Genomic Diversity of Tomato Brown Rugose Fruit Virus in Canadian Greenhouse Production Systems
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material, RNA Extraction, and Sequencing
2.2. Phylogenetics
2.3. Protein Modeling
3. Results
3.1. Evolutionary Relationships of Canadian ToBRFV Isolates
3.2. Diversity of Individual ToBRFV ORFs
3.3. Genomic Diversity of Canadian ToBRFV Isolates
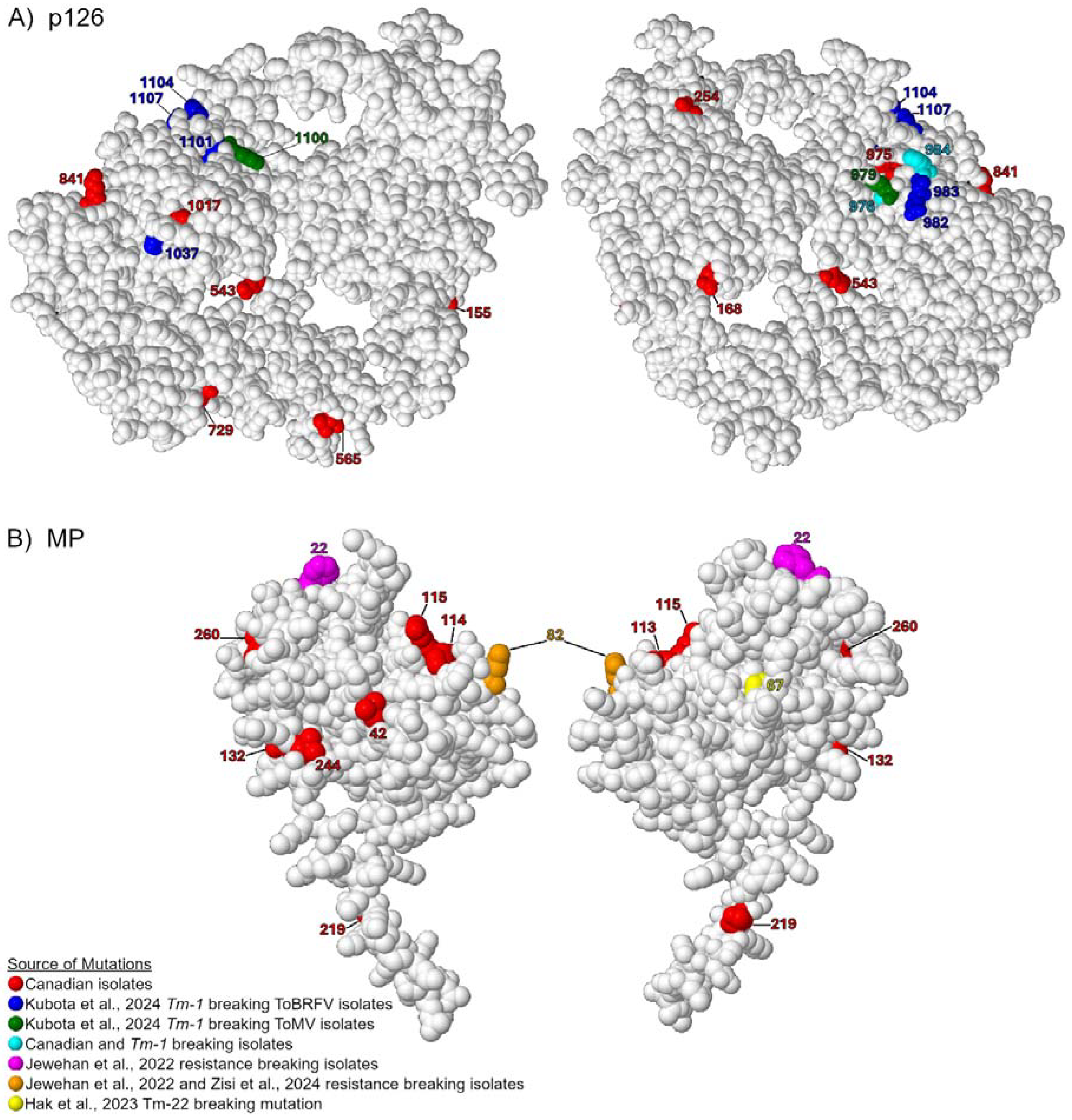
3.4. Comparing Canadian ToBRFV Polymorphisms to Resistance-Breaking Polymorphisms
4. Discussion
4.1. Canadian Sequence Diversity
4.2. Selective Forces on ToBRFV
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salem, N.; Mansour, A.; Ciuffo, M.; Falk, B.W.; Turina, M. A new tobamovirus infecting tomato crops in Jordan. Arch. Virol. 2016, 161, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Luria, N.; Smith, E.; Reingold, V.; Bekelman, I.; Lapidot, M.; Levin, I.; Elad, N.; Tam, Y.; Sela, N.; Abu-Ras, A.; et al. A new Israeli tobamovirus isolate infects tomato plants harouring Tm-22 resistance genes. PLoS ONE 2017, 12, e0170429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Griffiths, J.S.; Marchand, G.; Bernards, M.A.; Wang, A. Tomato brown rugose fruit virus: An emerging and rapidly spreading plant RNA virus that threatens tomato production worldwide. Mol. Plant Pathol. 2022, 23, 1262–1277. [Google Scholar] [CrossRef]
- Salem, N.M.; Jewehan, A.; Aranda, M.A.; Fox, A. Tomato brown rugose fruit virus pandemic. Annu. Rev. Phytopathol. 2023, 5, 137–164. [Google Scholar] [CrossRef]
- Lanfermeijer, F.X.; Dijkhuis, J.; Sturre, M.J.G.; de Haan, P.; Hille, J. Cloning and characterization of the durable tomato mosaic virus resistance gene Tm-2(2) from Lycopersicon esculentum. Plant Mol. Biol. 2003, 52, 1037–1049. [Google Scholar] [CrossRef]
- Yan, Z.Y.; Ma, H.Y.; Wang, L.; Tettey, C.; Zhao, M.S.; Geng, C.; Tian, Y.P.; Li, X.D. Identification of genetic determinants of tomato brown rugose fruit virus that enables infection of plants harbouring the Tm-22 resistance gene. Mol. Plant Pathol. 2021, 22, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Hak, H.; Raanan, H.; Schwarz, S.; Sherman, Y.; Dinesh-Kumar, S.P.; Spiegelman, Z. Activation of Tm-22 resistance is mediated by a conserved cysteine essential for tobacco mosaic virus movement. Mol. Plant Pathol. 2023, 24, 838–848. [Google Scholar] [CrossRef]
