Development of a Multiplex TaqMan Assay for Rapid Detection of Groundnut Bud Necrosis Virus: A Quarantine Pathogen in the USA
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Infected Plant Materials
2.2. Total Plant RNA Extraction and RNA Assessment
2.3. Primer Design for Conventional RT-PCR and Primers and Probes for TaqMan RT-qPCR Assays
2.4. Conventional RT-PCR and Sanger Sequencing
2.5. TaqMan Reverse Transcription-Quantitative PCR (RT-qPCR)
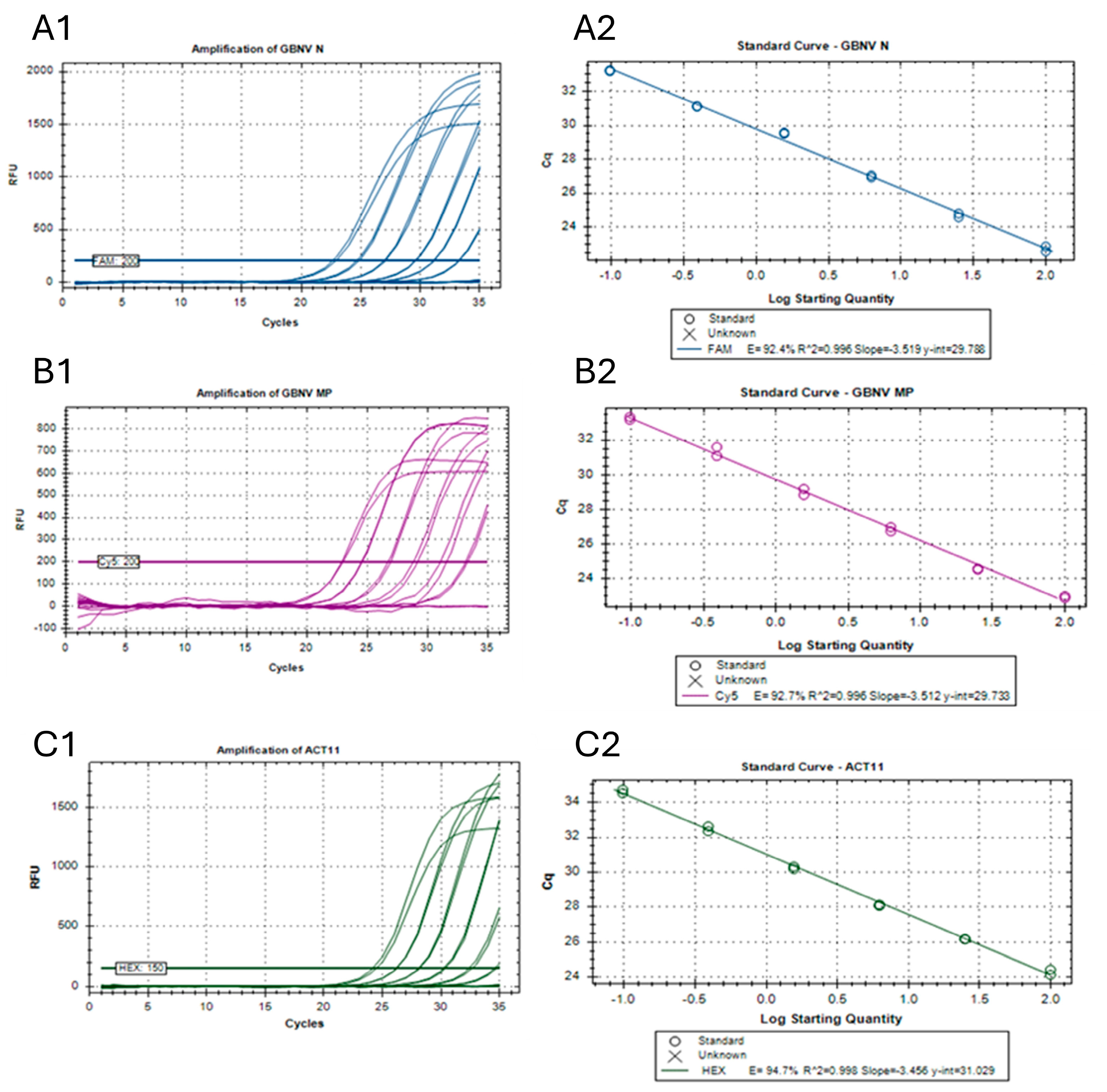
2.6. Standard Curves
2.7. External Validation
3. Results
3.1. In Silico Analysis for Primer and Probe Selection
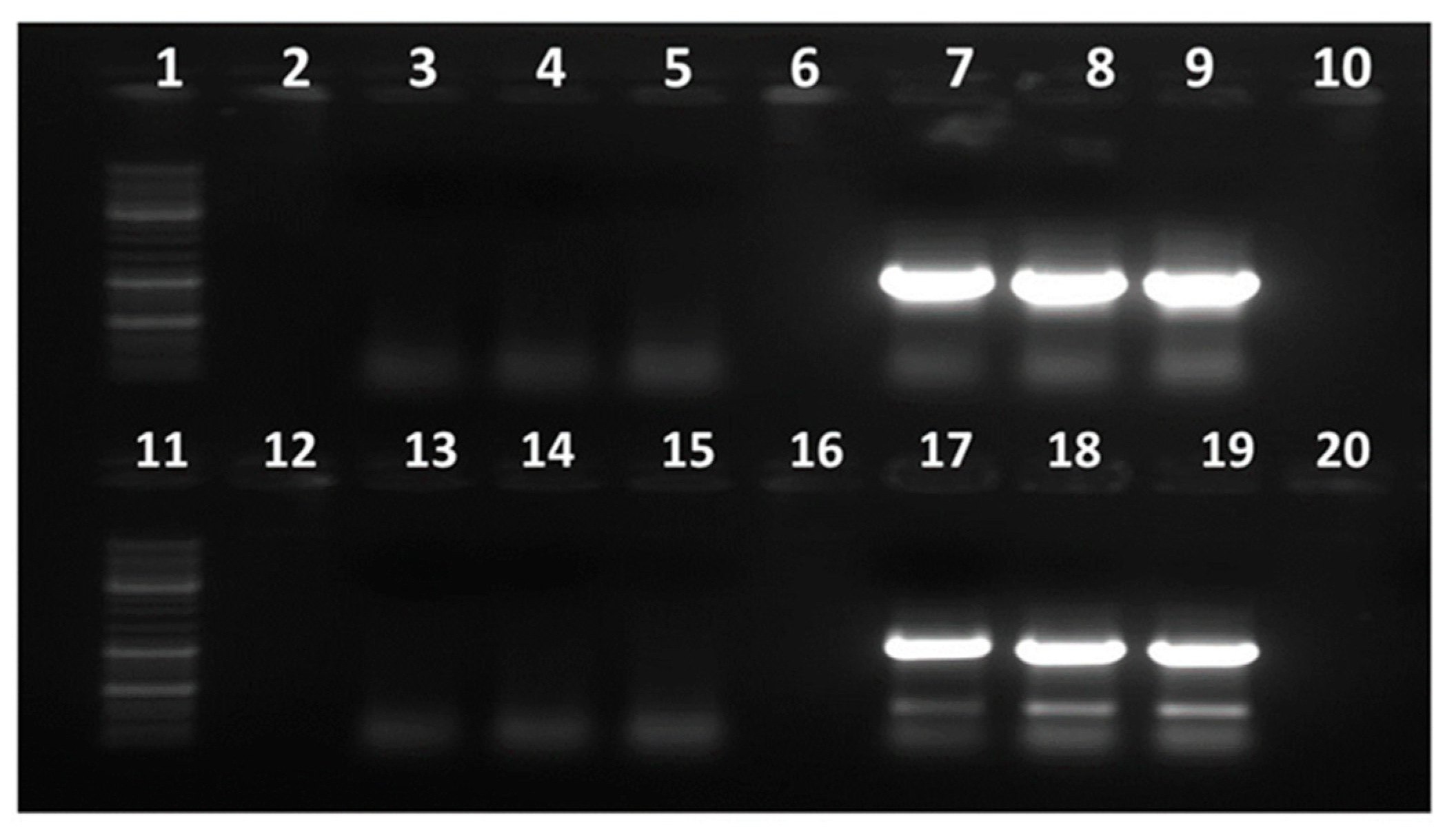
3.2. Development of Conventional RT-PCR Assay for Detection of GBNV
3.3. Development of Singleplex TaqMan RT-qPCR Assay
3.4. Efficiencies, Sensitivities, and Detection Limits of the Singleplex Reactions
3.5. Development of TaqMan-Based Multiplex RT-qPCR
3.6. Comparisons of Efficiency of Singleplex and Multiplex RT-qPCR Assays
3.7. Specificity of the TaqMan Assay Developed for GBNV
3.8. External Validation of the Assay
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reddy, M.; Reddy, D.V.R.; Appa Rao, A. A new record of virus disease on peanut. Plant Dis. Rep. 1968, 52, 494–495. [Google Scholar]
