Adapting Next-Generation Sequencing to in Process CRISPR-Cas9 Genome Editing of Recombinant AcMNPV Vectors: From Shotgun to Tiled-Amplicon Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Line and Maintenance
2.2. Baculovirus Amplification and Quantification
2.3. Plasmid Design and Construction
2.4. Shotgun Sequencing of the p6.9GFP rBEV
2.5. Transfection in Multi-Well Plates
2.6. Infection in Multi-Well Plates
2.7. Harvesting from Multi-Well Plates
2.8. Flow Cytometry Analysis
2.9. Tiled-Amplicon Sequencing Assay for rBEV Genomes
2.9.1. Primer Design and Multiplex PCR
2.9.2. DNA Amplicon Cleanup and Quantification
2.9.3. DNA Library Construction
2.9.4. Sequencing DNA Library Preparation
2.10. Bioinformatics Pipeline for Major Species
3. Results
3.1. Consensus Sequence of p6.9GFP rBEV from Shotgun Sequencing
3.2. Tiled-Amplicon Sequencing of p6.9GFP rBEV
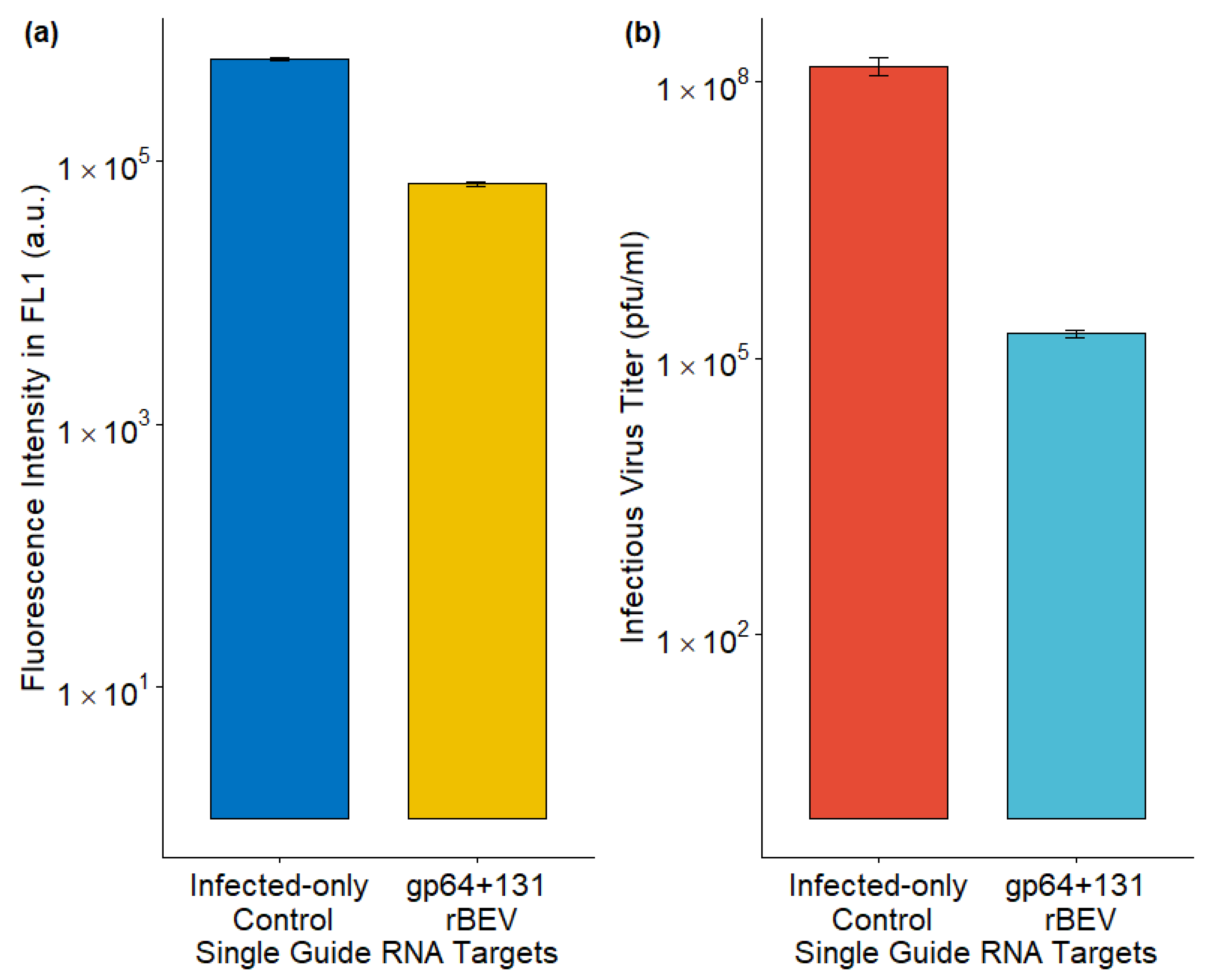
3.3. Lowering Infectious Baculovirus from Sf9-Cas9 Cells Through Co-Expressing a sgRNA Targeting gp64
3.4. Tiled-Amplicon Sequencing of p6.9GFP rBEV upon T-I Assay
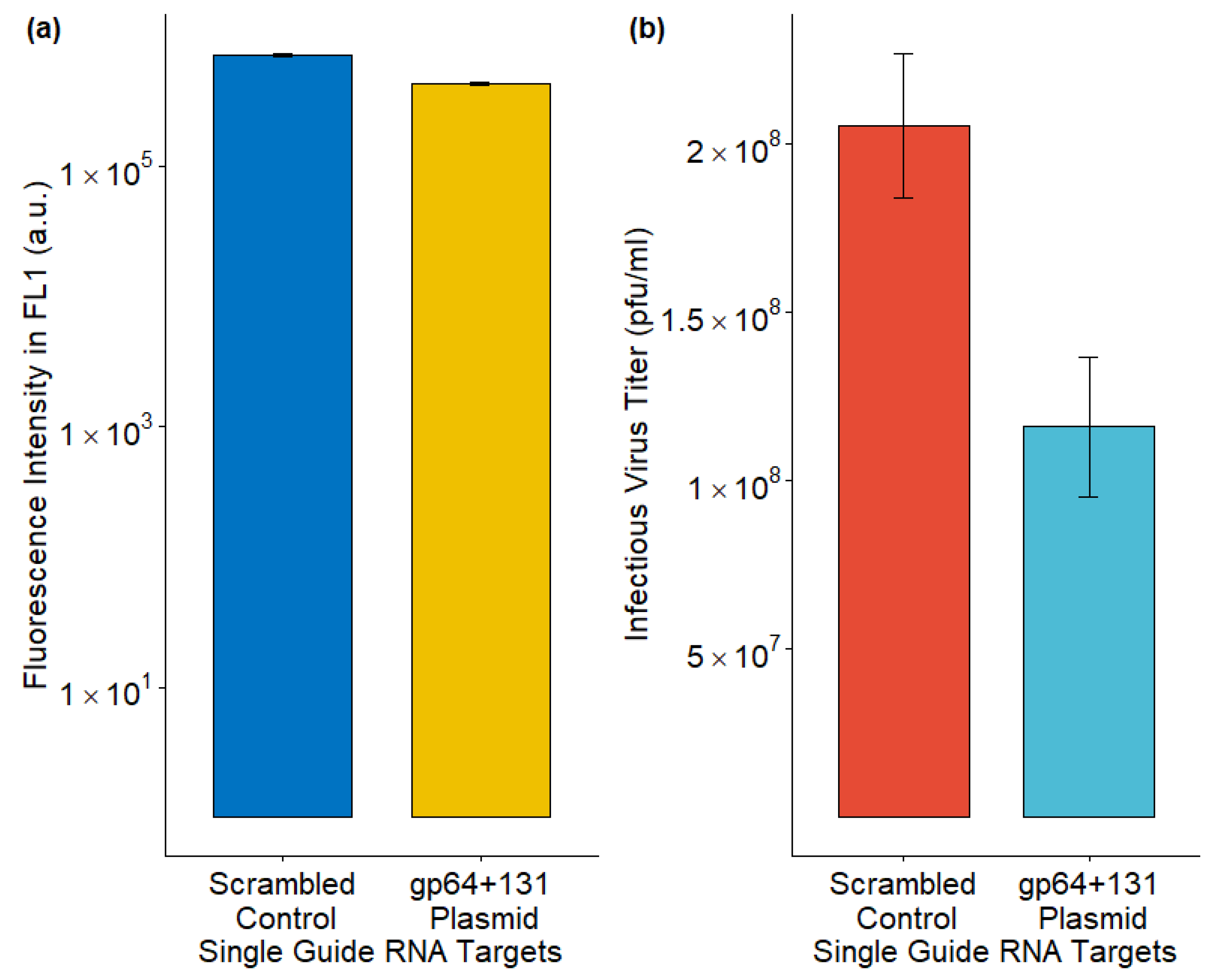
3.5. Lowering Infectious Budded Virus from Sf9-Cas9 Cells Through Plasmid-Based Delivery of sgRNA Targeting gp64
4. Discussion
4.1. Differences Compared to Reported Genomes from Shotgun Sequencing
4.2. Tiled-Amplicon Sequencing to Assess CRISPR-Cas9 Editing
4.2.1. Sequencing Infected-Only and Scrambled Controls
4.2.2. CRISPR-Cas9 Mutations Using p6.9GFP_sgRNA_gp64+131 rBEV in Sf9-Cas9 Cells
4.2.3. CRISPR-Cas9 Mutations Using a gp64+131 Plasmid in Sf9-Cas9 Cells with the T-I Assay
4.3. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AcMNPV | Autographa californica multiple nucleopolyhedrovirus |
| au | arbitrary units |
| BmNPV | Bombyx mori nucleopolyhedrovirus |
| BEVS | baculovirus expression vector system |
| BLT | bead-linked transposomes |
| EPDA | end-point dilution assay |
| eYFP | enhanced yellow fluorescent protein |
| GFP | green fluorescent protein |
| HIV-1 | human immunodeficiency virus type 1 |
| hpi | hours post infection |
| hpt | hours post-transfection |
| hrs | homologous repeat regions |
| HT1 | hybridization buffer |
| IVT | infectious virus titer |
| mAG | monomeric Azami Green |
| MOI | multiplicity of infection |
| NGS | next-generation sequencing |
| ORF | open reading frame |
| ori | origin of replication |
| PBS | phosphate-buffered saline |
| pfu/mL | plaque-forming units per mL |
| rBEV | recombinant baculovirus expression vector |
| SFM | serum-free media |
