Novel Orthohantavirus Associated with Hantavirus Pulmonary Syndrome in Northern Argentina
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Definition
2.2. Viral Characterization
2.3. Epidemiological Investigation and Environmental Survey
2.4. Georeferencing
2.5. Rodent Trapping and Characterization
3. Results
3.1. HPS Case Confirmation
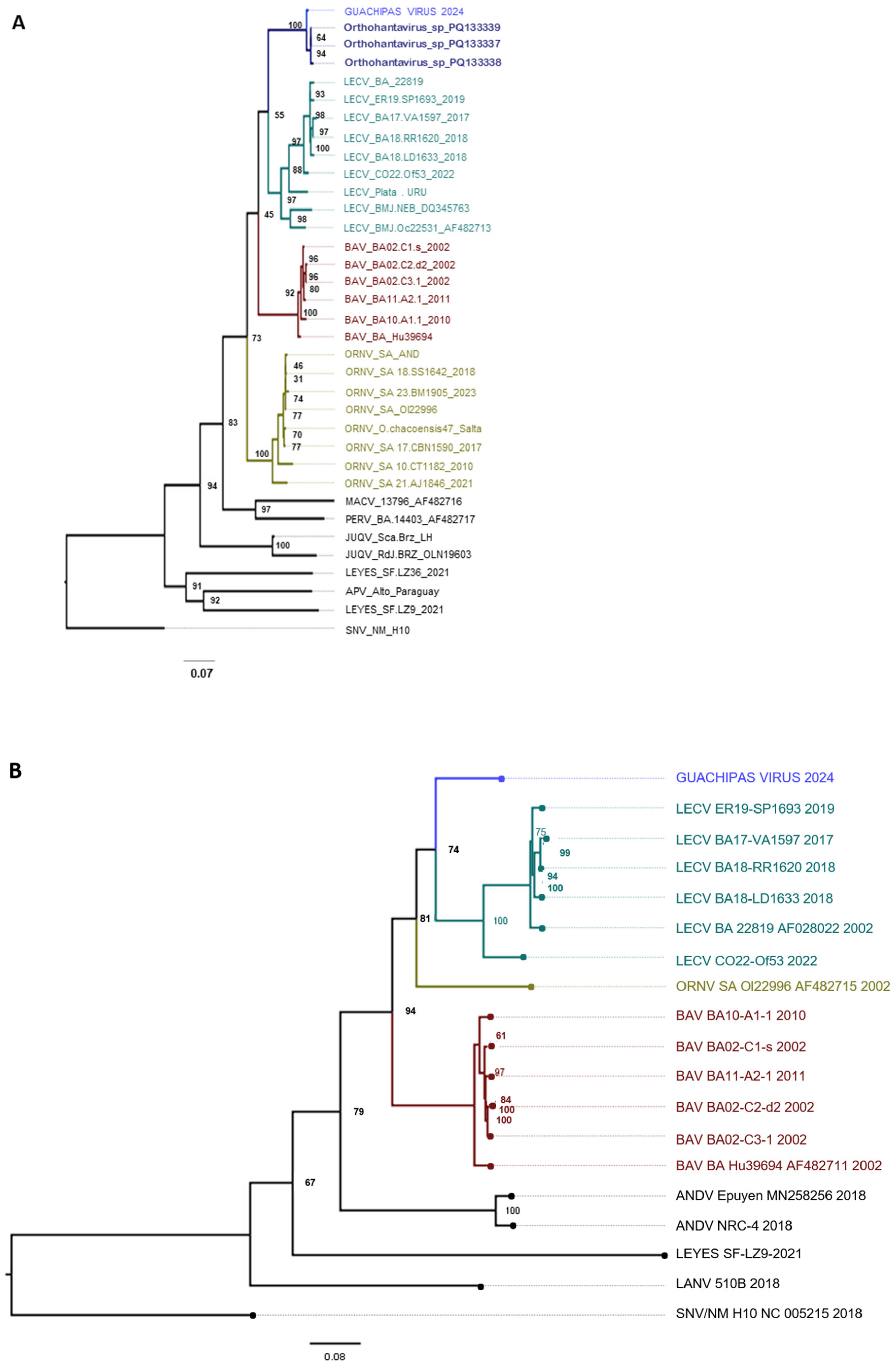
3.2. Genetic Characterization
3.3. Eco-Epidemiological Features
3.4. Rodent Community Description
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HPS | Hantavirus pulmonary syndrome |
| NRLH | National Reference Laboratory for Hantavirus |
| RNA | Ribonucleic acid |
| DNA | Desoxyribonucleic acid |
| RT-PCR | Reverse transcription–polymerase chain reaction |
| ELISA | Enzyme-linked immunosorbent assay |
| ANDV | Andes virus |
| BERV | Bermejo virus |
| BAV | Buenos Aires virus |
| LECV | Lechighuanas virus |
| ORNV | Oran virus |
| LNV | Laguna Negra virus |
| NWA | Northwest Argentina |
| NGS | Next-generation sequencing |
| NCBI | National Center for Biotechnology Information |
| BLAST | Basic Local Alignment Search Tool |
References
- López, N.; Padula, P.; Rossi, C.; Lázaro, M.E.; Franze-Fernández, M.T. Genetic identification of a new hantavirus causing severe pulmonary syndrome in Argentina. Virology 1996, 220, 223–226. [Google Scholar] [CrossRef]
- Padula, P.J.; Colavecchia, S.B.; Martínez, V.P.; Della Valle, M.O.G.; Edelstein, A.; Miguel, S.D.L.; Russi, J.; Riquelme, J.M.; Colucci, N.; Almirόn, M.; et al. Genetic diversity, distribution, and serological features of hantavirus infection in five countries in South America. J. Clin. Microbiol. 2000, 38, 3029–3035. [Google Scholar] [CrossRef] [PubMed]
- Ciancaglini, M.; Bellomo, C.M.; Cabreros, C.L.T.; Alonso, D.; Bassi, S.C.; Iglesias, A.; Martínez, V.P. Hantavirus pulmonary syndrome in Tucumán province associated to an unexpected viral genotype. Medicina 2017, 77, 81–84. [Google Scholar]
