Theobroma cacao Virome: Exploring Public RNA-Seq Data for Viral Discovery and Surveillance
Abstract
1. Introduction
2. Materials and Methods
2.1. Acquisition of RNA-Seq Libraries
2.2. RNA-Seq Processing and Assembly
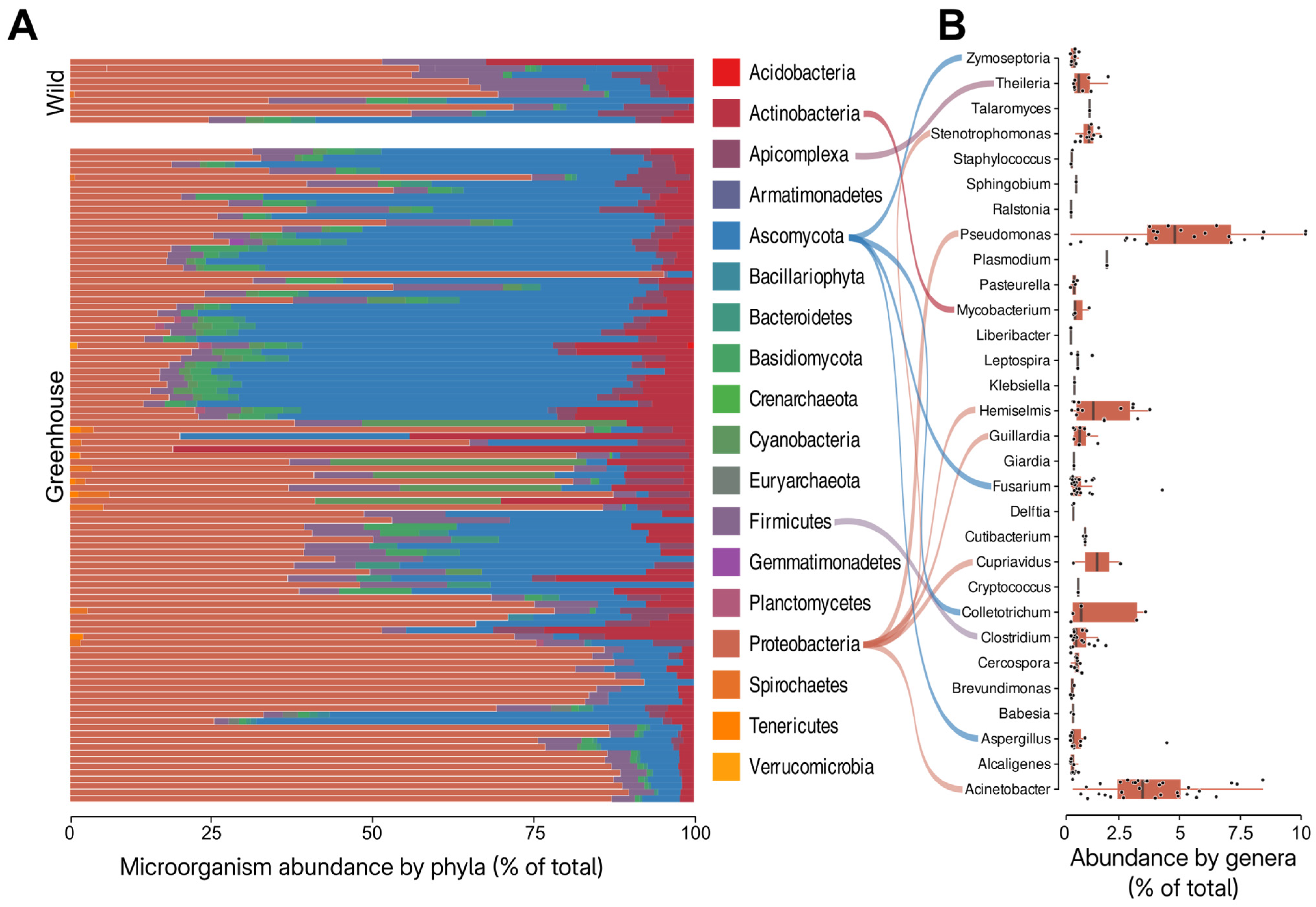
2.3. RNA-Seq-Based Assessment of the Cocoa Microbiome
2.4. Metaviromic Analysis
2.5. Novel Virus Definition
2.6. Integrative Genome Assembly
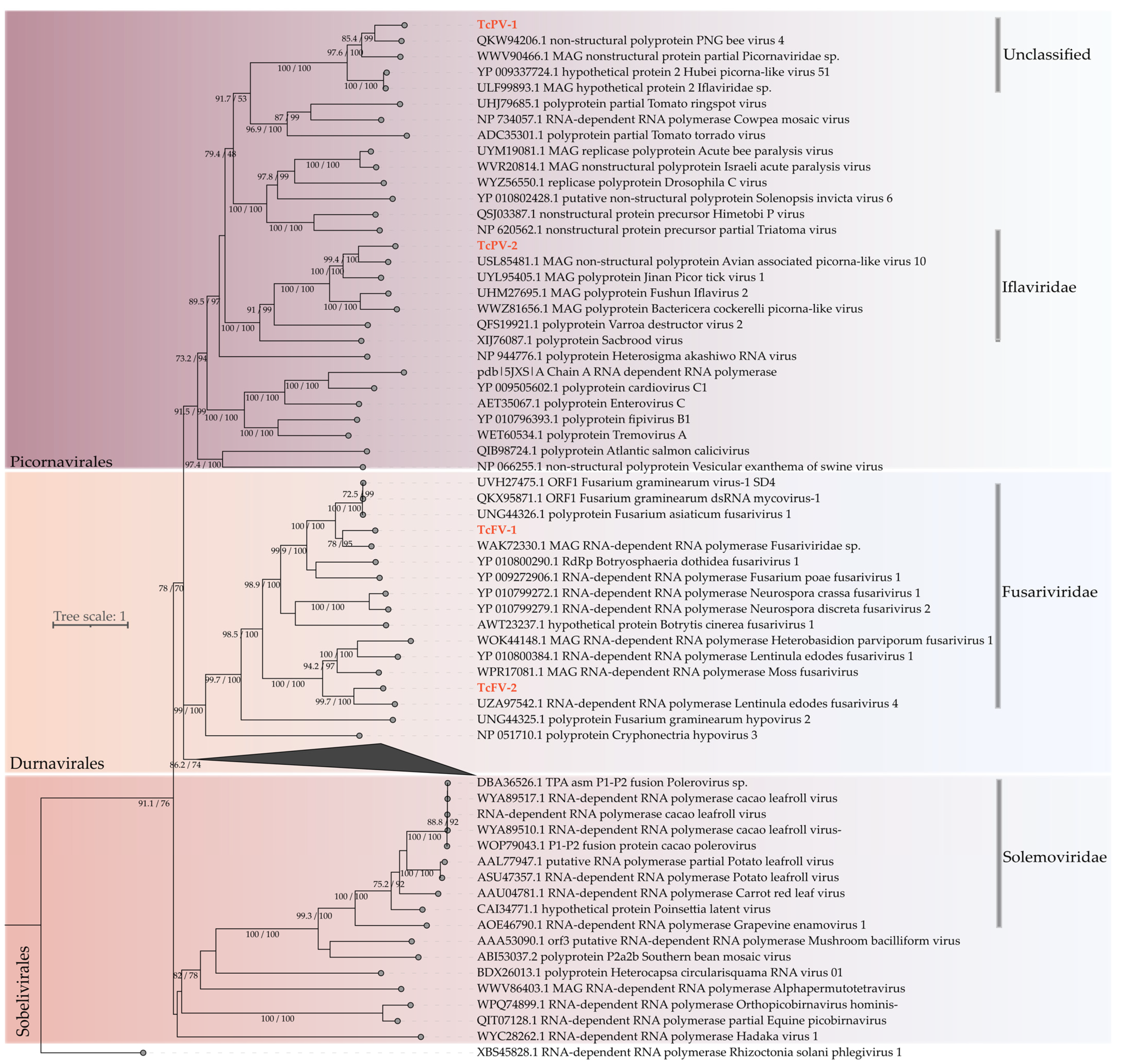
2.7. Molecular Phylogeny of Virus-Derived Sequences
2.8. RNA Abundance and the Widespread Presence of Viral Segments
3. Results
3.1. Theobroma cacao Microbiome Assessment
3.2. Theobroma cacao-Associated Virome
3.2.1. Characterization of Known Viral Species
3.2.2. Characterization of Novel Viral Species
- Kitrinoviricota
- Pisuviricota
- Caulimoviridae
3.3. Wide Spread of Known Pathogens and Novel Theobroma cacao-Associated Viruses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nair, K.P.P. Cocoa (Theobroma cacao L.). In The Agronomy and Economy of Important Tree Crops of the Developing World; Elsevier: Amsterdam, The Netherlands, 2010; pp. 131–180. [Google Scholar] [CrossRef]
- Kongor, J.E.; Hinneh, M.; de Walle, D.V.; Afoakwa, E.O.; Boeckx, P.; Dewettinck, K. Factors influencing quality variation in cocoa (Theobroma cacao) bean flavour profile—A review. Food Res. Int. 2016, 82, 44–52. [Google Scholar] [CrossRef]
- Kongor, J.E.; Owusu, M.; Oduro-Yeboah, C. Cocoa production in the 2020s: Challenges and solutions. CABI Agric. Biosci. 2024, 5, 102. [Google Scholar] [CrossRef]
- Paisic-Ramirez, R.; Hernández-Amasifuen, A.D.; Sánchez-Aguilar, W.D.; Corazon-Guivin, M.A.; Bobadilla, L.G.; Mansilla-Córdova, P.J.; Caetano, A.C.; Zuta, M.Z.S.; Guerrero-Abad, J.C. Effect of osmoregulatory on the secondary somatic embryogenesis of cocoa (Theobroma cacao L.). J. Appl. Biol. Biotechnol. 2024, 12, 177–183. [Google Scholar] [CrossRef]
- Alain, B.K. Economic impact of cocoa culture in Ivory Coast and in Ghana from 1980 to 2015. J. Hist. Archaeol. Anthr. Sci. 2024, 9, 62–67. [Google Scholar] [CrossRef]
- Obodai, J.; Asamoah, P.K.B.; Edusei, J. Cocoa Purchasing and the Issue of Insecurity in the Akontombra District in the Western Region of Ghana. Pelita Perkeb. 2018, 34, 128–136. [Google Scholar] [CrossRef]
- Widhiyoga, G.; Wijayati, H. Challenges Faced by Cocoa-based Industries from Indonesia in Global Value Chains. Husnayain Bus. Rev. 2022, 2, 1–10. [Google Scholar] [CrossRef]
