Covariation of Amino Acid Substitutions in the HIV-1 Envelope Glycoprotein gp120 and the Antisense Protein ASP Associated with Coreceptor Usage
Abstract
1. Introduction
2. Materials and Methods
2.1. Assembly of a Training Dataset of V3 Sequences with Known Tropism
2.2. Linear Discriminant Analysis (LDA) of the Training Dataset
2.3. Prediction of the Tropism in a Dataset of Viral Strains of Genotype B or C Containing an Antisense Gene asp
2.4. Comparative Analysis of the Amino Acid Composition in the V3 Region Between R5-Tropic and X4-Tropic Viral Strains
2.5. Comparative Analysis of the Amino Acid Composition in the env/asp Overlap Between R5-Tropic and X4-Tropic Viral Strains
2.6. Identification of Significant Patterns of Pairwise Associations Between Amino Acid Substitutions in ASP and Amino Acid Substitutions in V3 Loop Region
3. Results
3.1. Linear Discriminant Analysis (LDA) Accurately Predicts HIV-1 Coreceptor Usage
3.2. Prediction of Tropism in a Dataset of V3 Sequences and Detection of Amino Acid Positions Associated with Coreceptor Usage
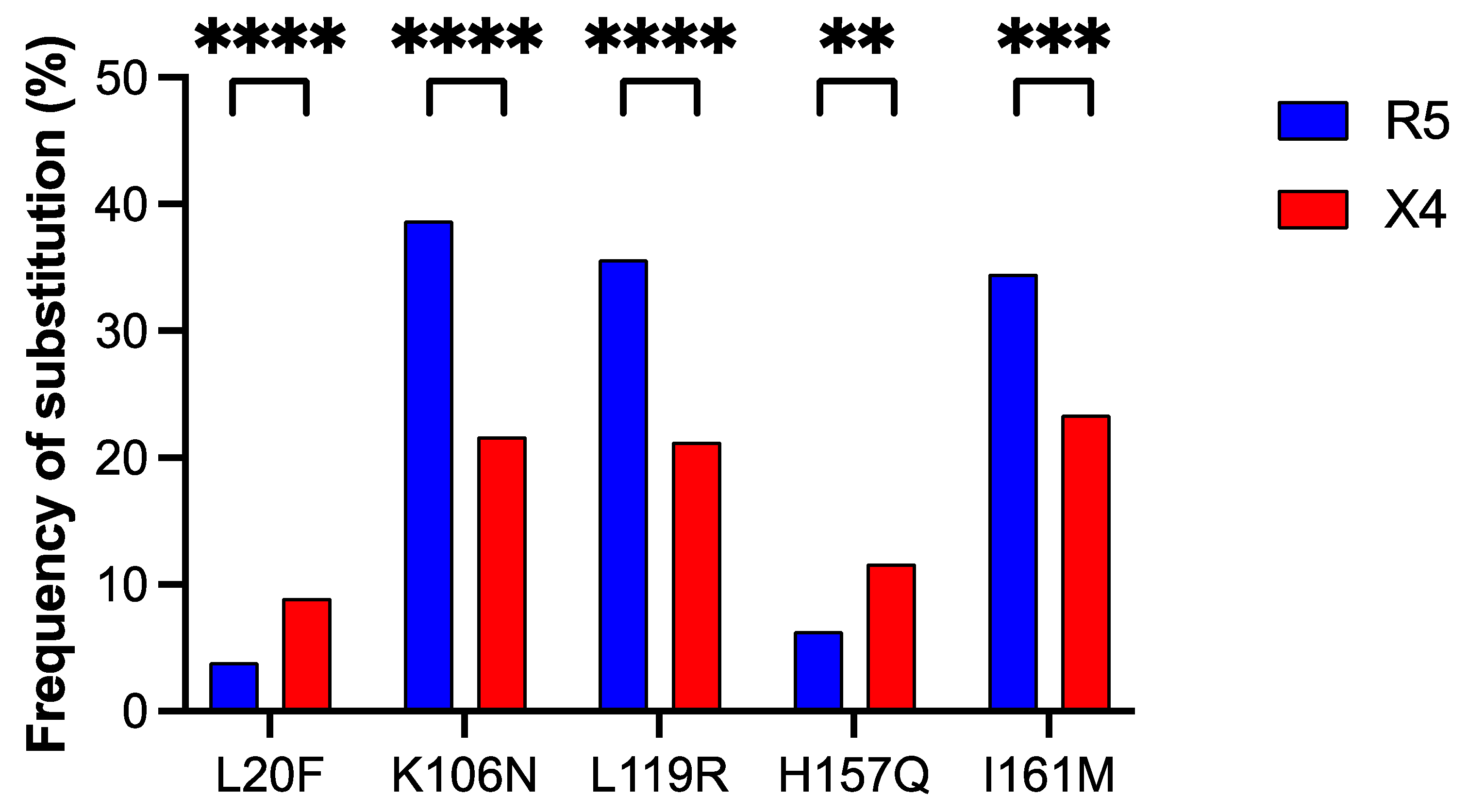
3.3. Detection of Amino Acid Positions in the ASP Protein Associated with Coreceptor Usage
3.4. Detection of Significant Patterns of Association Between V3 and ASP Amino Acid Substitutions
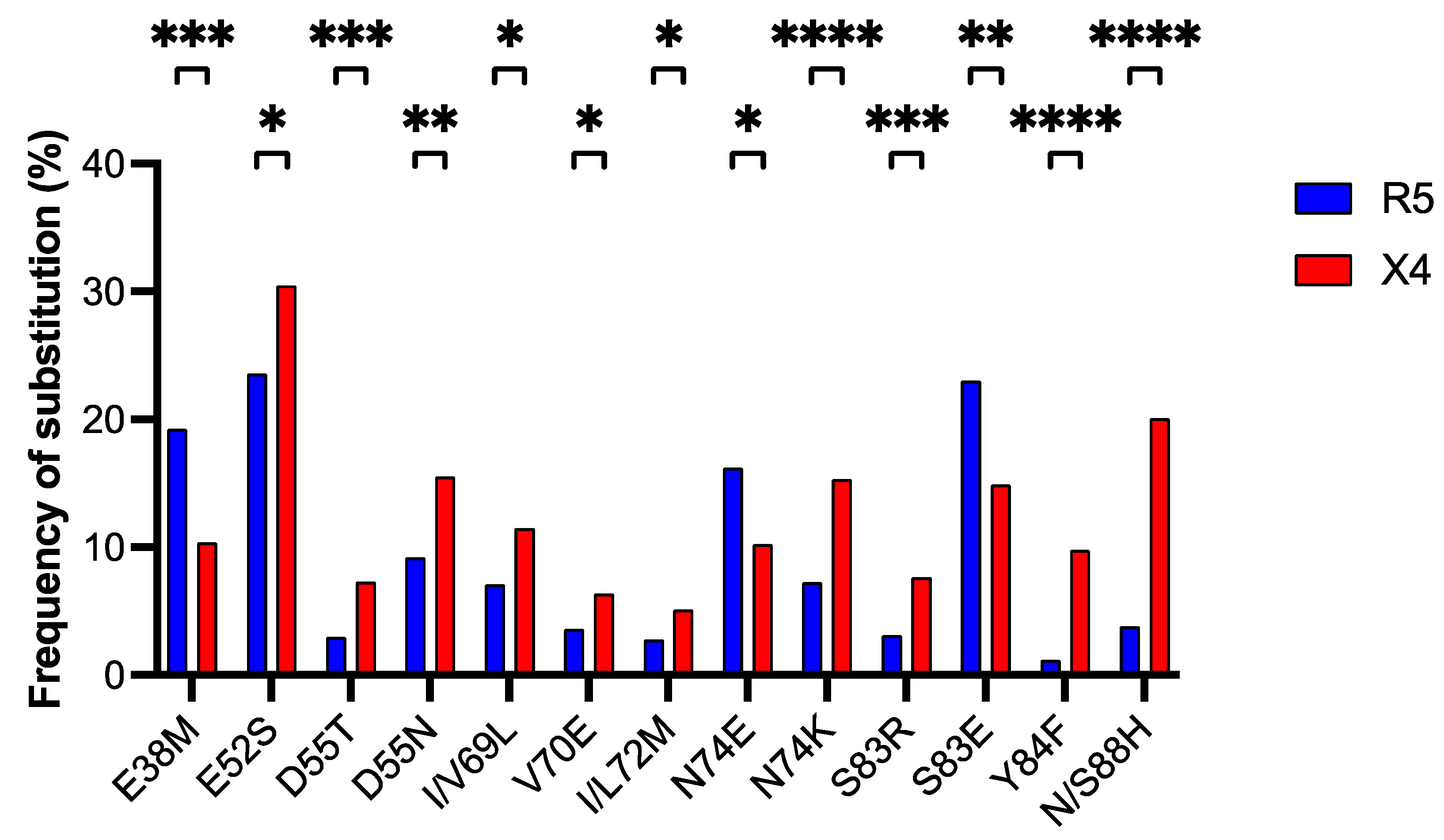
3.5. Detection of Amino Acid Positions in the V1/V2 Region Associated with Coreceptor Usage and Detection of Significant Patterns of Association Between V1/V2 and ASP Substitutions
3.6. Extension of Statistical Analysis to the Regions of gp120 Protein Outside V3 and V1/V2
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kwong, P.D.; Wyatt, R.; Robinson, J.; Sweet, R.W.; Sodroski, J.G.; Hendrickson, W.A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 1998, 393, 648–659. [Google Scholar] [CrossRef] [PubMed]
