Discovery and Genome Characterization of Three New Rhabdoviruses Infecting Passiflora spp. in Brazil. Viruses 2025, 17, 725. https://doi.org/10.3390/v17050725
Vidal AH, Abreu ACR, Dantas-Filho JFS, Vianna MJX, Lacorte C, Abreu EFM, Felix GP, Alves-Freitas DMT, Pinheiro-Lima B, Nogueira I, et al. Discovery and Genome Characterization of Three New Rhabdoviruses Infecting Passiflora spp. in Brazil. Viruses. 2025; 17(5):725. https://doi.org/10.3390/v17050725
Chicago/Turabian StyleVidal, Andreza Henrique, Ana Clara Rodrigues Abreu, Jorge Flávio Sousa Dantas-Filho, Monique Jacob Xavier Vianna, Cristiano Lacorte, Emanuel Felipe Medeiros Abreu, Gustavo Pereira Felix, Dione Mendes Teixeira Alves-Freitas, Bruna Pinheiro-Lima, Isadora Nogueira, and et al. 2025. "Discovery and Genome Characterization of Three New Rhabdoviruses Infecting Passiflora spp. in Brazil" Viruses 17, no. 5: 725. https://doi.org/10.3390/v17050725
APA StyleVidal, A. H., Abreu, A. C. R., Dantas-Filho, J. F. S., Vianna, M. J. X., Lacorte, C., Abreu, E. F. M., Felix, G. P., Alves-Freitas, D. M. T., Pinheiro-Lima, B., Nogueira, I., Faleiro, F. G., Rosa, R. C. C., Jesus, O. N., Sanches, M. M., Santos, Y. S., Blawid, R., Jiménez, J. L. S., Vaslin, M. F. S., Kitajima, E. W., ... Ribeiro, S. G. (2025). Discovery and Genome Characterization of Three New Rhabdoviruses Infecting Passiflora spp. in Brazil. Viruses, 17(5), 725. https://doi.org/10.3390/v17050725






