Classification of the LC4 Primarily-like Cell Line—Recapitulating a CDK4 Overexpressing Immune Evasive HIV-HCV-Induced HCC
Abstract
1. Introduction
2. Materials and Methods
3. Results
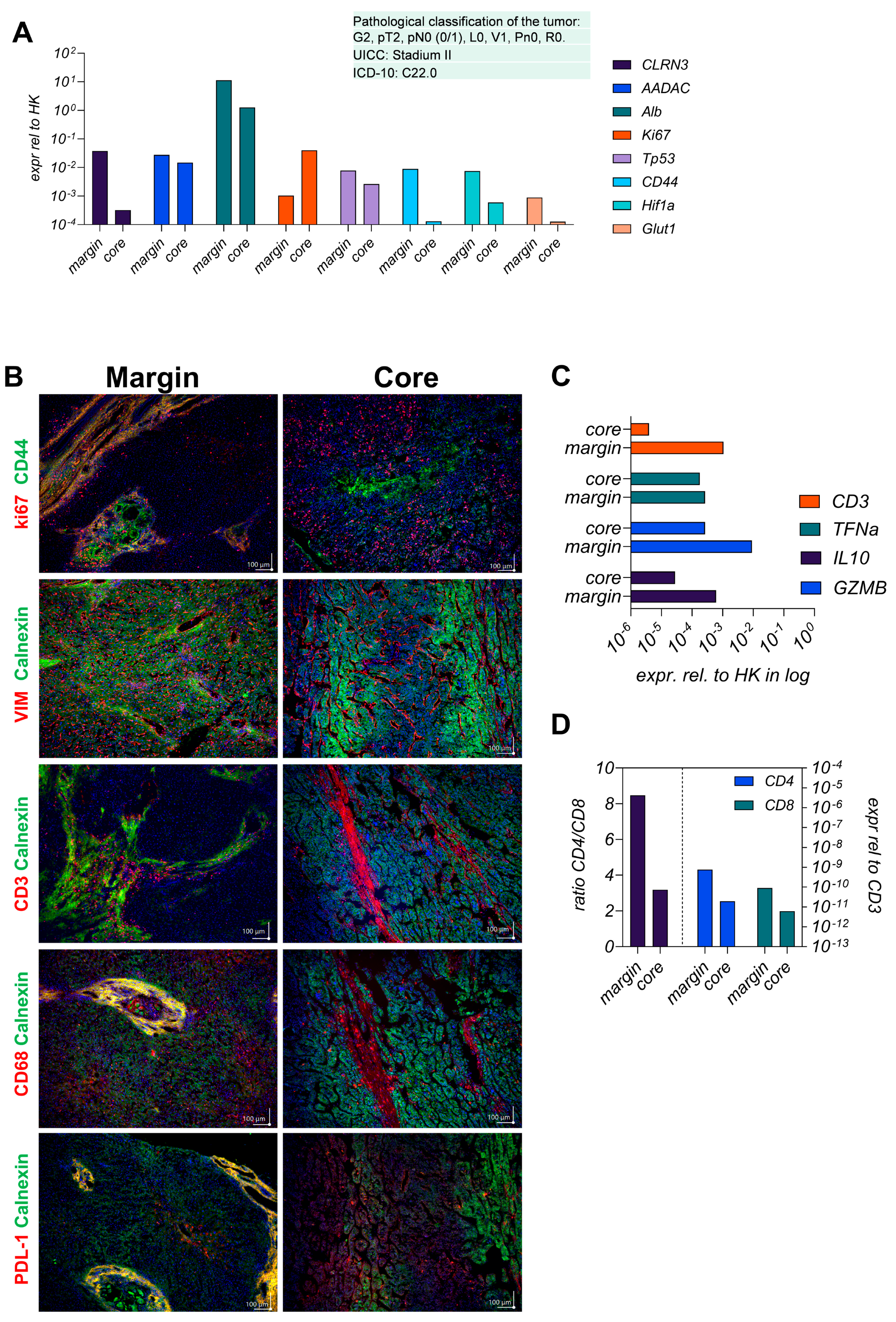
3.1. The Parental Tumor Regions Are Classified as Immunogenic Margins and Immunosuppressive Core
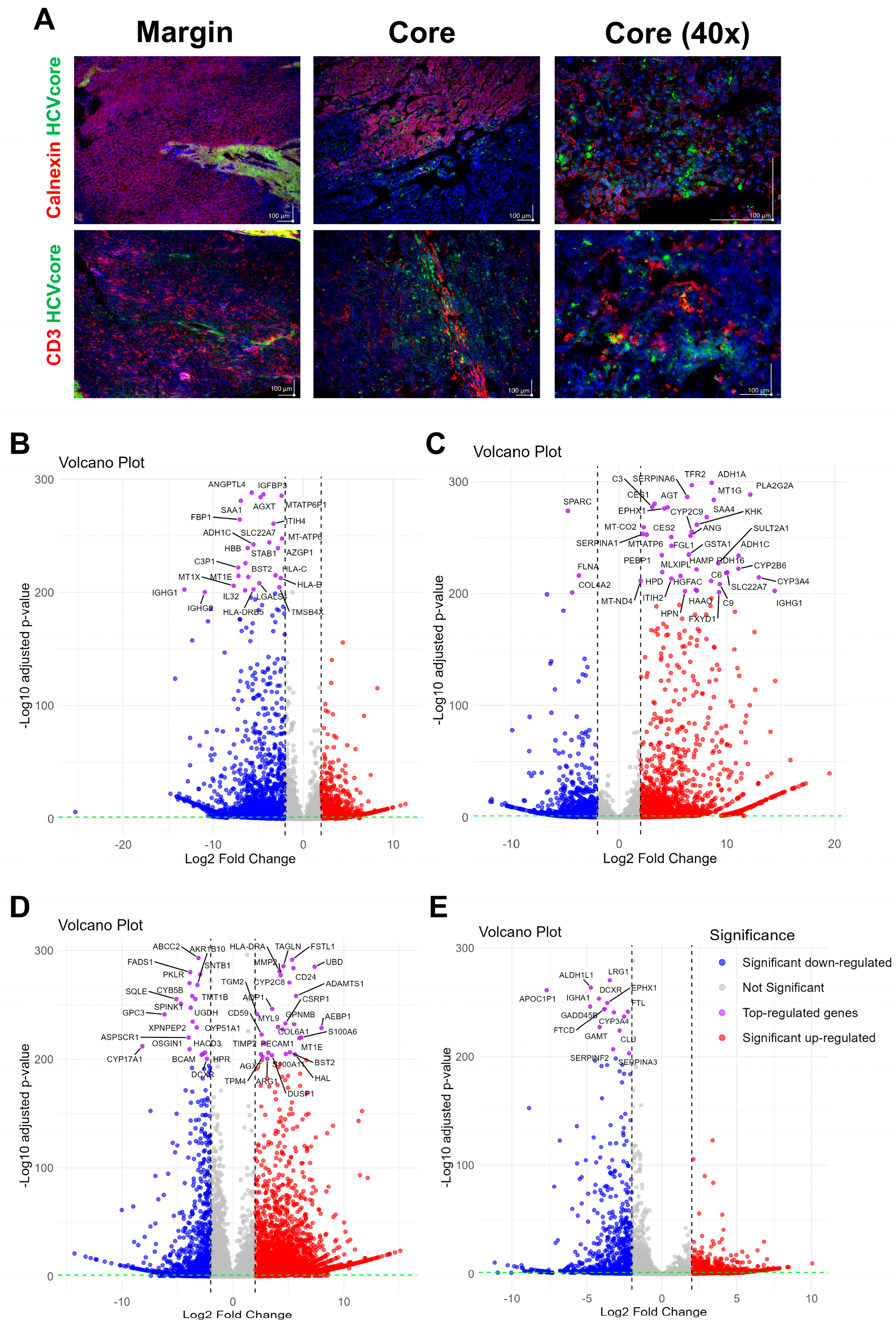
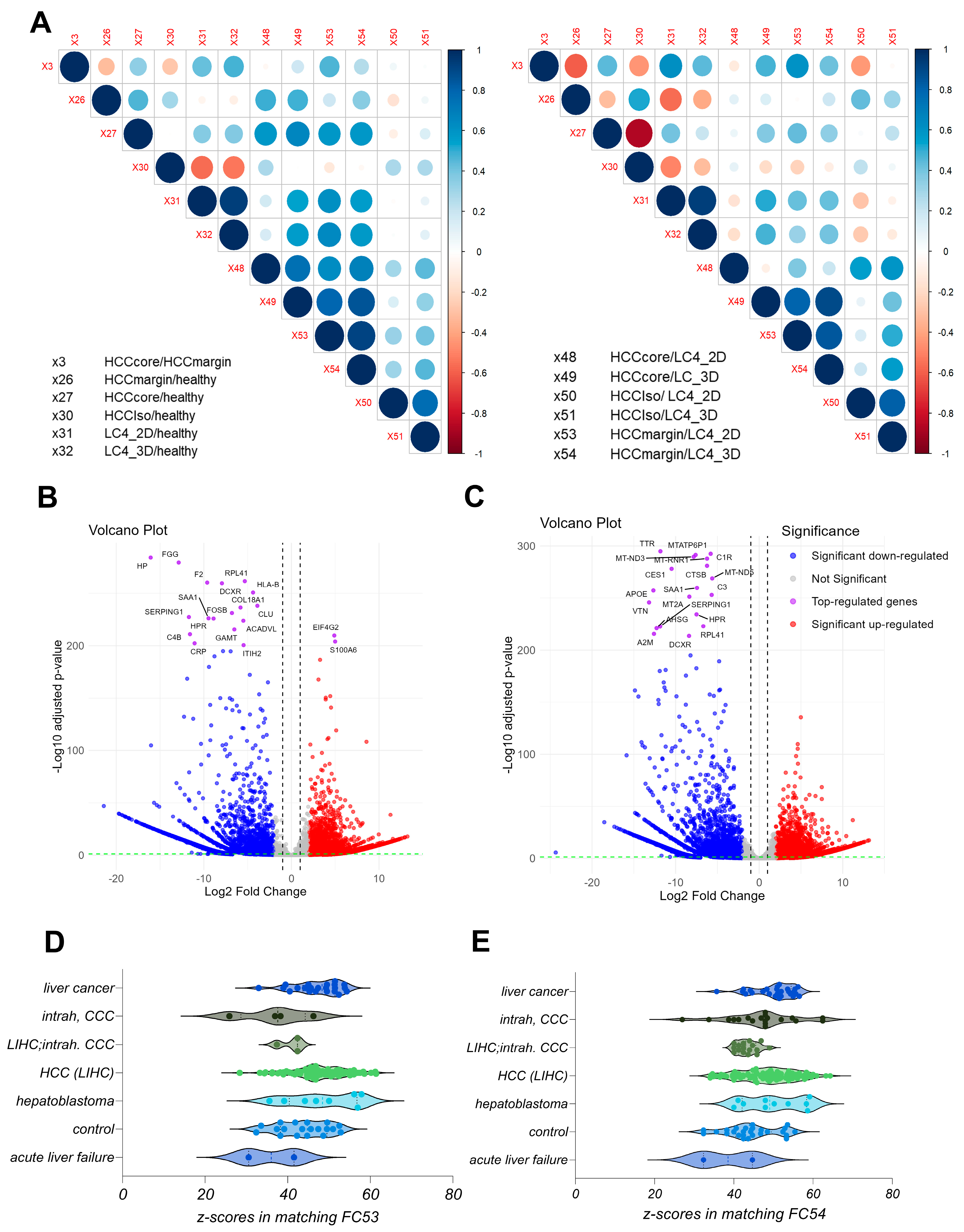
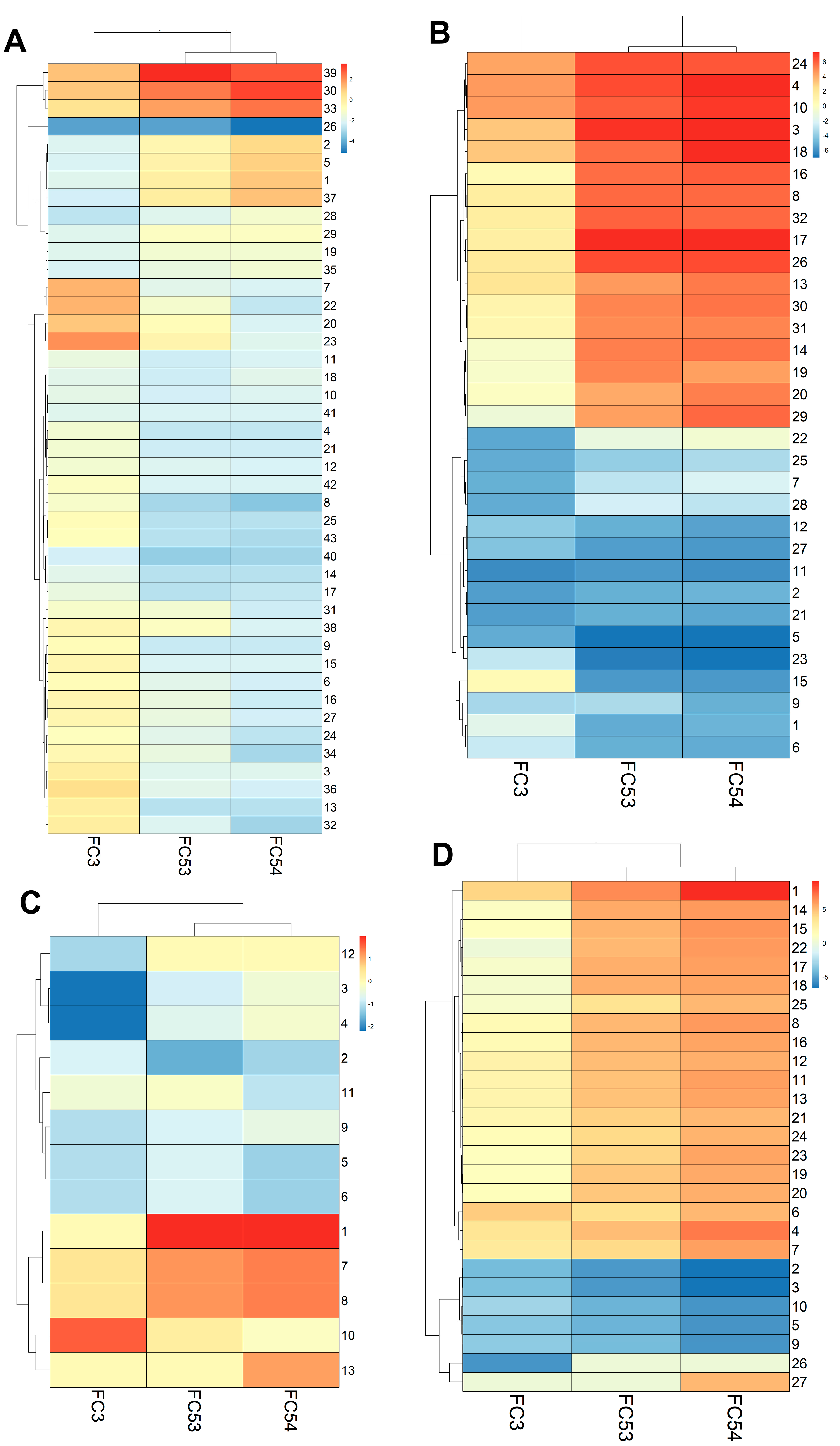
3.2. The Generated LC4 Cell Line Positively Correlates with the Parental HCC Core Region in Pathway Analysis and Shows High Similarity to Liver-Cancer-Related Disease
3.3. LC4 Cell Cines Retain Core Tumor Traits but Exhibit Enhanced Proliferation and Reduced Stress-Related Pathways Compared to Parental HCC Core
3.4. LC4 Patient-Derived Cells Retain Immune Evasion and Proliferative Traits but Show Reduced Immune Marker Expression Compared to Parental HCC Core
3.5. LC4 Cells in 3D Culture Exhibit Enhanced Proliferation and Immune Evasion
3.6. LC4 Cells Exhibit Stronger Transcriptomic Changes Compared to Common Liver Cancer Cell Lines, with 3D Culture Retaining More Tumor-like Characteristics
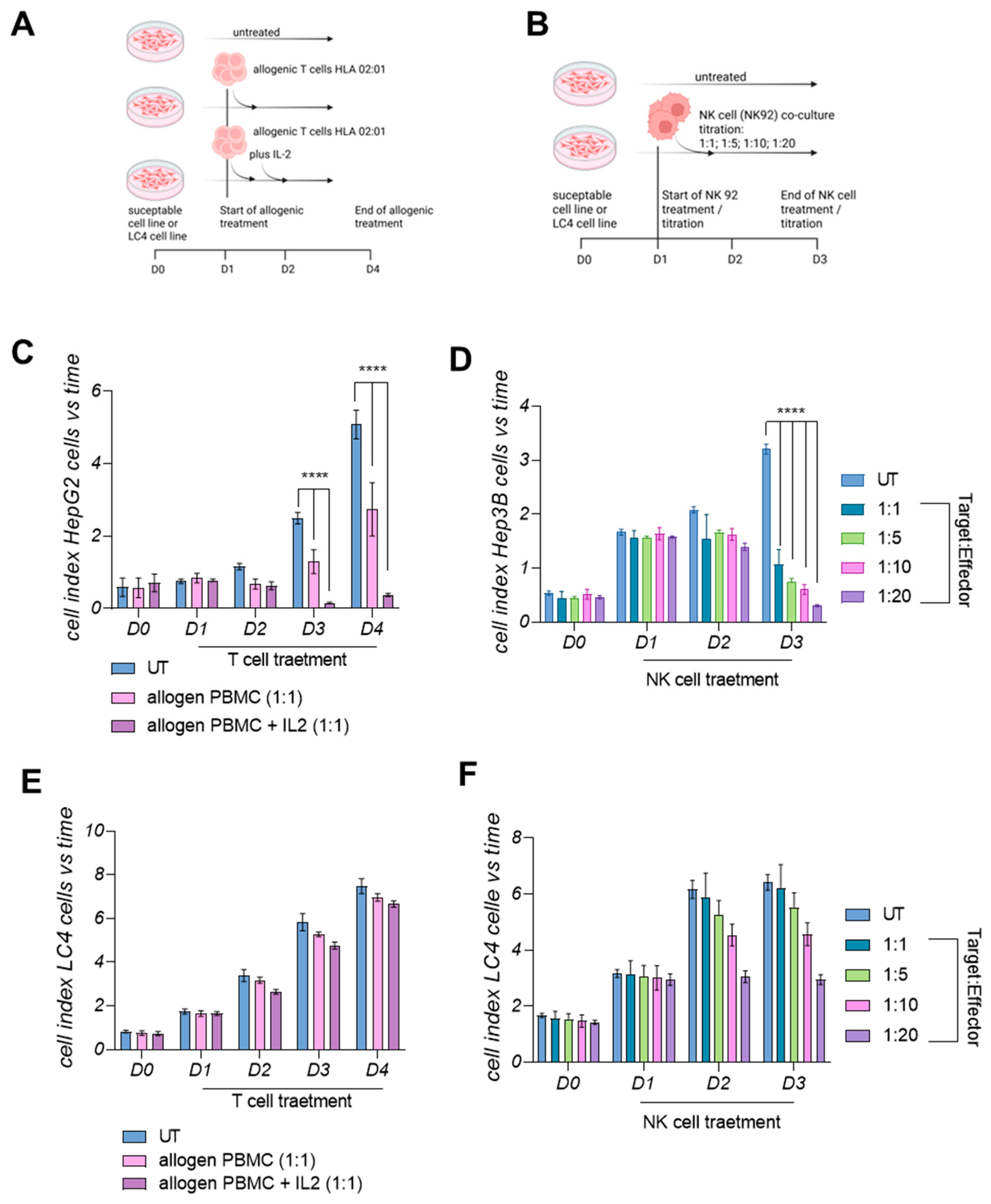
3.7. LC4 Cells Present a Lower Susceptibility Towards Active Allogenic T-Cells and Cytolytic NK Cells In Vitro