- Ghanekar, A.M.; Reddy, D.V.R.; Iizuka, N.; Amin, P.W.; Gibbons, R.W. Bud necrosis of groundnut (Arachis hypogaea) in India caused by tomato spotted wilt virus. Ann. Appl. Biol. 1979, 93, 173–179. [Google Scholar] [CrossRef]
- Reddy, D.V.R.; Wightman, J.A.; Beshear, R.J.; Highland, B.H.; Black, M.; Sreenivasulu, P.; Dwivedi, S.L.; Demski, J.W.; Mcdonald, D.; Smith, J.J.W.; et al. Bud Necrosis: A Disease of Groundnut Caused by Tomato Spotted Wilt Virus; 31; ICRISAT: Patancheru, India, 1991; Available online: https://oar.icrisat.org/id/eprint/1052 (accessed on 6 February 2025).
- Bhat, A.I.; Jain, R.K.; Varma, A.; Lal, S.K. Nucleocapsid protein gene sequence studies suggest that soybean bud blight is caused by a strain of groundnut bud necrosis virus. Curr. Sci. 2002, 82, 1389–1392. [Google Scholar]
- Gowda, S.; Tatineni, S.; Rayapati, N.; Mushegian, A.; Dawson, W.; Reddy, D. Characterization of the large (L) RNA of peanut bud necrosis tospovirus. Arch. Virol. 1998, 143, 2381–2390. [Google Scholar] [CrossRef] [PubMed]
- Satyanarayana, T.; Mitchell, S.E.; Reddy, D.V.; Brown, S.; Kresovich, S.; Jarret, R.; Naidu, R.A.; Demski, J.W. Peanut bud necrosis tospovirus S RNA: Complete nucleotide sequence, genome organization and homology to other tospoviruses. Arch. Virol. 