- Weissenhorn, W.; Dessen, A.; Harrison, S.C.; Skehel, J.J.; Wiley, D.C. Atomic structure of the ectodomain from HIV-1 gp41. Nature 1997, 387, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Liu, R.; Ellmeier, W.; Choe, S.; Unutmaz, D.; Burkhart, M.; Di Marzio, P.; Marmon, S.; Sutton, R.E.; Hill, C.M.; et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature 1996, 381, 661–666. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Berson, J.F.; Chen, Y.; Turner, J.D.; Zhang, T.; Sharron, M.; Jenks, M.H.; Wang, Z.; Kim, J.; Rucker, J.; et al. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc. Natl. Acad. Sci. USA 1997, 94, 6426–6431. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, R.; Sodroski, J.G. The HIV-1 envelope glycoproteins: Fusogens, antigens, and immunogens. Science 1998, 280, 1884–1888. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.S.; Boyle, T.J.; Lyerly, H.K.; Cullen, B.R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 1991, 253, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Regoes, R.R.; Bonhoeffer, S. The HIV coreceptor switch: A population dynamical perspective. Trends Microbiol. 2005, 13, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Whitcomb, J.M.; Huang, W.; Fransen, S.; Limoli, K.; Toma, J.; Wrin, T.; Chappey, C.; Kiss, L.D.; Paxinos, E.E.; Petropoulos, C.J. Development and characterization of a novel single-cycle recombinant-virus assay to determine human immunodeficiency virus type 1 coreceptor tropism. Antimicrob. Agents Chemother. 2007, 51, 566–575. [Google Scholar] [CrossRef]
- De Jong, J.J.; De Ronde, A.; Keulen, W.; Tersmette, M.; Goudsmit, J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: Analysis by single amino acid substitution. J. Virol. 1992, 66, 6777–6780. [Google Scholar] [CrossRef]
- Lengauer, T.; Sander, O.; Sierra, S.; Thielen, A.; Kaiser, R. Bioinformatics prediction of HIV coreceptor usage. Nat. Biotechnol. 2007, 25, 1407–1410. [Google Scholar] [CrossRef] [PubMed]
- Resch, W.; Hoffman, N.; Swanstrom, R. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology 2001, 288, 51–62. [Google Scholar] [CrossRef]
- Pillai, S.; Good, B.; Richman, D.; Corbeil, J. A new perspective on V3 phenotype prediction. AIDS Res. Hum. Retroviruses 2003, 19, 145–149. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Raghava, G.P. Hybrid approach for predicting coreceptor used by HIV-1 from its V3 loop amino acid sequence. PLoS ONE 2013, 8, e61437. [Google Scholar] [CrossRef]