- Spiegelman, Z.; Dinesh-Kumar, S.P. Breaking boundaries: The perpetual interplay between tobamoviruses and plant immunity. Annu. Rev. Virol. 2023, 10, 455–476. [Google Scholar] [CrossRef]
- Hamelink, R.; Kalisvaart, J.; Rashidi, H. TBRFV Resistant Tomato Plant. Patent Cooperation Treaty WIPO. 2019, p. 34. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2019110130&tab=PCTBIBLIO&_cid=P10-KPB4P1-64395-1 (accessed on 12 November 2024).
- Ashkenazi, V.; Rotem, Y.; Ecker, R.; Nashilevitz, S.; Barom, N. Resistance in Plants of Solanum lycopersicum to the Tobamovirus Tomato Brown Rugose Fruit Virus. Patent Cooperation Treaty WIPO, 2020, p. 60. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2020249798&tab=SEARCHREPORT (accessed on 12 November 2024).
- Ykema, M.; Verweij, C.W.; De La Bentem, F. Tomato Plant Resistant to Tomato Brown Rugose Fruit Virus. Patent Cooperation Treaty WIPO, 2020 p. 60, 143p. Available online: https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2020147921&tab=PCTBIBLIO (accessed on 12 November 2024).
- Zinger, A.; Lapidot, M.; Harel, A.; Doron-Faigenboim, A.; Gelbart, D.; Levin, I. Identification and mapping of tomato genome loci controlling tolerance and resistance to tomato brown rugose fruit virus. Plants 2021, 10, 179. [Google Scholar] [CrossRef]
- Jewehan, A.; Salem, N.; Toth, Z.; Salomon, P.; Szabo, Z. Screening of Solanum (section Lycopersicon and Juglandifolia) germplasm for reactions to the tomato brown rugose fruit virus (ToBRFV). J. Plant Dis. Prot. 2022, 129, 117–123. [Google Scholar] [CrossRef]
- Kabas, A.; Fidan, H.; Kucukaydin, H.; Atan, H.N. Screening of wild tomato species and interspecific hybrids for resistance/tolerance to Tomato brown rugose fruit virus (ToBRFV). Chil. J. Agric. Res. 2022, 82, 189–196. [Google Scholar] [CrossRef]
- Zisi, Z.; Ghijeselings, L.; Vogel, E.; Vos, C.; Matthijnssens, J. Single amino acid change in tomato brown rugose fruit virus breaks virus-specific resistance in new resistant tomato cultivar. Front. Plant Sci. 2024, 15, 1382862. [Google Scholar] [CrossRef]