1996, 141, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Mandal, B.; Jain, R.K.; Krishnareddy, M.; Krishna Kumar, N.K.; Ravi, K.S.; Pappu, H.R. Emerging problems of Tospoviruses (Bunyaviridae) and their management in the Indian subcontinent. Plant Dis. 2012, 96, 468–479. [Google Scholar] [CrossRef]
- Sujitha, A.; Bhaskara Reddy, B.V.; Sivaprasad, Y.; Usha, R.; Sai Gopal, D.V.R. First report of groundnut bud necrosis virus infecting onion (Allium cepa). Australas. Plant Dis. Notes 2012, 7, 183–187. [Google Scholar] [CrossRef][Green Version]
- Ansar, M.; Akram, M.; Singh, R.; Pundhir, V. Epidemiological studies of stem necrosis disease in potato caused by groundnut bud necrosis virus. Indian Phytopathol. 2015, 68, 321–325. [Google Scholar]
- Sastry, K.S. Impact of virus and viroid diseases on crop yields. In Plant Virus and Viroid Diseases in the Tropics; Springer: New York, NY, USA, 2013; Volume 1, pp. 99–160. [Google Scholar] [CrossRef]
- Reddy, D.; Buiel, A.; Tatineni, S.; Dwivedi, S.; Reddy, A.; Ratna, S.; Vijayalakshmi, K.; Rao, G.; Rayapati, N.; Wightman, J. Peanut bud necrosis disease: An overview. In Recent Studies on Peanut Bed Necrosis Disease; ICRISAT: Andhra Pradesh, India, 1995; pp. 3–7. Available online: https://oar.icrisat.org/2548/ (accessed on 6 February 2025).