- Edet, E.O.; Udoe, P.O.; Abang, S.O. Economic impact of climate change on cocoa production among South-Western states, Nigeria: Results from ricardian analysis. Glob. J. Pure Appl. Sci. 2018, 24, 171–180. [Google Scholar] [CrossRef]
- Förste, F.; Bauer, L.; Streeck, C.; Radtke, M.; Reinholz, U.; Kadow, D.; Keil, C.; Mantouvalou, I. Quantitative Analysis and 2D/3D Elemental Imaging of Cocoa Beans Using X-ray Fluorescence Techniques. Anal. Chem. 2023, 95, 5627–5634. [Google Scholar] [CrossRef]
- Rojo-Poveda, O.; Barbosa-Pereira, L.; Zeppa, G.; Stévigny, C. Cocoa Bean Shell—A By-Product with Nutritional Properties and Biofunctional Potential. Nutrients 2020, 12, 1123. [Google Scholar] [CrossRef]
- Forbes, S.J.; Northfield, T.D. Increased pollinator habitat enhances cacao fruit set and predator conservation. Ecol. Appl. 2017, 27, 887–899. [Google Scholar] [CrossRef]
- Kadio, A.K.C. Challenges and Perspectives of Local Cocoa Transformation in Côte D’Ivoire: A Case Study on the Cocoa Industry in Different Production Areas. Open J. Bus. Manag. 2023, 11, 2849–2867. [Google Scholar] [CrossRef]
- Lopes, M.; Junior, B.H.; Dias, C.; Santos, G.; Gramacho, K.; Cascardo, J.; Gesteira, A.; Micheli, F. Expression analysis of transcription factors from the interaction between cacao and Moniliophthora perniciosa (Tricholomataceae). Genet. Mol. Res. 2010, 9, 1279–1297. [Google Scholar] [CrossRef] [PubMed]
- Sena, K.; Alemanno, L.; Gramacho, K.P. The infection process of Moniliophthora perniciosa in cacao. Plant Pathol. 2014, 63, 1272–1281. [Google Scholar] [CrossRef]
- Litholdo, C.G.; Leal, G.A.; Albuquerque, P.S.B.; Figueira, A. Differential expression of jasmonate biosynthesis genes in cacao genotypes contrasting for resistance against Moniliophthora perniciosa. Plant Cell Rep. 2015, 34, 1747–1759. [Google Scholar] [CrossRef]
- Rubini, M.R.; Silva-Ribeiro, R.T.; Pomella, A.W.V.; Maki, C.S.; Araújo, W.L.; dos Santos, D.R.; Azevedo, J.L. Diversity of endophytic fungal community of cacao (Theobroma cacao L.) and biological control of Crinipellis perniciosa, causal agent of Witches’ Broom Disease. Int. J. Biol. Sci. 2005, 1, 24–33. [Google Scholar] [CrossRef]
- Phillips-Mora, W.; Wilkinson, M.J. Frosty Pod of Cacao: A Disease with a Limited Geographic Range but Unlimited Potential for Damage. Phytopathology 2007, 97, 1644–1647. [Google Scholar] [CrossRef]
- Ploetz, R.C. Cacao Diseases: Important Threats to Chocolate Production Worldwide. Phytopathology 2007, 97, 1634–1639. [Google Scholar] [CrossRef]