| sgRNA | single guide RNA |
| SNPs | single nucleotide polymorphisms |
| T-I assay | transfection-infection assay |
| VLP | virus-like particle |
| WP10 | wild population 2010 |
References
- Van Oers, M.M.; Pijlman, G.P.; Vlak, J.M. Thirty years of baculovirus-insect cell protein expression: From dark horse to mainstream technology. J. Gen. Virol. 2015, 96, 6–23. [Google Scholar] [CrossRef] [PubMed]
- Ayres, M.D.; Howard, S.C.; Kuzio, J.; Lopez-Ferber, M.; Possee, R.D. The Complete DNA Sequence of Autographa californica Nuclear Polyhedrosis Virus. Virology 1994, 202, 586–605. [Google Scholar] [CrossRef] [PubMed]
- Miele, S.A.B.; Garavaglia, M.J.; Belaich, M.N.; Ghiringhelli, P.D. Baculovirus: Molecular Insights on Their Diversity and Conservation. Int. J. Evol. Biol. 2011, 2011, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Rohrmann, G.F. The AcMNPV genome: Gene content, conservation, and function. In Baculovirus Molecular Biology; National Center for Biotechnology Information: Bethesda, MD, USA, 2019; pp. 201–275. [Google Scholar]
- Bruder, M.R.; Aucoin, M.G. A sensitive assay for scrutiny of Autographa californica Mult. Nucleopolyhedrovirus Genes Using CRISPR-Cas9. Appl. Microbiol. Biotechnol. 2023, 107, 4323–4335. [Google Scholar] [CrossRef] [PubMed]
- Hitchman, R.B.; Possee, R.D.; Crombie, A.T.; Chambers, A.; Ho, K.; Siaterli, E.; Lissina, O.; Sternard, H.; Novy, R.; Loomis, K.; et al. Genetic modification of a baculovirus vector for increased expression in insect cells. Cell Biol. Toxicol. 2010, 26, 57–68. [Google Scholar] [CrossRef]
- Hitchman, R.B.; Possee, R.D.; Siaterli, E.; Richards, K.S.; Clayton, A.J.; Bird, L.E.; Owens, R.J.; Carpentier, D.C.J.; King, F.L.; Danquah, J.O.; et al. Improved expression of secreted and membrane-targeted proteins in insect cells. Biotechnol. Appl. Biochem. 2010, 56, 85–93. [Google Scholar] [CrossRef]
- Pijlman, G.P.; Van Den Born, E.; Martens, D.E.; Vlak, J.M. Autographa californica baculoviruses with large genomic deletions are rapidly generated in infected insect cells. Virology 2001, 283, 132–138. [Google Scholar] [CrossRef]
- George, S.; Jauhar, A.M.; Mackenzie, J.; Kie, S.; Aucoin, M.G. Temporal Characterization of Protein Production Levels From Baculovirus Vectors Coding for GFP and RFP Genes Under Non-Conventional Promoter Control. Biotechnol. Bioeng. 2015, 112, 1822–1831. [Google Scholar] [CrossRef]
- Bassett, A.R.; Tibbit, C.; Ponting, C.P.; Liu, J.L. Mutagenesis and homologous recombination in Drosophila cell lines using CRISPR/Cas9. Biol. Open 2014, 3, 42–49. [Google Scholar] [CrossRef]