- Jensen, M.A.; Li, F.S.; van’t Wout, A.B.; Nickle, D.C.; Shriner, D.; He, H.X.; McLaughlin, S.; Shankarappa, R.; Margolick, J.B.; Mullins, J.I. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 2003, 77, 13376–13388. [Google Scholar] [CrossRef] [PubMed]
- Bozek, K.; Lengauer, T.; Sierra, S.; Kaiser, R.; Domingues, F.S. Analysis of physicochemical and structural properties determining HIV-1 coreceptor usage. PLoS Comput. Biol. 2013, 9, e1002977. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.S.; Yin, J.; Leng, F.; Teng, R.F.; Xu, C.; Xia, X.Y.; Pan, X.M. HIV coreceptor tropism determination and mutational pattern identification. Sci. Rep. 2016, 6, 21280. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Z.X.; Pan, X.M. HIV-1 tropism prediction by the XGboost and HMM methods. Sci. Rep. 2019, 9, 9997. [Google Scholar] [CrossRef] [PubMed]
- Vandekerckhove, L.P.; Wensing, A.M.; Kaiser, R.; Brun-Vezinet, F.; Clotet, B.; De Luca, A.; Dressler, S.; Garcia, F.; Geretti, A.M.; Klimkait, T.; et al. European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect. Dis. 2011, 11, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Briggs, D.R.; Tuttle, D.L.; Sleasman, J.W.; Goodenow, M.M. Envelope V3 amino acid sequence predicts HIV-1 phenotype (co-receptor usage and tropism for macrophages). AIDS 2000, 14, 2937–2939. [Google Scholar] [CrossRef]
- Fisher, R.A. The use of multiple measurements in taxonomic problems. Ann. Eugen. 1936, 8, 376–386. [Google Scholar] [CrossRef]
- Morrison, D.F. Multivariate Statistical Methods; McGraw-Hill: New York, NY, USA, 1976. [Google Scholar]
- Pavesi, A.; Romerio, F. Creation of the HIV-1 antisense gene asp coincided with the emergence of the pandemic group M and is associated with faster disease progression. Microbiol. Spectr. 2024, 12, e0380223. [Google Scholar] [CrossRef]