- Jaiswal, N.; Chanda, B.; Gilliard, A.; Shi, A.; Ling, K.S. Evaluation of tomato germplasm against tomato brown rugose fruit virus and identification of resistance in Solanum pimpinellifolium. Plants 2024, 13, 581. [Google Scholar] [CrossRef] [PubMed]
- Topcu, Y.; Yildiz, K.; Kayikci, H.C.; Aydin, S.; Feng, Q.; Sapkota, M. Deciphering resistance to tomato brown rugose fruit virus (ToBRFV) using genome-wide association studies. Sci. Hortic. 2025, 341, 113968. [Google Scholar] [CrossRef]
- Conti, G.; Rodriguez, M.C.; Venturuzzi, A.L.; Asurmendi, S. Modulation of host plant immunity by tobamovirus proteins. Ann. Bot. 2017, 119, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Jewehan, A.; Kiemo, F.; Salem, N.; Toth, Z.; Salamon, P.; Szabo, Z. Isolation and molecular characterization of a tomato brown rugose fruit virus mutant breaking the tobamovirus resistance found in wild Solanum species. Arch. Virol. 2022, 167, 1559–1563. [Google Scholar] [CrossRef]
- Ishibashi, K.; Ishikawa, M. Replication of Tobamovirus RNA. Annu. Rev. Phytopathol. 2016, 54, 55–78. [Google Scholar] [CrossRef] [PubMed]
- Vogler, H.; Akbergenov, R.; Shivaprasad, P.V.; Dang, V.; Fasler, M.; Kwon, M.; Zhanybekova, S.; Hohn, T.; Heinlein, M. Modification of small RNAs associated with suppression of RNA silencing by Tobamovirus replicase protein. J. Virol. 2007, 81, 10379–10388. [Google Scholar] [CrossRef]
- Malpica-Lopez, N.; Rajeswaran, R.; Deknazariants, D.; Seguin, J.; Golyaev, V.; Farinelli, L.; Pooggin, M.M. Revisiting the roles of tobamovirus replicase complex proteins in viral replication and silencing suppression. Mol. Plant-Microbe Interact. 2018, 31, 125–144. [Google Scholar] [CrossRef]
- Ishibashi, K.; Kezuka, Y.; Kobayashi, C.; Kato, M.; Inoue, T.; Nonaka, T.; Ishikawa, M.; Matsumura, H.; Katoh, E. Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1. Proc. Natl. Acad. Sci. USA 2014, 111, E3486–E3495. [Google Scholar] [CrossRef]
- Kubota, K.; Takeyama, S.; Matsushita, Y.; Ishibashi, K. Isolation of spontaneous mutants of tomato brown rugose fruit virus that efficiently infects Tm-1 homozygous tomato plants. J. Gen. Plant Pathol. 2024, 90, 187–195. [Google Scholar] [CrossRef]
- Ishibashi, K.; Ishikawa, M. The resistance protein Tm-1 inhibits formation of a tomato mosaic virus replication protein-host membrane protein complex. J. Virol. 2013, 87, 7933–7939. [Google Scholar] [CrossRef]