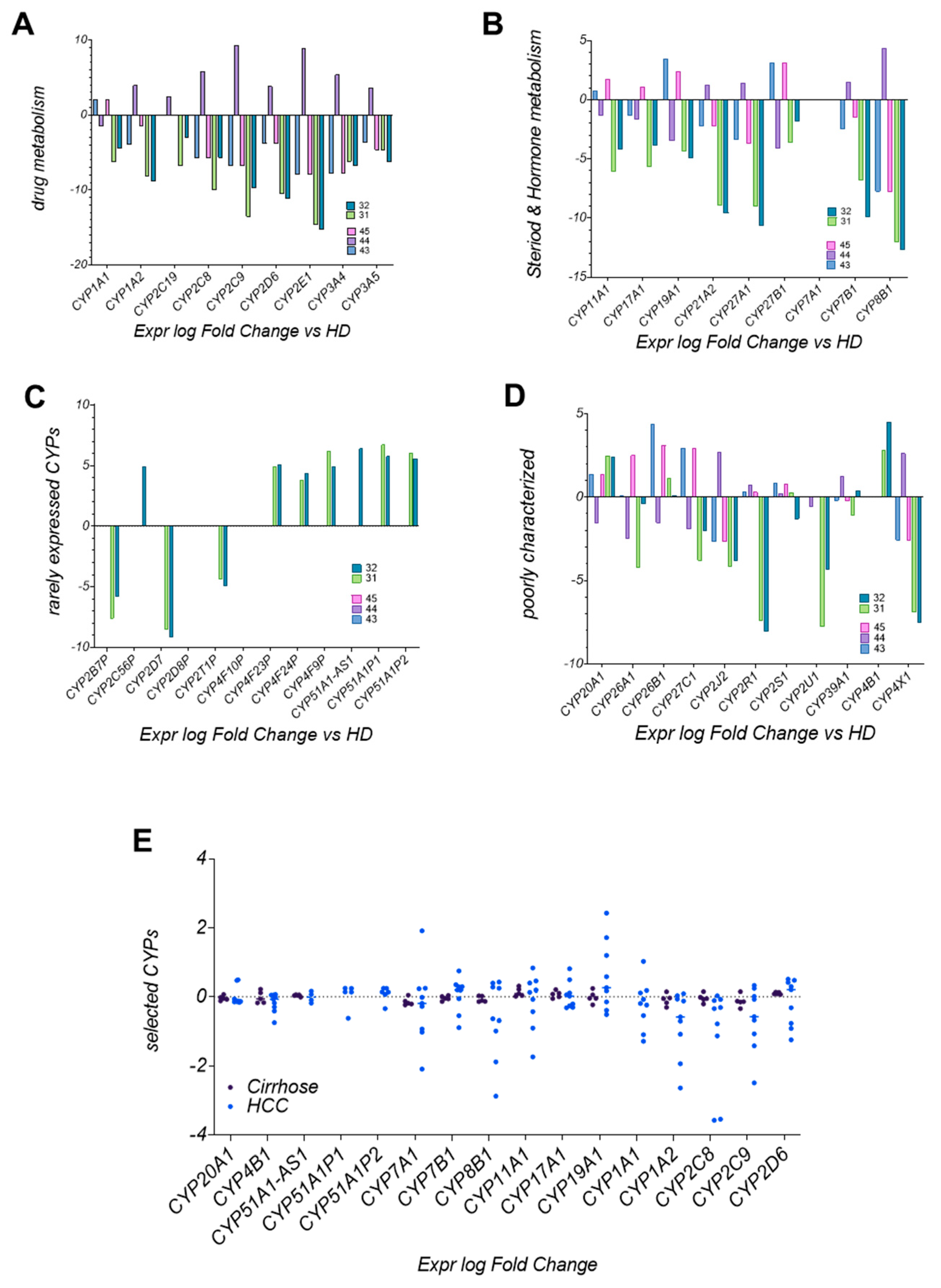
3.8. CYP Enzyme Expression in LC4 Reflects Metabolic Reprogramming in HCC
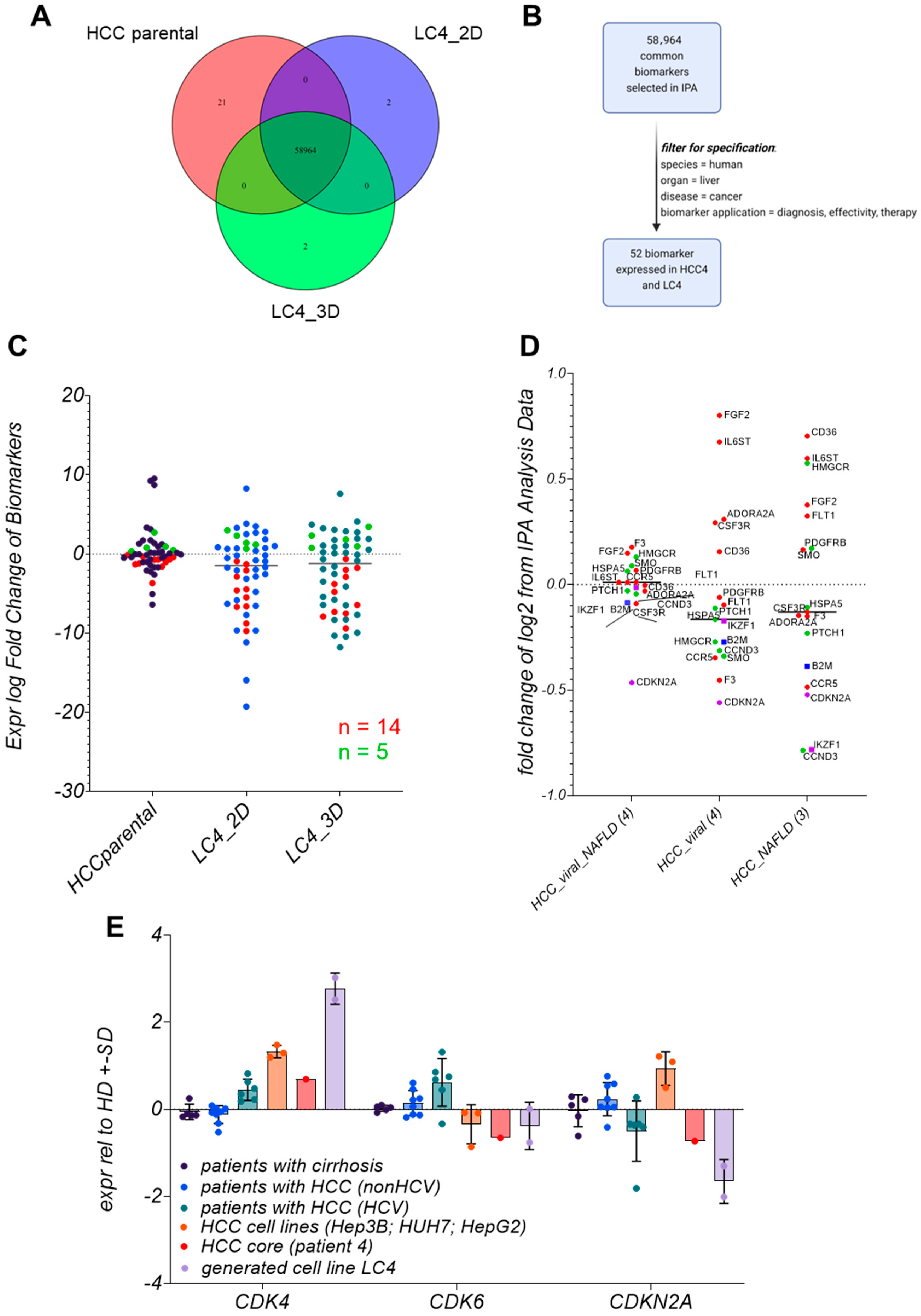
3.9. Biomarker Identification in LC4 Cells Reveals That CDKN2A, IKFZ1, and B2M Are Consistently Regulated
3.10. LC4 Cells Offer a Model for CDK6-Independent Regulation of CDKN2A Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HCC | Hepatocellular carcinoma |
| MASH | Metabolic steatohepatitis |
| HCV | Hepatitis C virus |
| HIV | Human Immunodeficiency Virus |
| TME | Tumor microenvironment |
| PD-L1 | Programmed death-ligand 1 |
| VEGF | Vascular endothelial growth factor |
| CTLA-4 | Cytotoxic T-lymphocyte-associated protein 4 |
| PDX | Patient-derived xenografts |
| RNA-seq | RNA sequencing |
| DEGs | Differentially expressed genes |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A |
| CYP | Cytochrome P450 |
| B2M | Beta-2-microglobulin |
| IKZF1 | Ikaros family zinc finger protein 1 |
| EMT | Epithelial–mesenchymal transition |
| TAM | Tumor-associated macrophages |
| MDSC | Myeloid-derived suppressor cells |
| IPA | Ingenuity Pathway Analysis |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Burra, P.; Zanetto, A.; Germani, G. Liver Transplantation for Alcoholic Liver Disease and Hepatocellular Carcinoma. Cancers 2018, 10, 46. [Google Scholar] [CrossRef]