- Hebbar, P.K. Cacao Diseases: A Global Perspective from an Industry Point of View. Phytopathology 2007, 97, 1658–1663. [Google Scholar] [CrossRef]
- Adomako, D.; Hutcheon, W.V. Carbohydrate Metabolism and Translocation in Healthy and Cocoa Swollen Shoot Virus-Infected Cocoa Plants. Physiol. Plant. 1974, 30, 90–96. [Google Scholar] [CrossRef]
- Ploetz, R. The Impact of Diseases on Cacao Production: A Global Overview. In Cacao Diseases; Bailey, B.A., Meinhardt, L.W., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 33–59. [Google Scholar] [CrossRef]
- Marelli, J.-P.; Guest, D.I.; Bailey, B.A.; Evans, H.C.; Brown, J.K.; Junaid, M.; Barreto, R.W.; Lisboa, D.O.; Puig, A.S. Chocolate Under Threat from Old and New Cacao Diseases. Phytopathology 2019, 109, 1331–1343. [Google Scholar] [CrossRef]
- Dzahini-Obiatey, H.; Fox, R.T.V. Early signs of infection in Cacao swollen shoot virus (CSSV) inoculated cocoa seeds and the discovery of the cotyledons of the resultant plants as rich sources of CSSV. Afr. J. Biotechnol. 2010, 9, 593–603. [Google Scholar] [CrossRef]
- Ramos-Sobrinho, R.; Chingandu, N.; Gutierrez, O.A.; Marelli, J.-P.; Brown, J.K. A Complex of Badnavirus Species Infecting Cacao Reveals Mixed Infections, Extensive Genomic Variability, and Interspecific Recombination. Viruses 2020, 12, 443. [Google Scholar] [CrossRef] [PubMed]
- Obok, E.; Aikpokpodion, P.; Ani, O.; Allainguillaume, J.; Wetten, A. Cacao swollen shoot virus detection and DNA barcoding of its vectors and putative vectors in Theobroma cacao L. by using polymerase chain reaction. BioTechnologia 2021, 102, 229–244. [Google Scholar] [CrossRef] [PubMed]
- Andres, C.; Hoerler, R.; Home, R.; Joerin, J.; Dzahini-Obiatey, H.K.; Ameyaw, G.A.; Domfeh, O.; Blaser, W.J.; Gattinger, A.; Offei, S.K.; et al. Social network to inform and prevent the spread of cocoa swollen shoot virus disease in Ghana. Agron. Sustain. Dev. 2018, 38, 53. [Google Scholar] [CrossRef]
- Muller, E. Cacao Swollen Shoot Virus (CSSV): History, Biology, and Genome. In Cacao Diseases; Bailey, B.A., Meinhardt, L.W., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 337–358. [Google Scholar] [CrossRef]
- Abrokwah, F.; Dzahini-Obiatey, H.; Galyuon, I.; Osae-Awuku, F.; Muller, E. Geographical Distribution of Cacao swollen shoot virus Molecular Variability in Ghana. Plant Dis. 2016, 100, 2011–2017. [Google Scholar] [CrossRef]