- Chang, J.; Wang, R.; Yu, K.; Zhang, T.; Chen, X.; Liu, Y.; Shi, R.; Wang, X.; Xia, Q.; Ma, S. Genome-wide CRISPR screening reveals genes essential for cell viability and resistance to abiotic and biotic stresses in Bombyx mori. Genome Res. 2020, 30, 757–767. [Google Scholar] [CrossRef]
- Dong, Z.; Chen, T.; Zhang, J.; Hu, N.; Cao, M.; Dong, F.; Jiang, Y.; Chen, P.; Lu, C.; Pan, M. Establishment of a highly efficient virus-inducible CRISPR/Cas9 system in insect cells. Antivir. Res. 2016, 130, 50–57. [Google Scholar] [CrossRef]
- Dong, Z.; Huang, L.; Dong, F.; Hu, Z.; Qin, Q.; Long, J.; Cao, M.; Chen, P.; Lu, C.; Pan, M. Establishment of a baculovirus-inducible CRISPR/Cas9 system for antiviral research in transgenic silkworms. Appl. Microbiol. Biotechnol. 2018, 102, 9255–9265. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, D.; Zhang, X.; Chen, S.; Yang, D.; Tang, L.; Yang, X.; Wang, Y.; Luo, X.; Wang, M.; et al. Construction of baculovirus-inducible CRISPR/Cas9 antiviral system targeting BmNPV in Bombyx mori. Viruses 2022, 14, 59. [Google Scholar] [CrossRef] [PubMed]
- Mabashi-Asazuma, H.; Jarvis, D.L. CRISPR-Cas9 vectors for genome editing and host engineering in the baculovirus–insect cell system. Proc. Natl. Acad. Sci. USA 2017, 114, 9068–9073. [Google Scholar] [CrossRef] [PubMed]
- Bruder, M.R.; Walji, S.D.; Aucoin, M.G. Comparison of CRISPR-Cas9 Tools for Transcriptional Repression and Gene Disruption in the BEVS. Viruses 2021, 13, 1925. [Google Scholar] [CrossRef]
- Bruder, M.R.; Aucoin, M.G. Evaluation of Virus-Free Manufacture of Recombinant Proteins Using CRISPR-Mediated Gene Disruption in Baculovirus-Infected Insect Cells. Vaccines 2023, 11, 225. [Google Scholar] [CrossRef]
- Hausjell, C.S.; Klausberger, M.; Ernst, W.; Grabherr, R. Evaluation of an inducible knockout system in insect cells based on co-infection and CRISPR/Cas9. PLoS ONE 2023, 18, 1–13. [Google Scholar] [CrossRef]
- Quick, J.; Grubaugh, N.D.; Pullan, S.T.; Claro, I.M.; Smith, A.D.; Gangavarapu, K.; Oliveira, G.; Robles-Sikisaka, R.; Rogers, T.F.; Beutler, N.A.; et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017, 12, 1261–1276. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method for estimating fifty per cent endpoints. Am. J. Hyg. 1938, 27, 493–497. [Google Scholar]
- O’Reilly, D.R.; Miller, L.K.; Luckow, V.A. Baculovirus Expression Vectors: A Laboratory Manual; W. H. Freeman and Company: New York, NY, USA, 1992. [Google Scholar]