- Calderón, G.E.; Brignone, J.; Martin, M.L.; Calleri, F.; Sen, C.; Casas, N.; Calli, R.; Sinchi, A.; Enria, D.; Levis, S. Outbreak of hantavirus pulmonary syndrome in Tucumán, Argentina. Medicina 2018, 78, 151–157. [Google Scholar] [PubMed]
- Padula, P.; Della Valle, M.G.; Alai, M.G.; Cortada, P.; Villagra, M.; Gianella, A. Andes virus and first case report of Bermejo virus causing fatal pulmonary syndrome. Emerg. Infect. Dis. 2002, 8, 437–439. [Google Scholar] [CrossRef] [PubMed]
- Della Valle, M.G.; Cortez, J.; Edelstein, A.; Cacace, M.L.; Miguel, S.; Jurgelenas, G.; Padula, P.; Martinez, V.; Estani, S.S. Andes virus associated with hantavirus pulmonary syndrome in northern Argentina and determination of the precise site of infection. Am. J. Trop. Med. Hyg. 2002, 66, 713–720. [Google Scholar] [CrossRef]
- Levis, S.; Pini, N.; Garcia, J.; Bego, M.; Barquez, R.; Lozano, E.; Ripoll, C.; Jeor, S.S.; Bravo, D.; Ramírez, J.; et al. Hantavirus pulmonary syndrome in northwestern Argentina: Circulation of Laguna Negra virus associated with Calomys callosus. Am. J. Trop. Med. Hyg. 2004, 71, 658–663. [Google Scholar] [CrossRef]
- Martinez, V.P.; Bellomo, C.M.; Cacace, M.L.; Suarez, P.; Bogni, L.; Padula, P.J. Hantavirus pulmonary syndrome in Argentina, 1995–2008. Emerg. Infect. Dis. 2010, 16, 1853–1860. [Google Scholar] [CrossRef]
- Alonso, D.O.; Iglesias, A.; Coelho, R.; Periolo, N.; Bruno, A.; Córdoba, M.T.; Filomarino, N.; Quipildor, M.; Biondo, E.; Fortunato, E.; et al. Epidemiological description, case-fatality rate, and trends of Hantavirus Pulmonary Syndrome: 9 years of surveillance in Argentina. J. Med. Virol. 2019, 91, 1173–1181. [Google Scholar] [CrossRef]
- Alonso, D.O.; Pérez-Sautu, U.; Bellomo, C.M.; Prieto, K.; Iglesias, A.; Coelho, R.; Periolo, N.; Domenech, I.; Talmon, G.; Hansen, R.; et al. Person-to-Person Transmission of Andes Virus in Hantavirus Pulmonary Syndrome, Argentina, 2014. Emerg. Infect. Dis. 2020, 26, 756–759. [Google Scholar] [CrossRef]
- Hantavirus|Argentina.gob.ar. [En línea]. Available online: https://www.argentina.gob.ar/salud/glosario/hantavirus (accessed on 2 October 2024).