- Vemana, K.; Jain, R.K. Biological similarities and differences between tobacco streak virus and groundnut bud necrosis virus infecting groundnut (Arachis hypogea). Indian Phytopathol. 2012, 65, 177–183. [Google Scholar]
- Reitz, S.; Gao, Y.; Lei, Z. Thrips: Pests of concern to China and the United States. Agric. Sci. China 2011, 10, 867–892. [Google Scholar] [CrossRef]
- Golnaraghi, A.R.; Pourrahim, R.; Shahraeen, N.; Farzadfar, S. First report of groundnut bud necrosis virus in Iran. Plant Dis. 2002, 86, 561. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.E.; Whitfield, A.E. The genus Tospovirus: Emerging Bunyaviruses that threaten food security. Annu. Rev. Virol. 2016, 3, 101–124. [Google Scholar] [CrossRef] [PubMed]
- Reddy, D.V.R.; Ratna, A.S.; Sudarshana, M.R.; Poul, F.; Kumar, I.K. Serological relationships and purification of bud necrosis virus, a tospovirus occurring in peanut (Arachis hypogaea L.) in India. Ann. Appl. Biol. 1992, 120, 279–286. [Google Scholar] [CrossRef]
- Saritha, R.K.; Jain, R.K. Nucleotide sequence of the S and M RNA segments of a groundnut bud necrosis virus isolate from Vigna radiata in India. Arch. Virol. 2007, 152, 1195–1200. [Google Scholar] [CrossRef]
- Akram, M.; Kamaal, N. Biological characterization and variability of the nucleocapsid protein gene of groundnut bud necrosis virus isolates infecting pea from India. Phytopathol. Mediterr. 2012, 51, 266–275. [Google Scholar]
- Balol, G.; Patil, M. Serological and molecular detection of groundnut bud necrosis virus (GBNV) causing bud blight disease in tomato. Int. J. Plant Prot. 2013, 6, 320–322. [Google Scholar]
- Singh, A.; Permar, V.; Basavaraj; Tomar, B.S.; Praveen, S. Effect of temperature on symptoms expression and viral RNA accumulation in groundnut bud necrosis virus infected Vigna unguiculata. Iran J. Biotech. 2018, 16, 227–234. [Google Scholar] [CrossRef]
- Goswami, S.; Sahana, N.; Pandey, V.; Doblas, P.; Jain, R.K.; Palukaitis, P.; Canto, T.; Praveen, S. Interference in plant defense and development by non-structural protein NSs of groundnut bud necrosis virus. Virus Res. 2012, 163, 368–373. [Google Scholar] [CrossRef]
- Daimei, G.; Raina, H.S.; Devi, P.P.; Saurav, G.K.; Renukadevi, P.; Malathi, V.G.; Senthilraja, C.; Mandal, B.; Rajagopal, R. Influence of groundnut bud necrosis virus on the life history traits and feeding preference of its vector, Thrips Palmi. Phytopathology 2017, 107, 1440–1445. [Google Scholar] [CrossRef]
- Raigond, B.; Pathania, S.; Verma, G.; Bhardwaj, P.; Kochhar, T.; Chakrabarti, S.K. Development and application of reverse transcription-loop mediated isothermal amplification assay for sensitive detection of groundnut bud necrosis virus infecting potato. Virology 2023, 587, 109872. [Google Scholar] [CrossRef]
- Gorbet, D.W.; Knauft, D.A. Registration of ‘SunOleic 97R’ peanut. Crop Sci. 2000, 40, 1190–1191. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Reddy, D.S.; Bhatnagar-Mathur, P.; Cindhuri, K.S.; Sharma, K.K. Evaluation and validation of reference genes for normalization of quantitative real-time PCR based gene expression studies in peanut. PLoS ONE 2013, 8, e78555. [Google Scholar] [CrossRef]