- Ameyaw, G.A.; Wetten, A.; Dzahini-Obiatey, H.; Domfeh, O.; Allainguillaume, J. Investigation on Cacao swollen shoot virus (CSSV) pollen transmission through cross-pollination. Plant Pathol. 2013, 62, 421–427. [Google Scholar] [CrossRef]
- Trebissou, C.I.; N’guessan, R.K.; Tahi, M.G.; Aïdara, S.; Akaffou, S.D.; N’guetta, S.-P.A. Response of Forty (40) High-Producing Cacao Genotypes to Cacao Swollen Shoot Disease. J. Exp. Agric. Int. 2020, 42, 1–10. [Google Scholar] [CrossRef]
- Ameyaw, G.; Dzahini-Obiatey, H.; Domfeh, O. Perspectives on cocoa swollen shoot virus disease (CSSVD) management in Ghana. Crop. Prot. 2014, 65, 64–70. [Google Scholar] [CrossRef]
- Friscina, A.; Chiappetta, L.; Jacquemond, M.; Tepfer, M. Infection of non-host model plant species with the narrow-host-range Cacao swollen shoot virus. Mol. Plant Pathol. 2017, 18, 293–297. [Google Scholar] [CrossRef]
- Chingandu, N.; Zia-Ur-Rehman, M.; Sreenivasan, T.N.; Surujdeo-Maharaj, S.; Umaharan, P.; Gutierrez, O.A.; Brown, J.K. Molecular Characterization of Previously Elusive Badnaviruses Associated with Symptomatic Cacao in the New World. Arch. Virol. 2017, 162, 1363–1371. [Google Scholar] [CrossRef]
- Chingandu, N.; Kouakou, K.; Aka, R.; Ameyaw, G.; Gutierrez, O.A.; Herrmann, H.-W.; Brown, J.K. The Proposed New Species, Cacao Red Vein Virus, and Three Previously Recognized Badnavirus Species Are Associated with Cacao Swollen Shoot Disease. Virol. J. 2017, 14, 199. [Google Scholar] [CrossRef] [PubMed]
- Kouakou, K.; Kébé, B.I.; Kouassi, N.; Aké, S.; Cilas, C.; Muller, E. Geographical Distribution of Cacao swollen shoot virus Molecular Variability in Côte d’Ivoire. Plant Dis. 2012, 96, 1445–1450. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.-W.; Mackenzie, A.; Torronen, M.; Gibbs, A. Nucleotide Sequence of the Virion Protein Gene of Cacao Yellow Mosaic Tymovirus. Nucleic Acids Res. 1990, 18, 5886. [Google Scholar] [CrossRef] [PubMed]
- Puig, A.S. Detection of Cacao Mild Mosaic Virus (CaMMV) Using Nested PCR and Evidence of Uneven Distribution in Leaf Tissue. Agronomy 2021, 11, 1842. [Google Scholar] [CrossRef]
- Guiraud, B.S.H.B.; Tahi, G.M.; Trebissou, I.C.; Coulibaly, K.; N’Guessan, W.P.; N’Guessan, K.F.; Muller, E.; Zoro, B.I. Genetic Diversity and Identification of Cocoa Swollen Shoot Virus Detected in Tolerant Cocoa Trees (Theobroma cacao L.) in West and Central West Department of Côte d’Ivoire 2023. Available online: https://www.researchsquare.com/article/rs-3307863/v1 (accessed on 10 April 2025).