- Alonso, D.O.; Kehl, S.D.; Coelho, R.M.; Periolo, N.; Poklépovich Caride, T.; Sanchez Loria, J.; Cuba, F.G.; Pérez-Sautu, U.; Sanchez-Lockhart, M.; Palacios, G.; et al. Orthohantavirus diversity in Central-East Argentina: Insights from complete genomic sequencing on phylogenetics, Geographic patterns and transmission scenarios. PLoS Neglected Trop. Dis. 2024, 18, e0012465. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Karger, D.N.; Conrad, O.; Böhner, J.; Kawohl, T.; Kreft, H.; Soria-Auza, R.W.; Zimmermann, N.E.; Linder, H.P.; Kessler, M. Climatologies at high resolution for the earth’s land surface areas. Sci. Data 2017, 4, 170122. [Google Scholar] [CrossRef]
- Oligoryzomys Chacoensis|Categorización de los Mamíferos de Argentina. [En línea]. Available online: https://cma.sarem.org.ar/index.php/es/especie-nativa/oligoryzomys-chacoensis (accessed on 1 October 2024).
- Oligoryzomys Occidentalis|Categorización de los Mamíferos de Argentina. [En línea]. Available online: https://cma.sarem.org.ar/index.php/es/especie-nativa/oligoryzomys-occidentalis (accessed on 1 October 2024).
- Calomys Boliviae/Fecundus|Categorización de los mamíferos de Argentina. [En línea]. Available online: https://cma.sarem.org.ar/es/especie-nativa/calomys-boliviae-fecundus (accessed on 1 October 2024).
- Olson, D.M.; Dinerstein, E.; Wikramanayake, E.D.; Burgess, N.D.; Powell, G.V.N.; Underwood, E.C.; D’Amico, J.A.; Itoua, I.; Strand, H.E.; Morrison, J.C.; et al. Terrestrial Ecoregions of the World: A New Map of Life on Earth: A new global map of terrestrial ecoregions provides an innovative tool for conserving biodiversity. BioScience 2001, 51, 933–938. [Google Scholar] [CrossRef]
- Sikes, R.S. Guidelines of the American Society of Mammalogists for the Use of Wild Mammals in Research and Education. J. Mammal. 2016, 90, 663–688. [Google Scholar] [CrossRef]
- Carroll, D.S.; Tack, D.; Calisher, C.H. Biosafety Guidelines for Working with Small Mammals in a Field Environment. In Biological Safety: Principles and Practices; Wooley, D.P., Byers, K.B., Eds.; ASM Press: Washington, DC, USA, 2016. [Google Scholar] [CrossRef]
- López, W.R.; Altamiranda-Saavedra, M.; Kehl, S.D.; Ferro, I.; Bellomo, C.; Martínez, V.P.; Simoy, M.I.; Gil, J.F. Modeling potential risk areas of Orthohantavirus transmission in Northwestern Argentina using an ecological niche approach. BMC Public Health 2023, 23, 1236. [Google Scholar] [CrossRef]
- Mills, J.N. Regulation of rodent-borne viruses in the natural host: Implications for human disease. In Infectious Diseases from Nature: Mechanisms of Viral Emergence and Persistence; Springer: Vienna, Austria, 2005. [Google Scholar] [CrossRef]