- Ghorbani, A. Genetic analysis of tomato brown rugose fruit virus reveals evolutionary adaptation and codon usage bias patterns. Sci. Rep. 2024, 14, 21281. [Google Scholar] [CrossRef] [PubMed]
- Statistics Canada. Area, Production and Farm Gate Value of Marketed Vegetables; Table 32-10-0365-01; Statistics Canada: Ottawa, ON, Canada, 2025. [Google Scholar] [CrossRef]
- Nash, D.; Ellmen, I.; Knapp, J.J.; Menon, R.; Overton, A.K.; Cheng, J.; Lynch, M.D.J.; Nissimov, J.I.; Charles, T.C. A novel tiled amplicon sequencing assay targeting the tomato brown rugose fruit virus (ToBRFV) genome reveals widespread distribution in municipal wastewater treatment systems in the province of Ontario, Canada. Viruses 2024, 16, 460. [Google Scholar] [CrossRef] [PubMed]
- Di Rosa, E.; Durand, A.A.; Provost, C.; Constant, P. Epidemiology of tomato brown rugose fruit virus (ToBRFV) in commercial greenhouses. Plant Dis. 2025, 109, 633–637. [Google Scholar] [CrossRef]
- Zinger, A.; Doron-Faigenboim, A.; Gelbart, D.; Levin, I.; Lapidot, M. Contributions of the Tobamovirus resistance gene Tm-1 to control of ToBRFV resistance in tomato. bioRxiv 2025. [Google Scholar] [CrossRef]
- Hak, H.; Spiegelman, Z. The tomato brown rugose fruit virus movement protein overcomes Tm-22 resistance in tomato while attenuating viral transport. Mol. Plant-Microbe Interact. 2021, 34, 990–1092. [Google Scholar] [CrossRef]
- Van Damme, M.; Zois, R.; Verbeek, M.; Bai, Y.; Wolters, A.M.A. Directions from nature: How to halt the tomato brown rugose fruit virus. Agronomy 2023, 13, 1300. [Google Scholar] [CrossRef]
- Fall, M.L.; Xu, D.; Lemoyne, P.; Ben Moussa, I.E.; Beaulieu, C.; Carisse, O. A diverse virome of leafroll-infected grapevine unveiled by dsRNA sequencing. Viruses 2020, 12, 1142. [Google Scholar] [CrossRef]
- Kubaa, R.A.; Amoia, S.S.; Altamura, G.; Minafra, A.; Chiumenti, M.; Cillo, F. Nanopore technology applied to targeted detection of tomato brown rugose fruit virus allows sequencing of related viruses and diagnosis of mixed infections. Plants 2023, 12, 999. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Evans, R.; O’Neill, M.; Pritzel, A.; Antropova, N.; Senior, A.; Green, T.; Hassabis, D. Protein complex prediction with AlphaFold-Multimer. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Hassabis, D. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Mirdita, M.; Schütze, K.; Moriwaki, Y.; Heo, L.; Ovchinnikov, S.; Steinegger, M. ColabFold: Making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. [Google Scholar] [CrossRef]