- Dai, C.Y.; Chuang, W.L.; Yu, M.L. EASL recommendations on treatment of hepatitis C: Final update of the series ☆. J. Hepatol. 2020, 73, 1170–1218. [Google Scholar]
- Goto, K.; Roca Suarez, A.A.; Wrensch, F.; Baumert, T.F.; Lupberger, J. Hepatitis C Virus and Hepatocellular Carcinoma: When the Host Loses Its Grip. Int. J. Mol. Sci. 2020, 21, 3057. [Google Scholar] [CrossRef]
- Curry, M.P. HIV and Hepatitis C Virus: Special Concerns for Patients with Cirrhosis. J. Infect. Dis. 2013, 207 (Suppl. S1), S40–S44. [Google Scholar] [CrossRef]
- Hernandez, M.D.; Sherman, K.E. HIV/hepatitis C coinfection natural history and disease progression. Curr. Opin. HIV AIDS 2011, 6, 478–482. [Google Scholar] [CrossRef]
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef]
- Abdelhamed, W.; El-Kassas, M. Hepatocellular carcinoma recurrence: Predictors and management. Liver Res. 2023, 7, 321–332. [Google Scholar] [CrossRef]
- Petitprez, F.; Meylan, M.; de Reyniès, A.; Sautès-Fridman, C.; Fridman, W.H. The Tumor Microenvironment in the Response to Immune Checkpoint Blockade Therapies. Front. Immunol. 2020, 11, 784. [Google Scholar] [CrossRef]
- Pathak, S.; Sonbol, M.B. Second-Line Treatment Options for Hepatocellular Carcinoma: Current Landscape and Future Direction. J. Hepatocell. Carcinoma 2021, 8, 1147–1158. [Google Scholar] [CrossRef]