- Ameyaw, G.A.; Dzahini-Obiatey, H.K.; Domfeh, O.; Oppong, F.K.; Abaka-Ewusie, K. History and Data Analyses of ‘Cutting Out’ Method for Cocoa Swollen Shoot Virus Disease (CSSVD) Control in Ghana. J. Plant Dis. Prot. 2015, 122, 200–206. [Google Scholar] [CrossRef]
- Ofori, A.; Padi, F.K.; Ameyaw, G.A.; Dadzie, A.M.; Opoku-Agyeman, M.; Domfeh, O.; Ansah, F.O. Field evaluation of the impact of cocoa swollen shoot virus disease infection on yield traits of different cocoa (Theobroma cacao L.) clones in Ghana. PLoS ONE 2022, 17, e0262461. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Argout, X.; Salse, J.; Aury, J.-M.; Guiltinan, M.J.; Droc, G.; Gouzy, J.; Allegre, M.; Chaparro, C.; Legavre, T.; Maximova, S.N.; et al. The Genome of Theobroma cacao. Nat. Genet. 2011, 43, 101–108. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Huang, X.; Madan, A. CAP3: A DNA Sequence Assembly Program. Genome Res. 1999, 9, 868–877. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and Sensitive Protein Alignment Using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Singh, U.; Wurtele, E.S. Orfipy: A Fast and Flexible Tool for Extracting ORFs. Bioinformatics 2021, 37, 3019–3020. [Google Scholar] [CrossRef]
- Wistrand, M.; Sonnhammer, E.L.L. Improved Profile HMM Performance by Assessment of Critical Algorithmic Features in SAM and HMMER. BMC Bioinform. 2005, 6, 99. [Google Scholar] [CrossRef]
- Bigot, T.; Temmam, S.; Pérot, P.; Eloit, M. RVDB-Prot, A Reference Viral Protein Database and Its HMM Profiles. F1000Research 2019, 8, 530. [Google Scholar] [CrossRef]
- Finn, R.D.; Bateman, A.; Clements, J.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Heger, A.; Hetherington, K.; Holm, L.; Mistry, J.; et al. Pfam: The Protein Families Database. Nucleic Acids Res. 2014, 42, D222–D230. [Google Scholar] [CrossRef]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An Ultra-Fast Single-Node Solution for Large and Complex Metagenomics Assembly via Succinct de Bruijn Graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-Length Transcriptome Assembly from RNA-Seq Data Without a Reference Genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Li, W.; Godzik, A. Cd-hit: A Fast Program for Clustering and Comparing Large Sets of Protein or Nucleotide Sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A. AliView: A Fast and Lightweight Alignment Viewer and Editor for Large Datasets. Bioinformatics 2014, 30, 3276–3278. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Xiao, J.; Wang, X.; Zheng, Z.; Wu, Y.; Wang, Z.; Li, H.; Li, P. Molecular Characterization of a Novel Deltaflexivirus Infecting the Edible Fungus Pleurotus Ostreatus. Arch. Virol. 2023, 168, 162. [Google Scholar] [CrossRef]
- Bejerman, N.; Debat, H. Exploring the Tymovirales Landscape Through Metatranscriptomics Data. Arch. Virol. 2022, 167, 1785–1803. [Google Scholar] [CrossRef]
- Ramos-González, P.L.; Arena, G.D.; Tassi, A.D.; Chabi-Jesus, C.; Kitajima, E.W.; Freitas-Astúa, J. Kitaviruses: A Window to Atypical Plant Viruses Causing Nonsystemic Diseases. Annu. Rev. Phytopathol. 2023, 61, 97–118. [Google Scholar] [CrossRef]
- Chiba, S.; Suzuki, N.; Velasco, L.; Ayllón, M.A.; Lee-Marzano, S.-Y.; Sun, L.; Sabanadzovic, S.; Turina, M. ICTV Virus Taxonomy Profile: Fusariviridae 2024. J. Gen. Virol. 2024, 105, 001973. [Google Scholar] [CrossRef]