- Rochsar, E.; Torgeman, S.; Bandel, K.; Koren, A.; Klap, C.; Dombrovsky, A.; Zamir, D. Tissue specific resistance and susceptibility to the tomato brown rugose fruit virus (ToBRFV) conferred by Solanum pennellii loci. BMC Plant Biol. 2025, 25, 51. [Google Scholar] [CrossRef]
- de Koning, P.P.M.; Fowkes, A.R.; Zisi, Z.; Beris, D.; Oplaat, C.; McGreig, S.; Skelton, A.; Adams, I.P.; van Gemert, J.; de Krom, C.; et al. Tomato brown rugose fruit virus nextstrain build version 4: Pathways of introduction and local spread. PhytoFrontiers 2025, 5, 2690–5442. [Google Scholar] [CrossRef]
- Meshi, T.; Motoyoshi, F.; Adachi, A.; Watanabe, Y.; Takamatsu, N.; Okada, Y. Two concomitant base substitutions in the putative replicase genes of tobacco mosaic virus confer the ability to overcome the effects of a tomato resistance gene, Tm-1. EMBO J. 1988, 7, 1575–1581. [Google Scholar] [CrossRef]
- Strasser, M.; Pfitzner, A.J.P. The double-resistance-breaking Tomao mosaic virus strain ToMV1-2 contains two independent single resistance-breaking domains. Arch. Virol. 2007, 152, 903–914. [Google Scholar] [CrossRef]
- Paudel, D.B.; Sanfacon, H. Exploring the diversity of mechanisms associated with plant tolerance to virus infection. Front. Plant Sci. 2018, 9, 1575. [Google Scholar] [CrossRef]
- Hughes, A.L.; Hughes, M.A.K. More effective purifying selection on RNA viruses than in DNA viruses. Gene 2009, 20, 117–125. [Google Scholar] [CrossRef]
- Abrahamian, P.; Cai, W.; Nunziata, S.O.; Ling, K.S.; Jaiswal, N.; Mavrodieva, V.A.; Rivera, Y.; Nakhla, M.K. Comparative analysis of tomato brown rugose fruit virus isolates shows limited genetic diversity. Viruses 2022, 14, 2816. [Google Scholar] [CrossRef] [PubMed]
- Carr, J.P. Engineered resistance to Tobamoviruses. Viruses 2024, 16, 1007. [Google Scholar] [CrossRef] [PubMed]
- Fraser, R.S.S.; Loughlin, S.A.R. Effects of temperature on the Tm-1 gene for resistance to tobacco mosaic virus in tomato. Physiol. Plant Pathol. 1982, 20, 109–117. [Google Scholar] [CrossRef]