- Pavesi, A.; Romerio, F. Extending the Coding Potential of Viral Genomes with Overlapping Antisense ORFs: A Case for the De Novo Creation of the Gene Encoding the Antisense Protein ASP of HIV-1. Viruses 2022, 14, 146. [Google Scholar] [CrossRef] [PubMed]
- Affram, Y.; Zapata, J.C.; Gholizadeh, Z.; Tolbert, W.D.; Zhou, W.; Iglesias-Ussel, M.D.; Pazgier, M.; Ray, K.; Latinovic, O.S.; Romerio, F. The HIV-1 Antisense Protein ASP Is a Transmembrane Protein of the Cell Surface and an Integral Protein of the Viral Envelope. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Gholizadeh, Z.; Iqbal, M.S.; Li, R.; Romerio, F. The HIV-1 Antisense Gene ASP: The New Kid on the Block. Vaccines 2021, 9, 513. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, S.; Pokarowski, P.; Pokarowska, M.; Kolinski, A.; Katayama, T.; Kanehisa, M. AAindex: Amino acid index database, progress report 2008. Nucleic Acids Res. 2008, 36, D202–D205. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Matthews, B.W. Comparison of the predicted and observed secondary structure of T4 phage lysozyme. Biochim. Et Biophys. Acta 1975, 405, 442–451. [Google Scholar] [CrossRef]
- Cardozo, T.; Kimura, T.; Philpott, S.; Weiser, B.; Burger, H.; Zolla-Pazner, S. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res. Hum. Retroviruses 2007, 23, 415–426. [Google Scholar] [CrossRef]
- Sander, O.; Sing, T.; Sommer, I.; Low, A.J.; Cheung, P.K.; Harrigan, P.R.; Lengauer, T.; Domingues, F.S. Structural descriptors of gp120 V3 loop for the prediction of HIV-1 coreceptor usage. PLoS Comput. Biol. 2007, 3, e58. [Google Scholar] [CrossRef]
- Montagna, C.; De Crignis, E.; Bon, I.; Re, M.C.; Mezzaroma, I.; Turriziani, O.; Graziosi, C.; Antonelli, G. V3 net charge: Additional tool in HIV-1 tropism prediction. AIDS Res. Hum. Retroviruses 2014, 30, 1203–1212. [Google Scholar] [CrossRef]
- Kieslich, C.A.; Shin, D.; Lopez de Victoria, A.; Gonzalez-Rivera, G.; Morikis, D. A predictive model for HIV type 1 coreceptor selectivity. AIDS Res. Hum. Retroviruses 2013, 29, 1386–1394. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, L.; Fried, U.; Clevestig, P.; Ehrnst, A. Charged amino acid patterns of coreceptor use in the major subtypes of human immunodeficiency virus type 1. J. Gen. Virol. 2011, 92, 1917–1922. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.W.; Lee, M.K.; Carney, M.C.; Berson, J.F.; Doms, R.W.; Martin, M.A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J. Virol. 1998, 72, 2509–2515. [Google Scholar] [CrossRef]