- Lytle, C.D.; Sagripanti, J.-L. Predicted inactivation of viruses of relevance to biodefense by solar radiation. J. Virol. 2005, 79, 14244–14252. [Google Scholar] [CrossRef]
- Blumthaler, M.; Ambach, W.; Ellinger, R. Increase in solar UV radiation with altitude. J. Photochem. Photobiol. B Biol. 1997, 39, 130–134. [Google Scholar] [CrossRef]
- Bohlman, M.C.; Morzunov, S.P.; Meissner, J.; Taylor, M.B.; Ishibashi, K.; Rowe, J.; Levis, S.; Enria, D.; Jeor, S.C.S. Analysis of hantavirus genetic diversity in Argentina: S segment-derived phylogeny. J. Virol. 2002, 76, 3765–3773. [Google Scholar] [CrossRef]
- Cruz, C.D.; Forshey, B.M.; Vallejo, E.; Agudo, R.; Vargas, J.; Blazes, D.L.; Guevara, C.; Laguna-Torres, V.A.; Halsey, E.S.; Kochel, T.J. Novel strain of Andes virus associated with fatal human infection, central Bolivia. Emerg. Infect. Dis. 2012, 18, 750–757. [Google Scholar] [CrossRef] [PubMed]
- Ferro, I.; Lopez, W.; Cassinelli, F.; Aguirre, S.; Cuyckens, G.A.E.; Kehl, S.; Abán-Moreyra, D.; Castillo, P.; Bellomo, C.; Gil, J.; et al. Hantavirus Pulmonary Syndrome Outbreak Anticipation by a Rapid Synchronous Increase in Rodent Abundance in the Northwestern Argentina Endemic Region: Towards an Early Warning System for Disease Based on Climate and Rodent Surveillance Data. Pathogens 2024, 13, 753. [Google Scholar] [CrossRef]
- Camp, J.V.; Spruill-Harrell, B.; Owen, R.D.; Solà-Riera, C.; Williams, E.P.; Eastwood, G.; Sawyer, A.M.; Jonsson, C.B. Mixed Effects of Habitat Degradation and Resources on Hantaviruses in Sympatric Wild Rodent Reservoirs within a Neotropical Forest. Viruses 2021, 13, 85. [Google Scholar] [CrossRef] [PubMed]
- Lowen, A.C. It’s in the mix: Reassortment of segmented viral genomes. PLoS Pathog. 2018, 14, e1007200. [Google Scholar] [CrossRef] [PubMed]
- Holmes, Z.C.; Zhang, Y.-Z. The evolution and emergence of hantaviruses. Curr. Op. Virol. 2015, 10, 27–33. [Google Scholar] [CrossRef]
- Klempa, B. Reassortment events in the evolution of hantaviruses. Virus Genes 2018, 54, 638–646. [Google Scholar] [CrossRef]
- Demirev, A.V.; Lee, S.; Park, S.; Kim, H.; Cho, S.; Lee, K.; Kim, K.; Song, J.-W.; Park, M.-S.; Kim, J.I. Exploring the Genetic Diversity and Molecular Evolution of Seoul and Hantaan Orthohantaviruses. Viruses 2024, 16, 105. [Google Scholar] [CrossRef]
- Kabwe, E.; Shamsutdinov, A.F.; Suleimanova, S.; Martynova, E.V.; Ismagilova, R.K.; Shakirova, V.G.; Savitskaya, T.A.; Isaeva, G.S.; Rizvanov, A.A.; Khaiboullina, S.F.; et al. Puumala Orthohantavirus Reassortant Genome Variants Likely Emerging in the Watershed Forests. Int. J. Mol. Sci. 2023, 24, 1018. [Google Scholar] [CrossRef]
- Longdon, B.; Brockhurst, M.A.; Russell, C.A.; Welch, J.J.; Jiggins, F.M. The Evolution and Genetics of Virus Host Shifts. PLoS Pathog 2014, 10, e1004395. [Google Scholar] [CrossRef]