- Le Gall, O.; Christian, P.; Fauquet, C.M.; King, A.M.Q.; Knowles, N.J.; Nakashima, N.; Stanway, G.; Gorbalenya, A.E. Picornavirales, a Proposed Order of Positive-Sense Single-Stranded RNA Viruses with a Pseudo-T = 3 Virion Architecture. Arch. Virol. 2008, 153, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Teycheney, P.-Y.; Geering, A.D.W.; Dasgupta, I.; Hull, R.; Kreuze, J.F.; Lockhart, B.; Muller, E.; Olszewski, N.; Pappu, H.; Pooggin, M.M.; et al. ICTV Virus Taxonomy Profile: Caulimoviridae. J. Gen. Virol. 2020, 101, 1025–1026. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.; Ullah, I.; Dunwell, J.M.; Daymond, A.J.; Richardson, M.; Allainguillaume, J.; Wetten, A. Identification and Distribution of Novel Badnaviral Sequences Integrated in the Genome of Cacao (Theobroma cacao). Sci. Rep. 2021, 11, 8270. [Google Scholar] [CrossRef] [PubMed]
- Wemheuer, F.; Berkelmann, D.; Wemheuer, B.; Daniel, R.; Vidal, S.; Bisseleua Daghela, H.B. Agroforestry Management Systems Drive the Composition, Diversity, and Function of Fungal and Bacterial Endophyte Communities in Theobroma cacao Leaves. Microorganisms 2020, 8, 405. [Google Scholar] [CrossRef]
- Santana, J.O.; Gramacho, K.P.; de Souza Eduvirgens Ferreira, K.T.; Rezende, R.P.; Mangabeira, P.A.O.; Dias, R.P.M.; Couto, F.M.; Pirovani, C.P. Witches’ Broom Resistant Genotype CCN51 Shows Greater Diversity of Symbiont Bacteria in Its Phylloplane than Susceptible Genotype Catongo. BMC Microbiol. 2018, 18, 194. [Google Scholar] [CrossRef]
- Illeghems, K.; De Vuyst, L.; Papalexandratou, Z.; Weckx, S. Phylogenetic Analysis of a Spontaneous Cocoa Bean Fermentation Metagenome Reveals New Insights into Its Bacterial and Fungal Community Diversity. PLoS ONE 2012, 7, e38040. [Google Scholar] [CrossRef]
- Agyirifo, D.S.; Wamalwa, M.; Otwe, E.P.; Galyuon, I.; Runo, S.; Takrama, J.; Ngeranwa, J. Metagenomics Analysis of Cocoa Bean Fermentation Microbiome Identifying Species Diversity and Putative Functional Capabilities. Heliyon 2019, 5, e02170. [Google Scholar] [CrossRef]
- Almeida, O.G.G.; De Martinis, E.C.P. Metagenome-Assembled Genomes Contribute to Unraveling of the Microbiome of Cocoa Fermentation. Appl. Environ. Microbiol. 2021, 87, e0058421. [Google Scholar] [CrossRef]
- Fernández-Niño, M.; Rodríguez-Cubillos, M.J.; Herrera-Rocha, F.; Anzola, J.M.; Cepeda-Hernández, M.L.; Mejía, J.L.A.; Chica, M.J.; Olarte, H.H.; Rodríguez-López, C.; Calderón, D.; et al. Dissecting Industrial Fermentations of Fine Flavour Cocoa Through Metagenomic Analysis. Sci. Rep. 2021, 11, 8638. [Google Scholar] [CrossRef]
- Mota-Gutierrez, J.; Ferrocino, I.; Giordano, M.; Suarez-Quiroz, M.L.; Gonzalez-Ríos, O.; Cocolin, L. Influence of Taxonomic and Functional Content of Microbial Communities on the Quality of Fermented Cocoa Pulp-Bean Mass. Appl. Environ. Microbiol. 2021, 87, e0042521. [Google Scholar] [CrossRef]
- Santos, J.P.N.; Rodrigues, G.V.P.; Ferreira, L.Y.M.; Monteiro, G.P.; Fonseca, P.L.C.; Lopes, Í.S.; Florêncio, B.S.; da Silva Junior, A.B.; Ambrósio, P.E.; Pirovani, C.P.; et al. The Virome of Cocoa Fermentation-Associated Microorganisms. Viruses 2024, 16, 1226. [Google Scholar] [CrossRef] [PubMed]
- Espinal, R.B.A.; de Santana, S.F.; Santos, V.C.; Lizardo, G.N.R.; Silva, R.J.S.; Corrêa, R.X.; Loguercio, L.L.; Góes-Neto, A.; Pirovani, C.P.; Fonseca, P.L.C.; et al. Uncovering a Complex Virome Associated with the Cacao Pathogens Ceratocystis Cacaofunesta and Ceratocystis Fimbriata. Pathogens 2023, 12, 287. [Google Scholar] [CrossRef] [PubMed]