- Wang, X.; Guo, S.; Hameed, M.; Zhang, J.; Pang, L.; Li, B.; Qiu, Y.; Liu, K.; Shao, D.; Ma, Z.; et al. Rapid differential detection of genotype I and III Japanese encephalitis virus from clinical samples by a novel duplex TaqMan probe-based RT-qPCR assay. J. Virol. Methods 2020, 279, 113841. [Google Scholar] [CrossRef]
- Huang, J.L.; Lin, H.T.; Wang, Y.M.; Weng, M.H.; Ji, D.D.; Kuo, M.D.; Liu, H.W.; Lin, C.S. Sensitive and specific detection of strains of Japanese encephalitis virus using a one-step TaqMan RT-PCR technique. J. Med. Virol. 2004, 74, 589–596. [Google Scholar] [CrossRef]
- Saettler, A.W.; Schaad, N.W.; Roth, D.A. Detection of Bacteria in Seed and Other Planting Material; APS Press: St. Paul, MN, USA, 1989. [Google Scholar]
- Schaad, N.W.; Frederick, R.D.; Shaw, J.; Schneider, W.L.; Hickson, R.; Petrillo, M.D.; Luster, D.G. Advances in molecular-based diagnostics in meeting crop biosecurity and phytosanitary issues. Annu. Rev. Phytopathol. 2003, 41, 305–324. [Google Scholar] [CrossRef]
- Eun, A.J.; Seoh, M.; Wong, S. Simultaneous quantitation of two orchid viruses by the TaqMan real-time RT-PCR. J. Virol. Methods 2000, 87, 151–160. [Google Scholar] [CrossRef]
- Boonham, N.; Kreuze, J.; Winter, S.; van der Vlugt, R.; Bergervoet, J.; Tomlinson, J.; Mumford, R. Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Res. 2014, 186, 20–31. [Google Scholar] [CrossRef]
- Boonham, N.; Smith, P.; Walsh, K.; Tame, J.; Morris, J.; Spence, N.; Bennison, J.; Barker, I. The detection of tomato spotted wilt virus (TSWV) in individual thrips using real time fluorescent RT-PCR (TaqMan). J. Virol. Methods 2002, 101, 37–48. [Google Scholar] [CrossRef]
- Korimbocus, J.; Coates, D.; Barker, I.; Boonham, N. Improved detection of sugarcane yellow leaf virus using a real-time fluorescent (TaqMan) RT-PCR assay. J. Virol. Methods 2002, 103, 109–120. [Google Scholar] [CrossRef]
- Boonham, N.; Walsh, K.; Preston, S.; North, J.; Smith, P.; Barker, I. The detection of tuber necrotic isolates of potato virus Y, and the accurate discrimination of PVY(O), PVY(N) and PVY(C) strains using RT-PCR. J. Virol. Methods 2002, 102, 103–112. [Google Scholar] [CrossRef]
- Acinas, S.G.; Sarma-Rupavtarm, R.; Klepac-Ceraj, V.; Polz, M.F. PCR-induced sequence artifacts and bias: Insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 2005, 71, 8966–8969. [Google Scholar] [CrossRef]
- Tremblay, J.; Singh, K.; Fern, A.; Kirton, E.S.; He, S.; Woyke, T.; Lee, J.; Chen, F.; Dangl, J.L.; Tringe, S.G. primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol. 2015, 6, 771. [Google Scholar] [CrossRef]
- Fouhy, F.; Clooney, A.G.; Stanton, C.; Claesson, M.J.; Cotter, P.D. 16S rRNA gene sequencing of mock microbial populations- impact of DNA extraction method, primer choice and sequencing platform. BMC Microbio. 2016, 16, 123. [Google Scholar] [CrossRef]