- Chakraborty, M.; Powichrowski, J.; Bruder, M.R.; Nielsen, L.; Sung, C.; Boegel, S.J.; Aucoin, M.G. Probing Baculovirus Vector Gene Essentiality for Foreign Gene Expression Using a CRISPR-Cas9 System. Methods Mol. Biol. 2024, 2829, 127–156. [Google Scholar]
- Port, F.; Chen, H.M.; Lee, T.; Bullock, S.L. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, E2967–E2976. [Google Scholar] [CrossRef] [PubMed]
- Claudi, B.; Spröte, P.; Chirkova, A.; Personnic, N.; Zankl, J.; Schürmann, N.; Schmidt, A.; Bumann, D. Phenotypic variation of salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell 2014, 158, 722–733. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge—Accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Eren, A.M.; Kiefl, E.; Shaiber, A.; Veseli, I.; Miller, S.E.; Schechter, M.S.; Fink, I.; Pan, J.N.; Yousef, M.; Fogarty, E.C.; et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat. Microbiol. 2021, 6, 3–6. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Briefings Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Pinello, L.; Canver, M.C.; Hoban, M.D.; Orkin, S.H.; Kohn, D.B.; Bauer, D.E.; Yuan, G.C. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016, 34, 695–697. [Google Scholar] [CrossRef]
- Clement, K.; Rees, H.; Canver, M.C.; Gehrke, J.M.; Farouni, R.; Hsu, J.Y.; Cole, M.A.; Liu, D.R.; Joung, J.K.; Bauer, D.E.; et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 2019, 37, 220–224. [Google Scholar] [CrossRef]
- Boezen, D.; Ali, G.; Wang, M.; Wang, X.; van der Werf, W.; Vlak, J.M.; Zwart, M.P. Empirical estimates of the mutation rate for an alphabaculovirus. PLoS Genet. 2022, 18, e1009806. [Google Scholar] [CrossRef] [PubMed]
- Maghodia, A.B.; Jarvis, D.L.; Geisler, C. Complete genome sequence of the Autographa californica multiple nucleopolyhedrovirus strain E2. Genome Announc. 2014, 2, 6–7. [Google Scholar] [CrossRef]
- Labun, K.; Montague, T.G.; Krause, M.; Torres Cleuren, Y.N.; Tjeldnes, H.; Valen, E. CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019, 47, W171–W174. [Google Scholar] [CrossRef] [PubMed]
- Pushko, P.; Tumpey, T.M.; Bu, F.; Knell, J.; Robinson, R.; Smith, G.E. Influenza virus-like particles comprised of the HA, NA, and M1 proteins of H9N2 influenza virus induce protective immune responses in BALB/c mice. Vaccine 2005, 23, 5751–5759. [Google Scholar] [CrossRef]
- Pushko, P.; Kort, T.; Nathan, M.; Pearce, M.B.; Smith, G.E.; Tumpey, T.M. Recombinant H1N1 virus-like particle vaccine elicits protective immunity in ferrets against the 2009 pandemic H1N1 influenza virus. Vaccine 2010, 28, 4771–4776. [Google Scholar] [CrossRef] [PubMed]