- Reyes, B.M.D.; Fonseca, P.L.C.; Heming, N.M.; Conceição, L.B.d.A.; Nascimento, K.T.d.S.; Gramacho, K.P.; Arevalo-Gardini, E.; Pirovani, C.P.; Aguiar, E.R.G.R. Characterization of the Microbiota Dynamics Associated with Moniliophthora roreri, Causal Agent of Cocoa Frosty Pod Rot Disease, Reveals New Viral Species. Front. Microbiol. 2024, 13, 1053562. [Google Scholar] [CrossRef]
- Li, F.; Wu, B.; Yan, L.; Qin, X.; Lai, J. Metabolome and Transcriptome Profiling of Theobroma cacao Provides Insights into the Molecular Basis of Pod Color Variation. J. Plant Res. 2021, 134, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Shujaei, K.; Gibbs, A.J.; Hajizadeh, M. On the Evolution and Biogeography of Apple Stem Grooving Capillovirus. AgriXiv 2020, 20203318783. [Google Scholar] [CrossRef]
- Wang, Y.; Zhuang, H.; Yang, Z.; Wen, L.; Wang, G.; Hong, N. Molecular Characterization of an Apple Stem Grooving Virus Isolate from Kiwifruit (Actinidia chinensis) in China. Can. J. Plant Pathol. 2018, 40, 76–83. [Google Scholar] [CrossRef]
- Chung, B.N.; Kwon, S.-J.; Yoon, J.-Y.; Cho, I.-S. First Report of Cnidium officinale as a Natural Host Plant of Apple Stem Grooving Virus in South Korea. Plant Dis. 2022, 106, 338. [Google Scholar] [CrossRef]
- EFSA Panel on Plant Health (PLH); Jeger, M.; Bragard, C.; Caffier, D.; Dehnen-Schmutz, K.; Gilioli, G.; Gregoire, J.; Miret, J.A.J.; MacLeod, A.; Navarro, M.N.; et al. Pest Categorisation of Tatter Leaf Virus. EFSA J. 2017, 15, e05033. [Google Scholar] [CrossRef]
- Chu, Y.-M.; Lim, W.-S.; Yea, S.-J.; Cho, J.-D.; Lee, Y.-W.; Kim, K.-H. Complexity of dsRNA Mycovirus Isolated from Fusarium Graminearum. Virus Genes 2004, 28, 135–143. [Google Scholar] [CrossRef]
- Li, P.; Bhattacharjee, P.; Wang, S.; Zhang, L.; Ahmed, I.; Guo, L. Mycoviruses in Fusarium Species: An Update. Front. Cell. Infect. Microbiol. 2019, 9, 257. [Google Scholar] [CrossRef]
- Marzano, S.-Y.L.; Nelson, B.D.; Ajayi-Oyetunde, O.; Bradley, C.A.; Hughes, T.J.; Hartman, G.L.; Eastburn, D.M.; Domier, L.L. Identification of Diverse Mycoviruses through Metatranscriptomics Characterization of the Viromes of Five Major Fungal Plant Pathogens. J. Virol. 2016, 90, 6846–6863. [Google Scholar] [CrossRef] [PubMed]
- Leandro-Muñoz, M.E.; Tixier, P.; Germon, A.; Rakotobe, V.; Phillips-Mora, W.; Maximova, S.; Avelino, J. Effects of Microclimatic Variables on the Symptoms and Signs Onset of Moniliophthora roreri, Causal Agent of Moniliophthora Pod Rot in Cacao. PLoS ONE 2017, 12, e0184638. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Ospina, J.; Molina-Hernández, J.B.; Chaves-López, C.; Romanazzi, G.; Paparella, A. The Role of Fungi in the Cocoa Production Chain and the Challenge of Climate Change. J. Fungi 2021, 7, 202. [Google Scholar] [CrossRef] [PubMed]
- Myers, J.M.; James, T.Y. Mycoviruses. Curr. Biol. 2022, 32, R150–R155. [Google Scholar] [CrossRef]
- Mu, F.; Li, B.; Cheng, S.; Jia, J.; Jiang, D.; Fu, Y.; Cheng, J.; Lin, Y.; Chen, T.; Xie, J. Nine Viruses from Eight Lineages Exhibiting New Evolutionary Modes That Co-Infect a Hypovirulent Phytopathogenic Fungus. PLOS Pathog. 2021, 17, e1009823. [Google Scholar] [CrossRef]
- Li, K.; Zheng, D.; Cheng, J.; Chen, T.; Fu, Y.; Jiang, D.; Xie, J. Characterization of a Novel Sclerotinia Sclerotiorum RNA Virus as the Prototype of a New Proposed Family Within the Order Tymovirales. Virus Res. 2016, 219, 92–99. [Google Scholar] [CrossRef]