- Lee, S.W.; Lee, T.Y.; Yang, S.S.; Huang, Y.J.; Peng, Y.C. The Impact of Sequential Therapies after First-Line Systemic Therapies in Unresectable Hepatocellular Carcinoma. J. Clin. Med. 2024, 13, 2612. [Google Scholar] [CrossRef]
- Yu, J.; Li, M.; Ren, B.; Cheng, L.; Wang, X.; Ma, Z.; Yong, W.P.; Chen, X.; Wang, L.; Goh, B.C. Unleashing the efficacy of immune checkpoint inhibitors for advanced hepatocellular carcinoma: Factors, strategies, and ongoing trials. Front. Pharmacol. 2023, 14, 1261575. [Google Scholar] [CrossRef]
- Blidisel, A.; Marcovici, I.; Coricovac, D.; Hut, F.; Dehelean, C.A.; Cretu, O.M. Experimental Models of Hepatocellular Carcinoma-A Preclinical Perspective. Cancers 2021, 13, 3651. [Google Scholar] [CrossRef]
- Manduca, N.; Maccafeo, E.; De Maria, R.; Sistigu, A.; Musella, M. 3D cancer models: One step closer to in vitro human studies. Front. Immunol. 2023, 14, 1175503. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, W.; Cai, C.; Zhang, H.; Shen, H.; Han, Y. Patient-derived xenograft models in cancer therapy: Technologies and applications. Signal Transduct. Target. Ther. 2023, 8, 160. [Google Scholar] [CrossRef]