- Ouyang, S.; Park, G.; Atamian, H.S.; Han, C.S.; Stajich, J.E.; Kaloshian, I.; Borkovich, K.A. MicroRNAs suppress NB domain genes in tomato that confer resistance to Fusarium oxysporum. PLoS Pathog. 2014, 10, e1004464. [Google Scholar] [CrossRef]
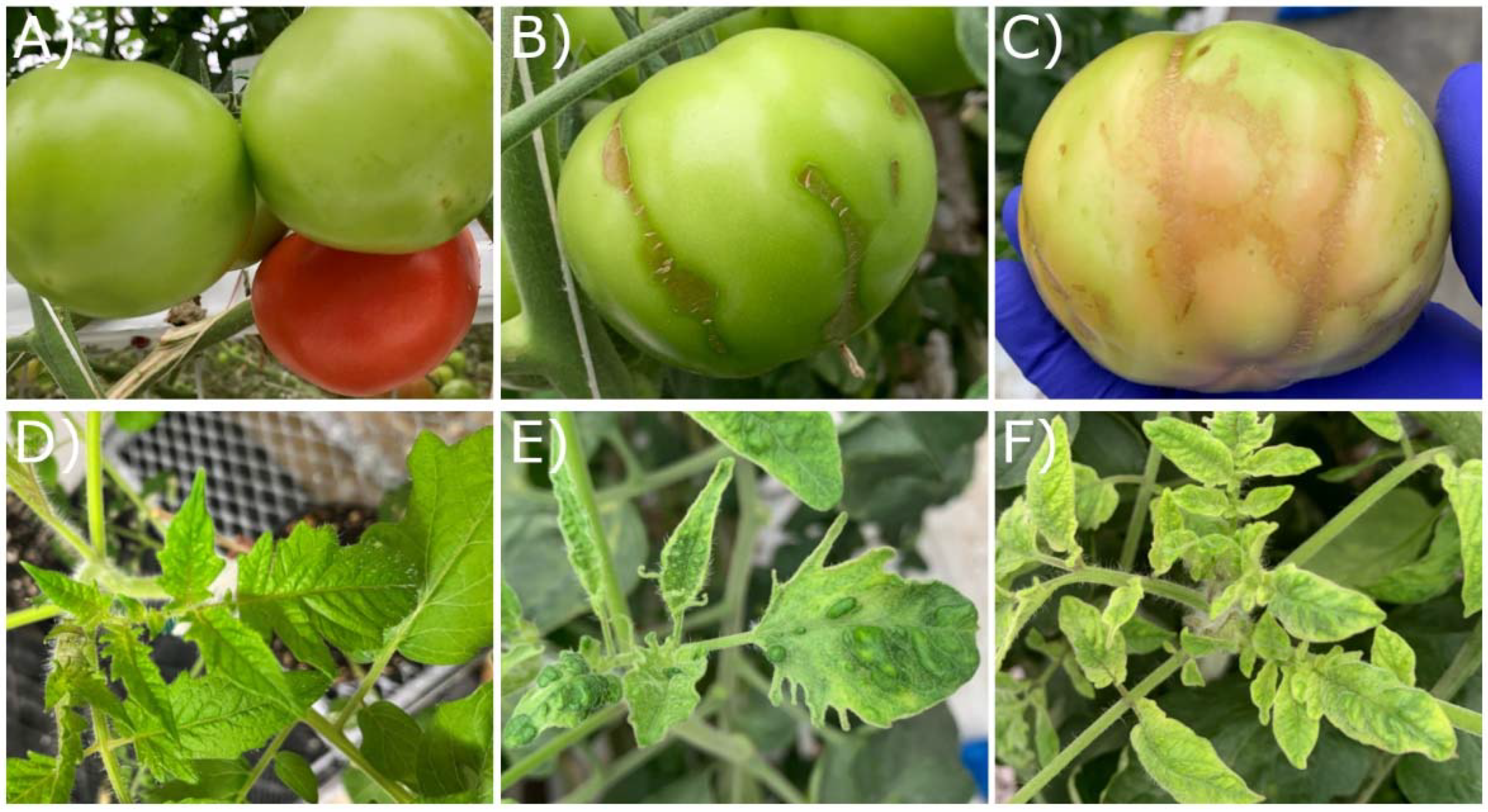
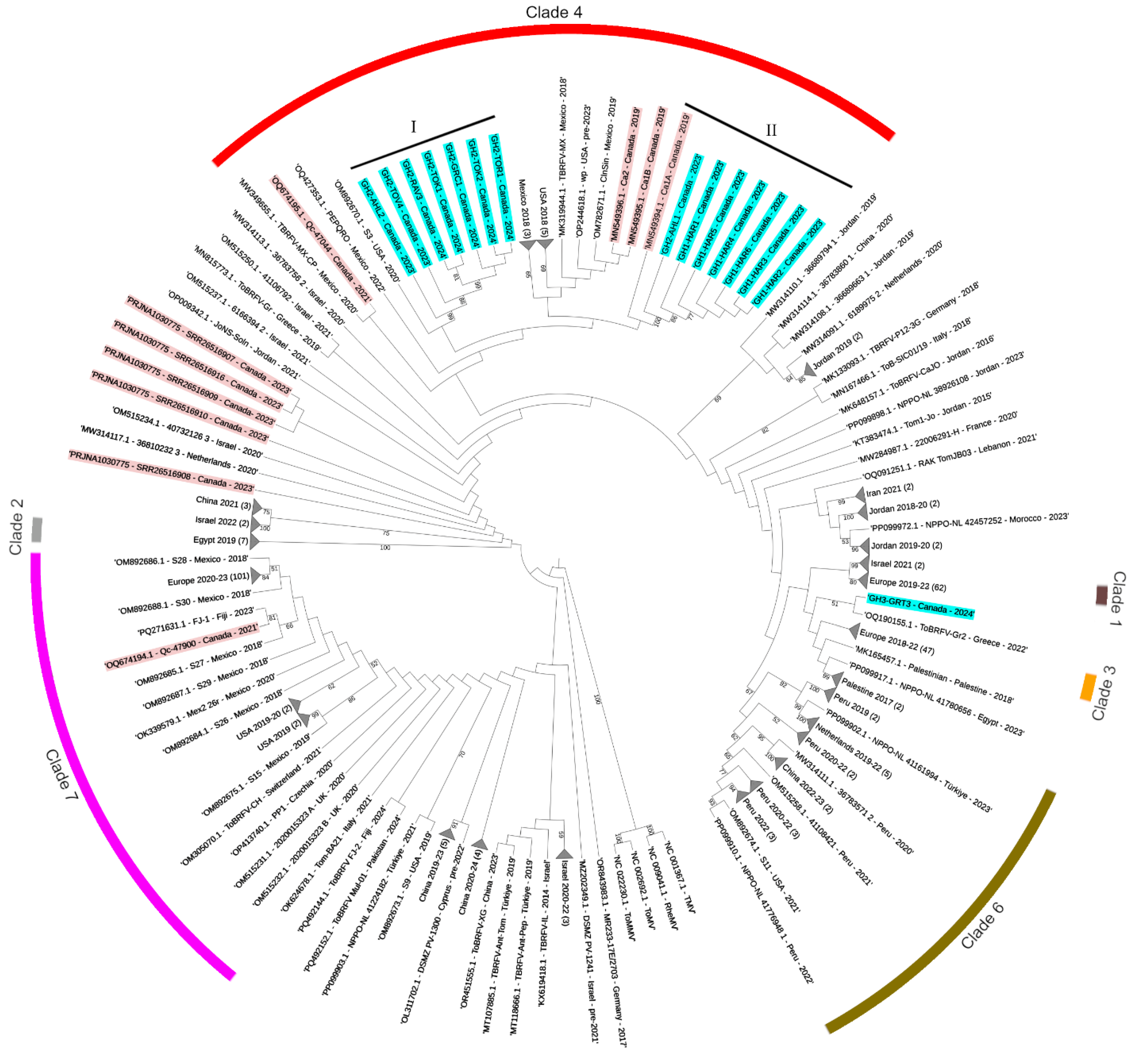
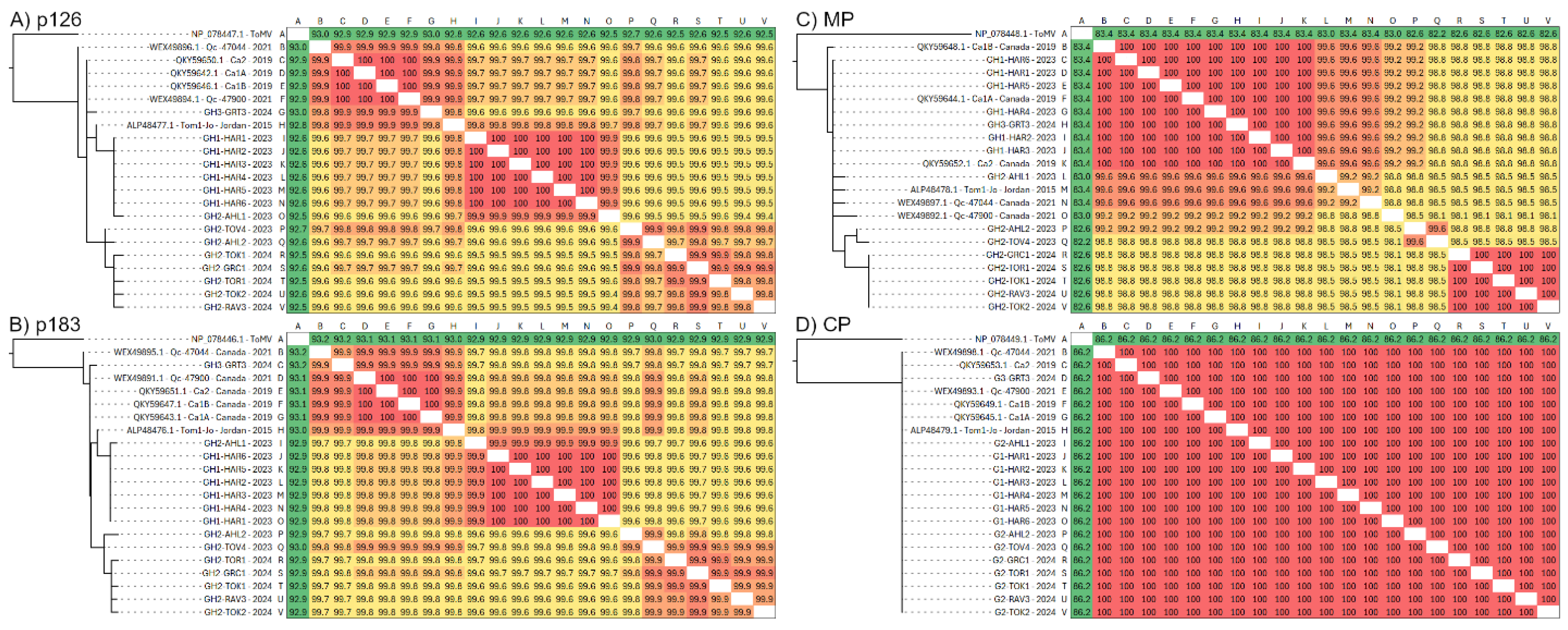
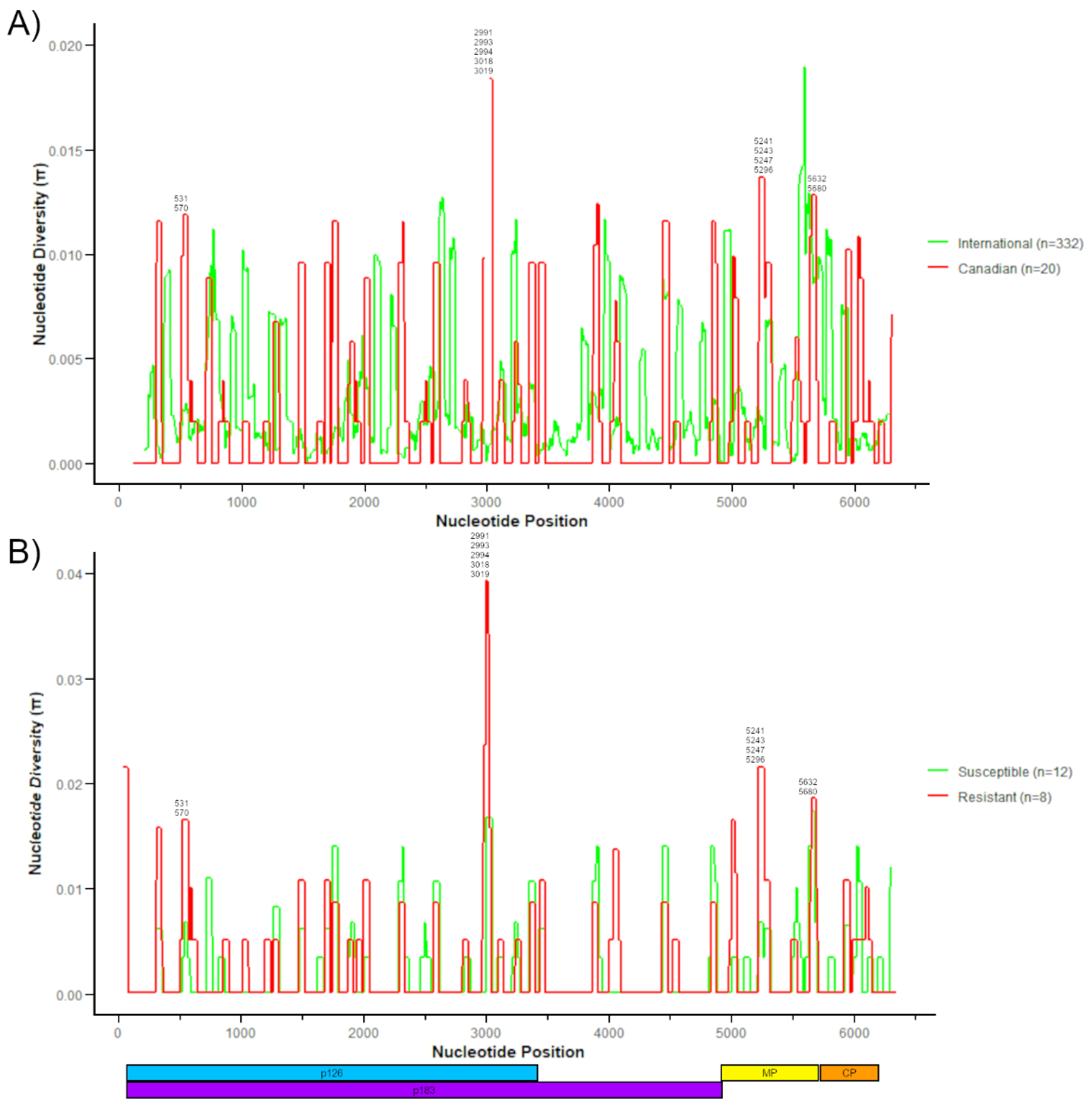
| Sample ID | Greenhouse | Unknown Variety * | Labeled Resistant | Tm-1 Allele Detection | Sampling Date | RT-PCR Detection | qPCR (ng/µL) |
|---|---|---|---|---|---|---|---|
| G1-HAR1 | 1 | 1 | Yes | Tm-1/tm-1 | 27 September 2023 | + | 2.52 × 109 |
| G1-HAR2 | 1 | 2 | No | tm-1/tm-1 | 27 September 2023 | + | 1.39 × 1010 |
| G1-HAR3 | 1 | 2 | No | tm-1/tm-1 | 27 September 2023 | + | 1.07 × 108 |
| G1-HAR4 | 1 | 2 | No | tm-1/tm-1 | 27 September 2023 | + | 3.76 × 106 |
| G1-HAR5 | 1 | 2 | No | tm-1/tm-1 | 27 September 2023 | + | 3.06 × 108 |
| G1-HAR6 | 1 | 2 | No | tm-1/tm-1 | 27 September 2023 | + | 8.09 × 108 |
| G2-AHL1 | 2 | 3 | Yes | tm-1/tm-1 | 27 June 2023 | − | 3.26 × 103 |
| G2-AHL2 | 2 | 4 | No | Tm-1/tm-1 | 27 June 2023 | − | 1.16 × 102 |
| G2-TOV4 | 2 | 3 | Yes | tm-1/tm-1 | 27 September 2023 | + | 1.45 × 108 |
| G2-RAV3 | 2 | 5 | Yes | tm-1/tm-1 | 24 July 2024 | + | 9.64 × 108 |
| G2-GRC1 | 2 | 4 | No | tm-1/tm-1 | 5 March 2024 | + | 5.36 × 106 |
| G2-TOR1 | 2 | 6 | Yes | Tm-1/tm-1 | 20 November 2024 | + | 4.79 × 106 |
| G2-TOK1 | 2 | 7 | Yes | Tm-1/Tm-1 | 20 November 2024 | + | 2.80 × 108 |
| G2-TOK2 | 2 | 7 | Yes | Tm-1/tm-1 | 20 November 2024 | + | 1.92 × 107 |
| G3-GRT3 | 3 | 8 | No | tm-1/tm-1 | 27 August 2024 | + | 2.84 × 109 |
| Qc-47044 | 4 | 9 | No | Unknown | 21 May 2021 | + | NT |
| Qc-47900 | 4 | 10 | No | Unknown | 21 May 2021 | + | NT |
| Sample ID | Total Reads | ToBRFV Reads | Average Depth | Average Read Length | %Coverage | Genbank Accession Number |
|---|---|---|---|---|---|---|
| G1-HAR1 | 1,793,953 | 6806 | 459.1 | 375.9 | 100 | PV446608.1 |
| G1-HAR2 | 740,414 | 4067 | 455.9 | 568.7 | 100 | PV446609.1 |
| G1-HAR3 | 239,752 | 4209 | 328.9 | 350.5 | 100 | PV446610.1 |
| G1-HAR4 | 114,079 | 4388 | 58.3 | 178.5 | 100 | PV446611.1 |