| S-Segment (1253 bp) Divergence | M-Segment (824 bp) Divergence | L-Segment (1735 bp) Divergence | Virus/Strain | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotides | Aminoacid | Nucleotides | Aminoacid | Nucleotides | Aminoacid | ||||||||
| Acc. No. | (%) | Acc. No. | (%) | Acc. No. | (%) | Acc. No. | (%) | Acc. No. | (%) | Acc. No. | (%) | ||
| ARG-OF.53 | OR890439 | 10.9 | WZP32982 | 1.5 | OR890440 | 13.4 | WZP32983 | 4.8 | UP | - | UP | - | Lechiguanas |
| ER19-SP1693 | OR908896 | 11.5 | WPW61495 | 1.3 | OR987856 | 14.7 | XGD07576 | 5.8 | XGD07567 | 15.1 | XGD07565 | 1.1 | Lechiguanas |
| Oc22531 | AF482713 | 11.8 | AAL82648 | 1.2 | AF028025 | NA | AAB87911 | NA | UP | - | UP | - | Lechiguanas -Bermejo |
| Arg-h484 | OP555730 | 12.8 | WNV26842 | 1 | OP555731 | 16.2 | WNV26843 | 6.8 | OP555736 | 17.7 | WNV26848 | 2 | Buenos Aires |
| Ol22996 | AF482715 | 13.3 | AAL82650 | 2 | AF028024 | 17.5 | AAB87910 | 10 | UP | - | UP | - | Orán |
| FBV554 | JF750419 | 15 | AEO51745 | 2.7 | JF750422 | NA | AEO51748 | NA | UP | - | UP | - | Tunari |
| TK184992 | OR184959 | 16.8 | WNR62054 | 5 | OR184986 | 21.5 | WNR62081 | 11.5 | OR184993 | 19.9 | WNR62163 | 3.9 | Juquitiva |
| ARG-Epuyen | MN258239 | 16.5 | QNQ18256 | 1.2 | MN258205 | 20.6 | QNQ18222 | 11.2 | MN258172 | 19 | QNQ18189 | 2.7 | Andes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bellomo, C.M.; Kehl, S.; Alonso, D.O.; López, W.; Cassinelli, F.; Coelho, R.M.; Bravo, G.; Aguirre, S.; Dib, M.; Periolo, N.; et al. Novel Orthohantavirus Associated with Hantavirus Pulmonary Syndrome in Northern Argentina. Viruses 2025, 17, 717. https://doi.org/10.3390/v17050717
Bellomo CM, Kehl S, Alonso DO, López W, Cassinelli F, Coelho RM, Bravo G, Aguirre S, Dib M, Periolo N, et al. Novel Orthohantavirus Associated with Hantavirus Pulmonary Syndrome in Northern Argentina. Viruses. 2025; 17(5):717. https://doi.org/10.3390/v17050717
Chicago/Turabian StyleBellomo, Carla M., Sebastian Kehl, Daniel Oscar Alonso, Walter López, Flavia Cassinelli, Rocío María Coelho, Gabriela Bravo, Sara Aguirre, Marcela Dib, Natalia Periolo, and et al. 2025. "Novel Orthohantavirus Associated with Hantavirus Pulmonary Syndrome in Northern Argentina" Viruses 17, no. 5: 717. https://doi.org/10.3390/v17050717
APA StyleBellomo, C. M., Kehl, S., Alonso, D. O., López, W., Cassinelli, F., Coelho, R. M., Bravo, G., Aguirre, S., Dib, M., Periolo, N., Toscano, C., Gil, J., García Campos, F., Ferro, I., & Martinez, V. P. (2025). Novel Orthohantavirus Associated with Hantavirus Pulmonary Syndrome in Northern Argentina. Viruses, 17(5), 717. https://doi.org/10.3390/v17050717