- Wei, Z.L.; Zhou, X.; Lan, C.L.; Huang, H.S.; Liao, X.W.; Mo, S.T.; Wei, Y.G.; Peng, T. Clinical implications and molecular mechanisms of Cyclin-dependent kinases 4 for patients with hepatocellular carcinoma. BMC Gastroenterol. 2022, 22, 77. [Google Scholar] [CrossRef]
- Staffeldt, L.; Mattert, G.; Riecken, K.; Rövenstrunk, G.; Volkmar, A.; Heumann, A.; Moustafa, M.; Jücker, M.; Fehse, B.; Schumacher, U.; et al. Generating Patient-Derived HCC Cell Lines Suitable for Predictive In Vitro and In Vivo Drug Screening by Orthotopic Transplantation. Cells 2023, 13, 82. [Google Scholar] [CrossRef]
- Bierwolf, J.; Volz, T.; Lütgehetmann, M.; Allweiss, L.; Riecken, K.; Warlich, M.; Fehse, B.; Kalff, J.C.; Dandri, M.; Pollok, J.M. Primary Human Hepatocytes Repopulate Livers of Mice After In Vitro Culturing and Lentiviral-Mediated Gene Transfer. Tissue Eng. Part A 2016, 22, 742–753. [Google Scholar] [CrossRef]
- Hösel, M.; Quasdorff, M.; Ringelhan, M.; Kashkar, H.; Debey-Pascher, S.; Sprinzl, M.F.; Bockmann, J.H.; Arzberger, S.; Webb, D.; von Olshausen, G.; et al. Hepatitis B Virus Activates Signal Transducer and Activator of Transcription 3 Supporting Hepatocyte Survival and Virus Replication. Cell. Mol. Gastroenterol. Hepatol. 2017, 4, 339–363. [Google Scholar] [CrossRef]