- Aucoin, M.G.; Mena, J.A.; Kamen, A.A. Bioprocessing of Baculovirus Vectors: A Review. Curr. Gene Ther. 2010, 10, 174–186. [Google Scholar] [CrossRef]
- Tomalski, M.D.; Wu, J.G.; Miller, L.K. The location, sequence, transcription, and regulation of a baculovirus DNA polymerase gene. Virology 1988, 167, 591–600. [Google Scholar] [CrossRef]
- Chateigner, A.; Bézier, A.; Labrousse, C.; Jiolle, D.; Barbe, V.; Herniou, E.A. Ultra deep sequencing of a baculovirus population reveals widespread genomic variations. Viruses 2015, 7, 3625–3646. [Google Scholar] [CrossRef]
- Harrison, R.L.; Bonning, B.C. Comparative analysis of the genomes of Rachiplusia ou and Autographa californica multiple nucleopolyhedroviruses. J. Gen. Virol. 2003, 84, 1827–1842. [Google Scholar] [CrossRef]
- Kumar, S.; Miller, L.K. Effects of serial passage of Autographa californica nuclear polyhedrosis virus in cell culture. Virus Res. 1987, 7, 335–349. [Google Scholar] [CrossRef]
- Cory, J.; Clarke, E.; Brown, M.; Hails, R.; O’Reilly, D. Microparasite manipulation of an insect: The influence of the egt gene on the interaction between a baculovirus and its lepidopteran host. Funct. Ecol. 2004, 18, 443–450. [Google Scholar] [CrossRef]
- Pazmiño-Ibarra, V.; Mengual-Martí, A.; Targovnik, A.M.; Herrero, S. Improvement of baculovirus as protein expression vector and as biopesticide by CRISPR/Cas9 editing. Biotechnol. Bioeng. 2019, 116, 2823–2833. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ma, S.; Wang, X.; Chang, J.; Gao, J.; Shi, R.; Zhang, J.; Lu, W.; Liu, Y.; Zhao, P.; et al. Highly efficient multiplex targeted mutagenesis and genomic structure variation in Bombyx mori cells using CRISPR/Cas9. Insect Biochem. Mol. Biol. 2014, 49, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.; Ye, Q.; Zhao, Z.; Wang, N.; Li, J.; Zou, M.; Lau, C.H.; Zhu, H.; Wang, S.; Ding, Y. Development of a baculoviral CRISPR/Cas9 vector system for beta-2-microglobulin knockout in human pluripotent stem cells. Mol. Genet. Genom. 2024, 299, 1–13. [Google Scholar] [CrossRef]
- Carstens, E.B.; Wu, Y. No single homologous repeat region is essential for DNA replication of the baculovirus Autographa californica multiple nucleopolyhedrovirus. J. Gen. Virol. 2007, 88, 114–122. [Google Scholar] [CrossRef]
- Bossert, M.; Carstens, E.B. Sequential deletion of AcMNPV homologous regions leads to reductions in budded virus production and late protein expression. Virus Res. 2018, 256, 125–133. [Google Scholar] [CrossRef]