| G1-HAR5 | 101,184 | 3866 | 306.7 | 353.0 | 100 | PV446612.1 |
| G1-HAR6 | 66,635 | 4840 | 425.5 | 413.7 | 100 | PV446613.1 |
| G2-TOV4 | 659,086 | 5425 | 432.6 | 414.7 | 100 | PV446618.1 |
| G2-RAV3 | 67,071 | 3200 | 40.8 | 110.1 | 100 | PV446614.1 |
| G2-GRC1 | 197,776 | 4565 | 329.3 | 372.0 | 100 | PV446606.1 |
| G2-GRT3 | 193,122 | 5366 | 396.3 | 367.1 | 100 | PV446607.1 |
| G2-AHL1 | 530,979 | 3841 | 15.9 | 81.2 | 100 | PV446604.1 |
| G2-AHL2 | 393,960 | 4304 | 135.2 | 239.2 | 100 | PV446605.1 |
| G2-TOR1 | 554,332 | 89 | 4.1 | 568.8 | 100 | PV446617.1 |
| G2-TOK1 | 223,423 | 287 | 23.5 | 712.5 | 100 | PV446615.1 |
| G2-TOK2 | 455,294 | 704 | 41.6 | 611.9 | 100 | PV446616.1 |
| Qc-47044 | 85,001 | 1264 | 27.3 | 138.1 | 97.7 | OQ674195.1 |
| Qc-47900 | 27,868 | 17,307 | 420 | 155.1 | 100 | OQ674194.1 |
| ORF | n | Size (bp) | S | Non-Syn | k | h | π | FLD | FLF | TD | dN/dS | %ID |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p126 | 20 | 3351 | 40 | 13 | 9.06 | 17 | 0.003 | −1.85 | −1.84 | −0.94 | 0.20 | 99.5–100 |
| p183 | 4848 | 51 | 14 | 11.8 | 17 | 0.002 | −1.79 | −1.61 | −0.85 | 0.14 | 99.4–100 | |
| MP | 801 | 15 | 8 | 2.90 | 10 | 0.004 | −1.89 | −1.95 | −1.16 | 0.59 | 99.3–100 | |
| CP | 480 | 6 | 0 | 1.35 | 7 | 0.003 | −1.83 | −1.85 | −1.03 | N/A | 99.4–100 | |
| Full | 6374 | 76 | 22 | 16.3 | 18 | 0.003 | −2.02 | −2.00 | −1.02 | 0.16 | 99.5–100 |
| NT Pos. | Ref. NT | Mut. NT | AA Pos. | Ref. AA | Mut. AA | ORF | Isolates | Canadian Isolates | All Isolates |
|---|---|---|---|---|---|---|---|---|---|
| 531 | G | A | 155 | G | S | p126/p183 | G2-RAV3 1, G2-GRC1, G2-TOK1 1, G2-TOK2 1, G2-TOR1 1 | 5 | 5 |
| 570 | G | A | 168 | D | N | p126/p183 | G2-RAV3 1 | 1 | 1 |
| 829 | A | T | 254 | D | V | p126/p183 | G2-AHL2 | 1 | 1 |
| 1696 | T | A | 543 | V | E | p126/p183 | G2-TOV4 1, G2-RAV3 1, G2-AHL2, G2-GRC1, G2-TOK1 1, G2-TOK2 1, G2-TOR1 1 | 7 | 7 |
| 1761 | G | A | 565 | V | M | p126/p183 | G1-HAR1 1, G1-HAR2-6, G2-AHL1 1 | 7 | 8 |
| 2338 | T | C | 729 | L | S | p126/p183 | 47044 | 1 | 5 |
| 2590 | A | G | 841 | K | R | p126/p183 | G1-HAR1 1, G1-HAR2-6, G2-AHL1 1 | 7 | 7 |
| 2991 | C | T | 975 | H | Y | p126/p183 | G2-AHL1 1, G2-TOR1 1 | 2 | 2 |
| 2993 | C | A | 975 | H | Q | p126/p183 | G2-TOK1 1 | 1 | 1 |