- Kah, J.; Wüstenberg, A.; Keller, A.D.; Sirma, H.; Montalbano, R.; Ocker, M.; Volz, T.; Dandri, M.; Tiegs, G.; Sass, G. Selective induction of apoptosis by HMG-CoA reductase inhibitors in hepatoma cells and dependence on p53 expression. Oncol. Rep. 2012, 28, 1077–1083. [Google Scholar] [CrossRef]
- Allweiss, L.; Volz, T.; Lütgehetmann, M.; Giersch, K.; Bornscheuer, T.; Lohse, A.W.; Petersen, J.; Ma, H.; Klumpp, K.; Fletcher, S.P.; et al. Immune cell responses are not required to induce substantial hepatitis B virus antigen decline during pegylated interferon-alpha administration. J. Hepatol. 2014, 60, 500–507. [Google Scholar] [CrossRef]
- Kah, J.; Volz, T.; Lütgehetmann, M.; Groth, A.; Lohse, A.W.; Tiegs, G.; Sass, G.; Dandri, M. Haem oxygenase-1 polymorphisms can affect HCV replication and treatment responses with different efficacy in humanized mice. Liver Int. 2017, 37, 1128–1137. [Google Scholar] [CrossRef]
- R Core Team, R. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer-Verlag: New York, NY, USA, 2016. [Google Scholar]
- Krämer, A.; Green, J.; Pollard Jr, J.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 1.0.12, 2022. Available online: https://github.com/raivokolde/pheatmap (accessed on 20 February 2025).
- Wei, T. R package ’corrplot’: Visualization of a Correlation Matrix. 2024. (Version 0.95). Available online: https://github.com/taiyun/corrplot (accessed on 20 February 2025).