- Fuks, G.; Elgart, M.; Amir, A.; Zeisel, A.; Turnbaugh, P.J.; Soen, Y.; Shental, N. Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome 2018, 6, 17. [Google Scholar] [CrossRef]
- Ding, S.W.; Howe, J.; Keese, P.; Mackenzie, A.; Meek, D.; Osorio-Keese, M.; Skotnicki, M.; Srifah, P.; Torronen, M.; Gibbs, A. The tymobox, a sequence shared by most Tymoviruses: Its use in molecular studies of Tymoviruses. Nucleic Acids Res. 1990, 18, 1181–1187. [Google Scholar] [CrossRef]
- Martin, R.R.; Constable, F.; Tzanetakis, I.E. Quarantine regulations and the impact of modern detection methods. Annu. Rev. Phytopathol. 2016, 54, 189–205. [Google Scholar] [CrossRef]
- Webster, C.G.; Reitz, S.R.; Perry, K.L.; Adkins, S. A natural M RNA reassortant arising from two species of plant- and insect-infecting bunyaviruses and comparison of its sequence and biological properties to parental species. Virology 2011, 413, 216–225. [Google Scholar] [CrossRef]

| RT-PCR Assay | Primer Name | Target | 5′-3′ Sequence | Tm (C) | Amplicon Size (bp) | Reference |
|---|---|---|---|---|---|---|
| Conventional | SB226F | GBNV nucleocapsid protein (NP) | CAAGGACTTTCTGTGTTCC | 52 | 1046 | This study |
| SB226R | AAGATTGCCTCTTTCGAGGTC | |||||
| SB227F | GBNV movement protein (MP) | GAAATAATGTCTCGCTTTTCTAA | 43 | 1119 | This study | |
| SB227R | TTTCAAGAAGATTATCCATCTC | |||||
| TaqMan | GBNV-NF | GBNV NP | TTCCTAATTTCTCTTTCTTCACA | 53 | 137 | This study |
| GBNV-NR | ATCTTTCGATACATGTGCTTTAA | |||||
| GBNV_N | 6-FAM AGGACCTCCAATGCAGAGCATCAT Iowa Black FQ | |||||
| GBNV-MPF | GBNV MP | GAACTGGTGGGAAACAGATA | 53 | 130 | This study | |
| GBNV-MPR | ATTTCAAGAAGATTATCCATCTC | |||||
| GBNV_MP | Cy5 TCTCATCATCATTTTCAGCTTCTAAT Iowa Black RQ-Sp | |||||
| ACT1F | βactin (ACT11) | ATGCTAGTGGTCGTACAACTGG | 50–60 | 108 | [27] | |
| ACT1R | CTAGACGAAGGATAGCATGTGG | |||||
| ACT1 | Hex TGGTGTCAGCCACACAGTCCCCAT Iowa Black FQ | This study |
| Standard | Concentration of Total RNA (ng/µL) in GBNV-Infected Dried Leaf Tissue | Total ng in 20 µL Reaction Mix | Cq Value for MP | Cq Value for NP | Cq Value for ACT11 |
|---|---|---|---|---|---|
| 1 | 100 | 200 | 22.95 ± 0.081 | 22.73 ± 0.204 | 24.48 ± 0.206 |
| 2 | 50 | 100 | 23.57 ± 0.006 | 23.51 ± 0.108 | 25.11 ± 0.061 |
| 3 | 25 | 50 | 24.56 ± 0.042 | 24.70 ± 0.152 | 26.17 ± 0.013 |
| 4 | 12.50 | 25 | 25.69 ± 0.733 | 25.89 ± 0.020 | 27.12 ± 0.057 |
| 5 | 6.25 | 12.5 | 26.86 ± 0.169 | 26.99 ± 0.086 | 28.10 ± 0.048 |
| 6 | 3.125 | 6.25 | 27.86 ± 0.008 | 28.03 ± 0.107 | 29.15 ± 0.013 |
| 7 | 1.563 | 3.125 | 29.01 ± 0.245 | 29.54 ± 0.050 | 30.25 ± 0.098 |
| 8 | 0.781 | 1.563 | 30.42 ± 0.258 | 29.88 ± 0.058 | 31.37 ± 0.073 |
| 9 | 0.391 | 0.781 | 31.34 ± 0.363 | 31.11 ± 0.012 | 32.48 ± 0.190 |
| 10 | 0.195 | 0.391 | 32.21 ± 0.105 | 32.15 ± 0.062 | 33.52 ± 0.111 |
| 11 | 0.098 | 0.195 | 33.25 ± 0.116 | 33.21 ± 0.036 | 34.63 ± 0.141 |
| 12 | 0.049 | 0.098 | 34.46 ± 0.009 | 34.48 ± 0.045 | - |