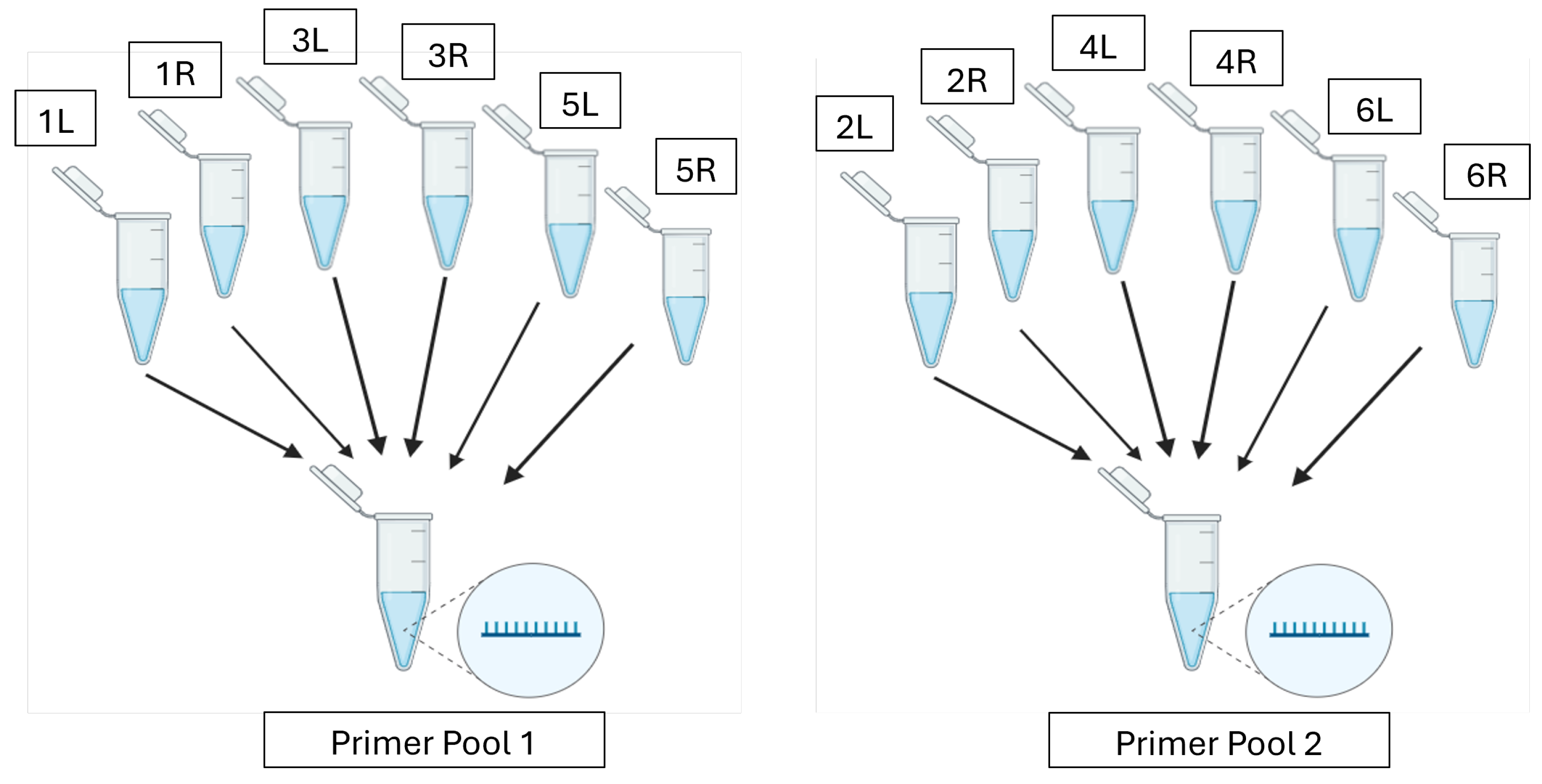

| PCR Component | PCR Pool 1 | PCR Pool 2 |
|---|---|---|
| Q5® High-Fidelity 2× Master Mix | 12.5 µL | 12.5 µL |
| Primer Pool 1 (10 µM) | 2.5 µL | - |
| Primer Pool 2 (10 µM) | - | 2.5 µL |
| DNA Template (1–10 ng) 1 | 10 µL | 10 µL |
| PCR Step Number | PCR Step | Temperature | Duration |
|---|---|---|---|
| 1 | Initial denaturation | 98 °C | 30 s |
| 2 | Denaturation | 95 °C | 15 s |
| 3 | Annealing/extension | 63 °C | 5 min |
| N/A | Repeat steps 2 and 3 for 35 cycles | N/A | N/A |
| 4 | Hold | 10 °C | Infinite |
| Mutation Region | Mutation Type | Mutation Length (bp) | AcMNPV C6 Sequence | Shotgun Sequence | a Amino Acid Change | Position on Shotgun Sequence |
|---|---|---|---|---|---|---|
| Homologous region 2 (hr2) | Deletion | 49 | ttaaact 1 | - | Not applicable | 27,333 |
| Homologous region 2 (hr2) | Deletion | 137 | aactcgc 2 | - | Not applicable | 27,371 |
| Homologous region 3 (hr3) | Deletion | 72 | aatcgtg 3 | - | Not applicable | 71,257 |
| Homologous region 3 (hr3) | Deletion | 72 | caagtag 4 | - | Not applicable | 71,341 |
| Homologous region 2 (hr2) | Insertion | 1 | - | C | Not applicable | 27,341 |
| Homologous region 2 (hr2) | SNPs | 1 | C | G | Not applicable | 27,416 |
| Homologous region 3 (hr3) | SNPs | 1 | A | C | Not applicable | 71,255 |
| Homologous region 3 (hr3) | SNPs | 1 | A | G | Not applicable | 71,294 |
| Homologous region 3 (hr3) | SNPs | 1 | A | G | Not applicable | 71,306 |
| Homologous region 3 (hr3) | SNPs | 1 | C | G | Not applicable | 71,378 |
| Homologous region 5 (hr5) | SNPs | 1 | T | C | Not applicable | 115,201 |
| Homologous region 5 (hr5) | SNPs | 1 | T | C | Not applicable | 115,250 |
| chiA/v-cath promoter | Deletion | 37 | tttaatt 5 | - | Not applicable | 105,599 |
| AcOrf-106 promoter | Insertion | 1 | - | A | Not applicable | 94,054 |
| AcOrf-106 promoter | Insertion | 1 | - | A | Not applicable | 94,124 |
| AcOrf-106 promoter | Insertion | 1 | - | C | Not applicable | 94,172 |
| fgf promoter | SNPs | 1 | G | T | Not applicable | 28,133 |
| AcOrf-57 promoter | SNPs | 1 | A | T | Not applicable | 47,546 |
| AcOrf-78 promoter | SNPs | 1 | G | C | Not applicable | 65,943 |
| chiA/v-cath promoter | SNPs | 7 | taaaaaa | cccgggc | Not applicable | 105,601 |
| ‡ lef2/AcOrf-603 recombination region | SNPs | 2 | CT | TC | Not applicable | 3743 |
| pk-1 | Deletion | 1 | T | - | - | 8157 |