- Sterling, R.K.; Sulkowski, M.S. Hepatitis C virus in the setting of HIV or hepatitis B virus coinfection. In Seminars in Liver Disease; Thieme Medical Publishers, Inc.: New York, NY, USA, 2004; Volume 24, pp. 61–68. [Google Scholar]
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef]
- Jo, H.; Park, Y.; Kim, T.; Kim, J.; Lee, J.S.; Kim, S.Y.; Chung, J.I.; Ko, H.Y.; Pyun, J.C.; Kim, K.S.; et al. Modulation of SIRT3 expression through CDK4/6 enhances the anti-cancer effect of sorafenib in hepatocellular carcinoma cells. BMC Cancer 2020, 20, 332. [Google Scholar] [CrossRef]
- Hofman, J.; Vagiannis, D.; Chen, S.; Guo, L. Roles of CYP3A4, CYP3A5 and CYP2C8 drug-metabolizing enzymes in cellular cytostatic resistance. Chem. Biol. Interact 2021, 340, 109448. [Google Scholar] [CrossRef]
- Ren, X.; Ji, Y.; Jiang, X.; Qi, X. Downregulation of CYP2A6 and CYP2C8 in Tumor Tissues Is Linked to Worse Overall Survival and Recurrence-Free Survival from Hepatocellular Carcinoma. Biomed. Res. Int. 2018, 2018, 5859415. [Google Scholar] [CrossRef] [PubMed]
- Abdelmonem, B.H.; Abdelaal, N.M.; Anwer, E.K.; Rashwan, A.A.; Hussein, M.A.; Ahmed, Y.F.; Khashana, R.; Hanna, M.M.; Abdelnaser, A. Decoding the Role of CYP450 Enzymes in Metabolism and Disease: A Comprehensive Review. Biomedicines 2024, 12, 1467. [Google Scholar] [CrossRef]
- Xiao, M.; Xu, J.; Wang, W.; Zhang, B.; Liu, J.; Li, J.; Xu, H.; Zhao, Y.; Yu, X.; Shi, S. Functional significance of cholesterol metabolism in cancer: From threat to treatment. Exp. Mol. Med. 2023, 55, 1982–1995. [Google Scholar] [CrossRef]
- Shklovskaya, E.; Rizos, H. MHC Class I Deficiency in Solid Tumors and Therapeutic Strategies to Overcome It. Int. J. Mol. Sci. 2021, 22, 6741. [Google Scholar] [CrossRef]
- Caraballo Cortés, K.; Osuch, S.; Perlejewski, K.; Radkowski, M.; Janiak, M.; Berak, H.; Rauch, A.; Fehr, J.S.; Hoffmann, M.; Günthard, H.F.; et al. T-Cell Exhaustion in HIV-1/Hepatitis C Virus Coinfection Is Reduced After Successful Treatment of Chronic Hepatitis C. In Open Forum Infectious Diseases; Oxford University Press: New York, NY, USA, 2023; Volume 10, p. ofad514. [Google Scholar]
- Wang, H.Y.; Cui, X.W.; Zhang, Y.H.; Chen, Y.; Lu, N.N.; Bai, L.; Duan, Z.P. Dynamic changes of phenotype and function of natural killer cells in peripheral blood before and after thermal ablation of hepatitis B associated hepatocellular carcinoma and their correlation with tumor recurrence. BMC Cancer 2023, 23, 486. [Google Scholar] [CrossRef]
- Haibe, Y.; El Husseini, Z.; El Sayed, R.; Shamseddine, A. Resisting Resistance to Immune Checkpoint Therapy: A Systematic Review. Int. J. Mol. Sci. 2020, 21, 6176. [Google Scholar] [CrossRef]
- Chen, J.C.; Perez-Lorenzo, R.; Saenger, Y.M.; Drake, C.G.; Christiano, A.M. IKZF1 Enhances Immune Infiltrate Recruitment in Solid Tumors and Susceptibility to Immunotherapy. Cell Syst. 2018, 7, 92–103.e4. [Google Scholar] [CrossRef]
- Rodrigues, J.; Heinrich, M.A.; Teixeira, L.M.; Prakash, J. 3D In Vitro Model (R)evolution: Unveiling Tumor–Stroma Interactions. Trends Cancer 2021, 7, 249–264. [Google Scholar] [CrossRef]
- Guldner, I.H.; Zhang, S. A journey to uncharted territory: New technical frontiers in studying tumor-stromal cell interactions. Integr. Biol. 2015, 7, 153–161. [Google Scholar] [CrossRef]


| Number | Pathway |
|---|---|
| 1 | 3-phosphoinositide Biosynthesis |
| 2 | 3-phosphoinositide Degradation |
| 3 | Acetate Conversion to Acetyl-CoA |
| 4 | Bile Acid Biosynthesis, Neutral Pathway |
| 5 | D-myo-inositol-5-phosphate Metabolism |
| 6 | Dopamine Degradation |
| 7 | Estrogen Biosynthesis |
| 8 | Ethanol Degradation II |
| 9 | Ethanol Degradation IV |
| 10 | Fatty Acid β-oxidation I |
| 11 | Glutaryl-CoA Degradation |
| 12 | Glutathione Redox Reactions I |
| 13 | Glutathione-mediated Detoxification |
| 14 | Glycine Betaine Degradation |
| 15 | Histamine Degradation |
| 16 | Histidine Degradation VI |
| 17 | Isoleucine Degradation I |
| 18 | Ketogenesis |
| 19 | Ketolytic |
| 20 | Melatonin Degradation I |
| 21 | NAD biosynthesis II (from tryptophan) |
| 22 | Nicotine Degradation II |
| 23 | Nicotine Degradation III |
| 24 | Noradrenaline and Adrenaline Degradation |
| 25 | Oxidative Ethanol Degradation III |
| 26 | Oxidative Phosphorylation |
| 27 | Phenylalanine Degradation IV (Mammalian, via Side Chain) |
| 28 | Phospholipases |
| 29 | Proteinoid Biosynthesis |
| 30 | Pyridoxal 5’-phosphate Salvage Pathway |
| 31 | Retinoate Biosynthesis I |
| 32 | Retinol Biosynthesis |
| 33 | Salvage Pathways of Pyrimidine Ribonucleotides |
| 34 | Serotonin Degradation |
| 35 | Sucrose Degradation V (Mammalian) |
| 36 | Super pathway of Cholesterol Biosynthesis |
| 37 | Super pathway of Inositol Phosphate Compounds |
| 38 | The Visual Cycle |
| 39 | tRNA Charging |
| 40 | Tryptophan Degradation III (Eukaryotic) |
| 41 | Tyrosine Degradation I |
| 42 | Urea Cycle |
| 43 | Valine Degradation I |
| Number | Pathway |
|---|---|
| 1 | Aspirin ADME |
| 2 | Atherosclerosis Signaling |
| 3 | Cell Cycle Checkpoints |
| 4 | Chromatin organization |
| 5 | Complement cascade |
| 6 | Complex I biogenesis |
| 7 | Dendritic Cell Maturation |
| 8 | Deubiquitinating |
| 9 | Eukaryotic Translation Elongation |
| 10 | Histone Modification Signaling Pathway |
| 11 | Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 12 | Interferon alpha/beta signaling |