| 2994 | T | C | 976 | Y | H | p126/p183 | G2-TOK2 1 | 1 | 3 |
| 3018 | C | T | 984 | H | Y | p126/p183 | G1-HAR1 1, G1-HAR2-6, G2-TOV4 1, G2-RAV3 1, G2-AHL1 1, G2-AHL2, G2-GRC1, G2-TOK1 1, G2-TOK2 1, G2-TOR1 1 | 14 | 32 |
| 3019 | A | T | 984 | H | F | p126/p183 | G2-TOV4 1, G2-RAV3 1, G2-AHL2, G2-GRC1, G2-TOK1 1, G2-TOK2 1, G2-TOR1 1 | 7 | 7 |
| 3117 | G | A | 1017 | V | I | p126/p183 | G2-GRT3 1 | 1 | 2 |
| 3924 | G | A | 1286 | A | T | p183 | G2-AHL2 | 1 | 1 |
| 5027 | T | C | 42 | L | S | MP | G2-RAV3 1, G2-GRC1, G2-TOK1 1, G2-TOK2 1, G2-TOR1 1 | 5 | 5 |
| 5241 | G | T | 113 | K | N | MP | G2-TOV4 1, G2-AHL2 | 2 | 2 |
| 5243 | A | G | 114 | K | R | MP | G2-TOV4 1 | 1 | 1 |
| 5247 | G | T | 115 | R | S | MP | G2-RAV3 1, G2-GRC1, G2-TOK1 1, G2-TOK2 1, G2-TOR1 1 | 5 | 5 |
| 5296 | G | A | 132 | E | K | MP | G2-TOV4 1, G2-RAV3 1, G2-AHL2, G2-GRC1, G2-TOK1 1, G2-TOK2 1, G2-TOR1 1 | 7 | 36 |
| 5558 | G | A | 219 | N | S | MP | 47044 | 1 | 1 |
| 5632 | G | A | 244 | V | I | MP | Ca1B | 1 | 7 |
| 5680 | G | A | 260 | V | I | MP | 47900 | 1 | 119 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fougere, G.C.; Xu, D.; Gaiero, J.R.; McCreary, C.; Marchand, G.; Despres, C.; Wang, A.; Fall, M.L.; Griffiths, J.S. Genomic Diversity of Tomato Brown Rugose Fruit Virus in Canadian Greenhouse Production Systems. Viruses 2025, 17, 696. https://doi.org/10.3390/v17050696
Fougere GC, Xu D, Gaiero JR, McCreary C, Marchand G, Despres C, Wang A, Fall ML, Griffiths JS. Genomic Diversity of Tomato Brown Rugose Fruit Virus in Canadian Greenhouse Production Systems. Viruses. 2025; 17(5):696. https://doi.org/10.3390/v17050696
Chicago/Turabian StyleFougere, Gregory C., Dong Xu, Jonathan R. Gaiero, Cara McCreary, Geneviève Marchand, Charles Despres, Aiming Wang, Mamadou Lamine Fall, and Jonathan S. Griffiths. 2025. "Genomic Diversity of Tomato Brown Rugose Fruit Virus in Canadian Greenhouse Production Systems" Viruses 17, no. 5: 696. https://doi.org/10.3390/v17050696
APA StyleFougere, G. C., Xu, D., Gaiero, J. R., McCreary, C., Marchand, G., Despres, C., Wang, A., Fall, M. L., & Griffiths, J. S. (2025). Genomic Diversity of Tomato Brown Rugose Fruit Virus in Canadian Greenhouse Production Systems. Viruses, 17(5), 696. https://doi.org/10.3390/v17050696