| AcOrf-17 | Deletion | 1 | A | - | - | 14,943 |
| AcOrf-21 | Deletion | 1 | C | - | - | 17,024 |
| AcOrf-112 | Deletion | 1 | A | - | - | 97,094 |
| ie-01 | Deletion | 2 | CG | - | - | 122,767 |
| ie-01 | Deletion | 1 | A | - | - | 123,928 |
| pk-1 | Insertion | 1 | - | C | His | 8143 |
| egt | Insertion | 40 | - | ctagaga 6 | † LEISRDL*RSLEIS | 12,428 |
| AcOrf-20 | Insertion | 3 | - | cgg | Pro | 16,922 |
| AcOrf-52 | Insertion | 1 | - | G | Ala | 44,907 |
| AcOrf-59 | Insertion | 1 | - | A | Asn | 48,451 |
| AcOrf-106 | Insertion | 2 | - | CA | ThrAsn | 94,260 |
| AcOrf-106 | Insertion | 1 | - | G | Glu | 94,293 |
| AcOrf-106 | Insertion | 2 | - | CG | Pro | 94,343 |
| AcOrf-107 | Insertion | 6 | - | atttgg | IleTrp | 94,422 |
| AcOrf-107 | Insertion | 1 | - | A | Arg | 94,433 |
| pe/pp34 | Insertion | 1 | - | G | Thr | 109,299 |
| AcOrf-1629 | SNPs | 1 | G | A | Phe | 6371 |
| AcOrf-11 | SNPs | 1 | G | A | Asp | 8685 |
| AcOrf-11 | SNPs | 1 | G | A | Trp | 9434 |
| AcOrf-20 | SNPs | 1 | A | G | Leu | 16,888 |
| AcOrf-20 | SNPs | 2 | GC | CA | Leu | 16,925 |
| AcOrf-20 | SNPs | 2 | AG | GA | ProPro | 16,930 |
| AcOrf-20 | SNPs | 2 | CG | GC | ProGlu | 16,933 |
| env-prot | SNPs | 1 | T | A | Val | 21,007 |
| p47 | SNPs | 1 | A | C | Val | 33,392 |
| gta | SNPs | 1 | G | A | Asn | 34,844 |
| gta | SNPs | 1 | G | T | Phe | 34,895 |
| odv-e66 | SNPs | 1 | C | G | Met | 37,998 |
| odv-e66 | SNPs | 1 | T | C | Ser | 38,304 |
| AcOrf-51 | SNPs | 1 | A | T | Phe | 44,182 |
| AcOrf-53 | SNPs | 1 | C | T | Phe | 45,551 |
| AcOrf-54 | SNPs | 1 | T | G | Val | 46,633 |
| AcOrf-57 | SNPs | 1 | C | T | Leu | 47,770 |
| AcOrf-58 | SNPs | 1 | T | G | Arg | 48,249 |
| lef9 | SNPs | 1 | G | A | Asn | 50,209 |
| lef9 | SNPs | 1 | G | T | Ser | 50,314 |
| tlp | SNPs | 1 | G | T | Glu | 68,066 |
| AcOrf-84 | SNPs | 1 | C | A | Arg | 71,492 |
| AcOrf-93 | SNPs | 1 | C | G | Glu | 79,916 |
| helicase | SNPs | 1 | G | C | Leu | 81,217 |
| AcOrf-106 | SNPs | 1 | A | C | Asn | 94,263 |
| AcOrf-114 | SNPs | 1 | C | G | Gln | 98,860 |
| AcOrf-120 | SNPs | 1 | A | G | Asp | 102,644 |
| AcOrf-120 | SNPs | 1 | A | G | Leu | 102,664 |
| pk-2 | SNPs | 1 | T | A | Phe | 103,878 |
| 94k | SNPs | 2 | TA | CC | GluVal | 113,302 |
| p10 | SNPs | 1 | C | G | Ser | 116,722 |
| p74 | SNPs | 2 | GT | TG | IleMet | 116,950 |
| p74 | SNPs | 1 | A | G | Gln | 118,441 |
| p74 | SNPs | 1 | A | T | Glu | 118,509 |
| me53 | SNPs | 1 | A | G | Arg | 120,034 |
| me53 | SNPs | 1 | A | G | Thr | 120,036 |
| ie-0/ie-01 | SNPs | 1 | G | A | Gln | 120,596 |