| 13 | Intra-Golgi and retrograde Golgi-to-ER traffic |
| 14 | L1CAM interactions |
| 15 | Mitochondrial translation |
| 16 | Mitotic G2-G2/M phases |
| 17 | Mitotic Metaphase and Anaphase |
| 18 | Mitotic Prometaphase |
| 19 | Neddylation |
| 20 | Nuclear Cytoskeleton Signaling Pathway |
| 21 | Pathogen Induced Cytokine Storm Signaling Pathway |
| 22 | Phagosome Formation |
| 23 | Phase I—Functionalization of compounds |
| 24 | Processing of Capped Intron-Containing Pre-mRNA |
| 25 | Production of Nitric Oxide and Reactive Oxygen Species in Macrophages |
| 26 | Protein Ubiquitination Pathway |
| 27 | Respiratory electron transport |
| 28 | Response to elevated platelet cytosolic Ca2+ |
| 29 | RHO GTPase cycle |
| 30 | RHO GTPases Activate Forming |
| 31 | Signaling by NTRK1 (TRKA) |
| 32 | TCF dependent signaling in response to WNT |
| Number | Pathway |
|---|---|
| 1 | Infection by hepatitis B virus |
| 2 | Apoptosis of liver cell lines |
| 3 | Proliferation of liver cells |
| 4 | Proliferation of hepatic stellate cells |
| 5 | Cell death of liver cells |
| 6 | Necrosis of liver |
| 7 | Inflammation of liver |
| 8 | Viral hepatitis |
| 9 | Proliferation of hepatocytes |
| 10 | Liver tumor |
| 11 | Increased Levels of LDH |
| 12 | Cell death of liver cell lines |
| 13 | Fibrosis of liver |
| Number | Pathway |
|---|---|
| 1 | Cell proliferation of tumor cell lines |
| 2 | Sensitivity of tumor cell lines |
| 3 | Sensitivity of cells |
| 4 | Cell proliferation of carcinoma cell lines |
| 5 | Cell death of tumor cell lines |
| 6 | Cell proliferation of hepatoma cell lines |
| 7 | Cell proliferation of adenocarcinoma cell lines |
| 8 | Infection by RNA virus |
| 9 | Necrosis |
| 10 | Apoptosis of tumor cell lines |
| 11 | Migration of carcinoma cell lines |
| 12 | Infection of tumor cell lines |
| 13 | Cell movement of carcinoma cell lines |
| 14 | Cell survival |
| 15 | Viral Infection |
| 16 | Infection of cells |
| 17 | Cell viability |
| 18 | Cell viability of tumor cell lines |
| 19 | Infection by Reoviridae |
| 20 | HIV infection |
| 21 | Cell proliferation of colorectal cancer cell lines |
| 22 | Infection by HIV-1 |
| 23 | Migration of tumor cell lines |
| 24 | Cell proliferation of liver cancer cell lines |
| 25 | Cell movement of tumor cell lines |
| 26 | Mobilization of Ca2+ |
| 27 | Viral life cycle |
| Number | Pathway |
|---|---|
| 1 | Neutrophil degranulation |
| 2 | Response to elevated platelet cytosolic Ca2+ |
| 3 | EIF2 Signaling |
| 4 | LXR/RXR Activation |
| 5 | Acute Phase Response Signaling |
| 6 | Post-translational protein phosphorylation |
| 7 | Regulation of Insulin-like Growth Factor (IGF) transport and uptake by IGFBPs |
| 8 | DHCR24 Signaling Pathway |
| 9 | Eukaryotic Translation Initiation |
| 10 | Seleno amino acid metabolism |
| 11 | Protein Ubiquitination Pathway |
| 12 | Response of EIF2AK4 (GCN2) to amino acid deficiency |
| 13 | Eukaryotic Translation Elongation |
| 14 | Eukaryotic Translation Termination |
| 15 | SRP-dependent co-translational protein targeting to membrane |
| 16 | RHO GTPase cycle |
| 17 | Nonsense-Mediated Decay (NMD) |
| 18 | Pulmonary Fibrosis Idiopathic Signaling Pathway |
| 19 | Production of Nitric Oxide and Reactive Oxygen Species in Macrophages |
| 20 | Mitotic Metaphase and Anaphase |
| 21 | Class I MHC mediated antigen processing and presentation |
| 22 | Extracellular matrix organization |
| 23 | Integrin cell surface interactions |
| 26 | PTEN Regulation |
| 31 | Nuclear Cytoskeleton Signaling Pathway |
| 32 | Degradation of beta-catenin by the destruction complex |
| 34 | Mitotic G2-G2/M phases |
| 38 | Deubiquitinating |
| 44 | Transcriptional regulation by RUNX3 |
| 53 | Cell Cycle Checkpoints |
| 58 | Neddylation |
| 70 | Complement cascade |
| 89 | Pathogen Induced Cytokine Storm Signaling Pathway |
| 110 | Cell surface interactions at the vascular wall |
| 123 | Binding and Uptake of Ligands by Scavenger Receptors |
| 141 | Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 148 | Fcgamma receptor (FCGR) dependent phagocytosis |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kah, J.; Staffeldt, L.; Volz, T.; Schulze, K.; Heumann, A.; Rövenstrunk, G.; Goebel, M.; Peine, S.; Dandri, M.; Lüth, S. Classification of the LC4 Primarily-like Cell Line—Recapitulating a CDK4 Overexpressing Immune Evasive HIV-HCV-Induced HCC. Viruses 2025, 17, 653. https://doi.org/10.3390/v17050653
Kah J, Staffeldt L, Volz T, Schulze K, Heumann A, Rövenstrunk G, Goebel M, Peine S, Dandri M, Lüth S. Classification of the LC4 Primarily-like Cell Line—Recapitulating a CDK4 Overexpressing Immune Evasive HIV-HCV-Induced HCC. Viruses. 2025; 17(5):653. https://doi.org/10.3390/v17050653
Chicago/Turabian StyleKah, Janine, Lisa Staffeldt, Tassilo Volz, Kornelius Schulze, Asmus Heumann, Götz Rövenstrunk, Meike Goebel, Sven Peine, Maura Dandri, and Stefan Lüth. 2025. "Classification of the LC4 Primarily-like Cell Line—Recapitulating a CDK4 Overexpressing Immune Evasive HIV-HCV-Induced HCC" Viruses 17, no. 5: 653. https://doi.org/10.3390/v17050653
APA StyleKah, J., Staffeldt, L., Volz, T., Schulze, K., Heumann, A., Rövenstrunk, G., Goebel, M., Peine, S., Dandri, M., & Lüth, S. (2025). Classification of the LC4 Primarily-like Cell Line—Recapitulating a CDK4 Overexpressing Immune Evasive HIV-HCV-Induced HCC. Viruses, 17(5), 653. https://doi.org/10.3390/v17050653