- Poimala, A.; Vainio, E.J. Complete Genome Sequence of a Novel Toti-like Virus from the Plant-Pathogenic Oomycete Phytophthora Cactorum. Arch. Virol. 2020, 165, 1679–1682. [Google Scholar] [CrossRef]
- Botella, L.; Jung, M.H.; Rost, M.; Jung, T. Natural Populations from the Phytophthora palustris Complex Show a High Diversity and Abundance of ssRNA and dsRNA Viruses. J. Fungi 2022, 8, 1118. [Google Scholar] [CrossRef]
- Raco, M.; Vainio, E.J.; Sutela, S.; Eichmeier, A.; Hakalová, E.; Jung, T.; Botella, L. High Diversity of Novel Viruses in the Tree Pathogen Phytophthora Castaneae Revealed by High-Throughput Sequencing of Total and Small RNA. Front. Microbiol. 2022, 13, 911474. [Google Scholar] [CrossRef]
- Quito-Avila, D.F.; Freitas-Astúa, J.; Melzer, M.J. Bluner-, cile-, and higreviruses (Kitaviridae). In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 247–251. ISBN 9780128145166. [Google Scholar]
- Escobar-Garcia, H.A.; de Andrade, D.J.; Carrillo, D.; Ochoa, R. Theobroma cacao, a New Host for Brevipalpus Yothersi (Acari: Tenuipalpidae) in Peru. Acarologia 2021, 61, 211–216. [Google Scholar] [CrossRef]
- Knorr, L.C.; Denmark, H.A. Injury to Citrus by the Mite Brevipalpus Phoenicis12. J. Econ. Èntomol. 1970, 63, 1996–1998. [Google Scholar] [CrossRef]
- Rodrigues, J.C.V.; Gallo-Meagher, M.; Ochoa, R.; Childers, C.C.; Adams, B.J. Mitochondrial DNA and RAPD Polymorphisms in the Haploid Mite Brevipalpus Phoenicis (Acari: Tenuipalpidae). Exp. Appl. Acarol. 2004, 34, 275–290. [Google Scholar] [CrossRef] [PubMed]
- Adegbola, R.O.; Keith, C.V.; Gutierrez, O.A.; Goenaga, R.; Brown, J.K. A Previously Undescribed Polerovirus (Solemoviridae) Infecting Theobroma cacao Germplasm. Plant Dis. 2022, 107, 975. [Google Scholar] [CrossRef] [PubMed]
- Adegbola, R.O.; Keith, C.V.; Gutierrez, O.; Goenaga, R.; Brown, J.K. Complete Genome Characterization of Cacao Leafroll Virus, a Newly Described Cacao-Infecting Polerovirus. Arch. Virol. 2024, 169, 83. [Google Scholar] [CrossRef]
- Ullah, I.; Dunwell, J.M. Bioinformatic, Genetic and Molecular Analysis of Several Badnavirus Sequences Integrated in the Genomes of Diverse Cocoa (Theobroma cacao L.) Germplasm. Saudi J. Biol. Sci. 2023, 30, 103648. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodrigues, G.V.P.; Santos, J.P.N.; Ferreira, L.Y.M.; Conceição, L.B.d.A.; Porto, J.A.M.; Aguiar, E.R.G.R. Theobroma cacao Virome: Exploring Public RNA-Seq Data for Viral Discovery and Surveillance. Viruses 2025, 17, 624. https://doi.org/10.3390/v17050624
Rodrigues GVP, Santos JPN, Ferreira LYM, Conceição LBdA, Porto JAM, Aguiar ERGR. Theobroma cacao Virome: Exploring Public RNA-Seq Data for Viral Discovery and Surveillance. Viruses. 2025; 17(5):624. https://doi.org/10.3390/v17050624
Chicago/Turabian StyleRodrigues, Gabriel Victor Pina, João Pedro Nunes Santos, Lucas Yago Melo Ferreira, Lucas Barbosa de Amorim Conceição, Joel Augusto Moura Porto, and Eric Roberto Guimarães Rocha Aguiar. 2025. "Theobroma cacao Virome: Exploring Public RNA-Seq Data for Viral Discovery and Surveillance" Viruses 17, no. 5: 624. https://doi.org/10.3390/v17050624
APA StyleRodrigues, G. V. P., Santos, J. P. N., Ferreira, L. Y. M., Conceição, L. B. d. A., Porto, J. A. M., & Aguiar, E. R. G. R. (2025). Theobroma cacao Virome: Exploring Public RNA-Seq Data for Viral Discovery and Surveillance. Viruses, 17(5), 624. https://doi.org/10.3390/v17050624