| 13 | 0.024 | 0.049 | - | - | - |
| 14 | 0.012 | 0.024 | - | - | - |
| 15 | 0.006 | 0.012 | - | - | - |
| Targeted Region | Replicate | Cq Values | |
|---|---|---|---|
| Multiplex | Singleplex | ||
| GBNV NP | 1 | 21.18 | 20.98 |
| 2 | 21.25 | 20.9 | |
| 3 | 21.22 | 20.87 | |
| 4 | 21.19 | 20.86 | |
| 5 | 21.21 | 21 | |
| 6 | 21.08 | 21.15 | |
| GBNV MP | 1 | 22.04 | 21.97 |
| 2 | 21.63 | 21.79 | |
| 3 | 21.93 | 21.7 | |
| 4 | 21.82 | 21.89 | |
| 5 | 21.38 | 21.55 | |
| 6 | 21.79 | 21.86 | |
| ACT11 | 1 | 27.08 | 27.04 |
| 2 | 26.84 | 26.58 | |
| 3 | 26.91 | 26.77 | |
| 4 | 27.02 | 26.87 | |
| 5 | 27.12 | 26.76 | |
| 6 | 27.07 | 27.04 | |
| Targeted Region | Cq Value (Singleplex)–Cq Value (Multiplex) | |
|---|---|---|
| Test Statistic S | Prob > |S| | |
| GBNV NP | −9.500 | 0.0625 |
| GBNV MP | 2.500 | 0.7188 |
| ACT11 | −10.500 | 0.0313 * |
| Sample | Targeted Region | Non-Reverse Transcriptase (RT Was Inactivated) | Non-Treated (No DNAse Treatment or RT Inactivation Was Performed) | DNase-Treated |
|---|---|---|---|---|
| Healthy peanut RNA | ACT11 | 24.91 ± 0.229 | 23.90 ± 0.222 | 27.75 ± 0.074 |
| GBNV-infected RNA | ACT11 | 24.88 ± 0.083 | 25.54 ± 0.238 | 30.72 ± 0.081 |
| GBNV NP | NA | 23.25 ± 0.149 | 23.40 ± 0.083 | |
| GBNV MP | NA | 21.35 ± 0.265 | 21.31 ± 0.401 |
| Sample | Targeted Region | Mean Cq Value |
|---|---|---|
| Healthy peanut RNA | ACT11 | 30.02 ± 0.164 |
| GBNV NP | NA | |
| GBNV MP | NA | |
| GBNV-infected (cowpea) | ACT11 | 29.06 ± 0.172 |
| GBNV NP | 19.99 ± 0.122 | |
| GBNV MP | 18.03 ± 0.276 | |
| GBNV-infected (peanut) | ACT11 | 28.56 ± 0.503 |
| GBNV NP | 16.46 ± 0.444 | |
| GBNV MP | 15.26 ± 0.716 | |
| GBNV-infected (unknown from the field) | ACT11 | 25.86 ± 0.177 |
| GBNV NP | 28.31 ± 0.130 | |
| GBNV MP | 25.56 ± 0.199 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deraniyagala, A.S.; Roy, A.; Tallury, S.; Sudini, H.K.; Culbreath, A.K.; Bag, S. Development of a Multiplex TaqMan Assay for Rapid Detection of Groundnut Bud Necrosis Virus: A Quarantine Pathogen in the USA. Viruses 2025, 17, 532. https://doi.org/10.3390/v17040532
Deraniyagala AS, Roy A, Tallury S, Sudini HK, Culbreath AK, Bag S. Development of a Multiplex TaqMan Assay for Rapid Detection of Groundnut Bud Necrosis Virus: A Quarantine Pathogen in the USA. Viruses. 2025; 17(4):532. https://doi.org/10.3390/v17040532
Chicago/Turabian StyleDeraniyagala, Anushi Suwaneththiya, Avijit Roy, Shyam Tallury, Hari Kishan Sudini, Albert K. Culbreath, and Sudeep Bag. 2025. "Development of a Multiplex TaqMan Assay for Rapid Detection of Groundnut Bud Necrosis Virus: A Quarantine Pathogen in the USA" Viruses 17, no. 4: 532. https://doi.org/10.3390/v17040532
APA StyleDeraniyagala, A. S., Roy, A., Tallury, S., Sudini, H. K., Culbreath, A. K., & Bag, S. (2025). Development of a Multiplex TaqMan Assay for Rapid Detection of Groundnut Bud Necrosis Virus: A Quarantine Pathogen in the USA. Viruses, 17(4), 532. https://doi.org/10.3390/v17040532