| ie-01 | SNPs | 1 | T | C | Ala | 122,767 |
| ie-01 | SNPs | 1 | T | G | Ala | 122,768 |
| pe38 | SNPs | 1 | G | C | Arg | 131,047 |
| Mutation Region | Mutation Type | Mutation Length (bp) | Reference/Targeted Genome 1 | Position on Targeted rBEV Sequence |
|---|---|---|---|---|
| Homologous region 2 | SNPs | 2 | CC/AT | 28,491 |
| Homologous region 3 | SNPs | 1 | C/G | 72,827 |
| AcOrf-20 promoter | SNPs | 1 | C/G | 17,961 |
| AcOrf-1629 | SNPs | 1 | A/G | 7427 |
| pk-1 | Deletion | 1 | T/- | 9210 |
| pk-1 | Insertion | 1 | -/C | 9195 |
| AcOrf-11 | SNPs | 1 | G/A | 9737 |
| AcOrf-84 | SNPs | 1 | C/A | 72,941 |
| Mutation Region | Mutation Type | Mutation Length (bp) | AcMNPV C6/Scrambled Sequence 1 |
|---|---|---|---|
| Homologous region 2 | Deletion | 35 | aaatgat 2/- |
| Homologous region 3 | SNPs | 1 | A/G |
| egt | Deletion | 20 | ctagaga 3/- |
| Read | Mutation Region | Mutation Type | Mutation Length (bp) | Reference/Targeted gp64 Sequence | Position on Targeted gDNA |
|---|---|---|---|---|---|
| Read 1 | gp64 | Deletion | 12 | gtccttttgcag/- | 108,669 |
| Read 2 | gp64 | Deletion | 2 | CC/- | 108,671 |
| Read 3 | gp64 | Deletion | 1 | C/- | 108,671 |
| Read 4 | gp64 | Deletion | 2 | GT/- | 108,669 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakraborty, M.; Nielsen, L.; Nash, D.; Nissimov, J.I.; Charles, T.C.; Aucoin, M.G. Adapting Next-Generation Sequencing to in Process CRISPR-Cas9 Genome Editing of Recombinant AcMNPV Vectors: From Shotgun to Tiled-Amplicon Sequencing. Viruses 2025, 17, 437. https://doi.org/10.3390/v17030437
Chakraborty M, Nielsen L, Nash D, Nissimov JI, Charles TC, Aucoin MG. Adapting Next-Generation Sequencing to in Process CRISPR-Cas9 Genome Editing of Recombinant AcMNPV Vectors: From Shotgun to Tiled-Amplicon Sequencing. Viruses. 2025; 17(3):437. https://doi.org/10.3390/v17030437
Chicago/Turabian StyleChakraborty, Madhuja, Lisa Nielsen, Delaney Nash, Jozef I. Nissimov, Trevor C. Charles, and Marc G. Aucoin. 2025. "Adapting Next-Generation Sequencing to in Process CRISPR-Cas9 Genome Editing of Recombinant AcMNPV Vectors: From Shotgun to Tiled-Amplicon Sequencing" Viruses 17, no. 3: 437. https://doi.org/10.3390/v17030437
APA StyleChakraborty, M., Nielsen, L., Nash, D., Nissimov, J. I., Charles, T. C., & Aucoin, M. G. (2025). Adapting Next-Generation Sequencing to in Process CRISPR-Cas9 Genome Editing of Recombinant AcMNPV Vectors: From Shotgun to Tiled-Amplicon Sequencing. Viruses, 17(3), 437. https://doi.org/10.3390/v17030437