- Lee, M.K.; Heaton, J.; Cho, M.W. Identification of determinants of interaction between CXCR4 and gp120 of a dual-tropic HIV-1DH12 isolate. Virology 1999, 257, 290–296. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Labrosse, B.; Treboute, C.; Brelot, A.; Alizon, M. Cooperation of the V1/V2 and V3 domains of human immunodeficiency virus type 1 gp120 for interaction with the CXCR4 receptor. J. Virol. 2001, 75, 5457–5464. [Google Scholar] [CrossRef]
- David, C.C.; Jacobs, D.J. Principal component analysis: A method for determining the essential dynamics of proteins. Methods Mol. Biol. 2014, 1084, 193–226. [Google Scholar] [CrossRef]
- Pavesi, A.; Romerio, F. Different Patterns of Codon Usage and Amino Acid Composition across Primate Lentiviruses. Viruses 2023, 15, 1580. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Dong, R.; He, R.L.; Yau, S.S. A novel alignment-free method for HIV-1 subtype classification. Infect. Genet. Evol. 2020, 77, 104080. [Google Scholar] [CrossRef] [PubMed]
- Mardia, K.V. Fisher’s pioneering work on discriminant analysis and its impact on Artificial Intelligence. J. Multivar. Anal. 2024, 203, 105341. [Google Scholar] [CrossRef]
- Nair, M.; Gettins, L.; Fuller, M.; Kirtley, S.; Hemelaar, J. Global and regional genetic diversity of HIV-1 in 2010-21: Systematic review and analysis of prevalence. Lancet Microbe 2024, 5, 100912. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.H. Human immunodeficiency virus may encode a novel protein on the genomic DNA plus strand. Science 1988, 239, 1420–1422. [Google Scholar] [CrossRef] [PubMed]
- Cassan, E.; Arigon-Chifolleau, A.M.; Mesnard, J.M.; Gross, A.; Gascuel, O. Concomitant emergence of the antisense protein gene of HIV-1 and of the pandemic. Proc. Natl. Acad. Sci. USA 2016, 113, 11537–11542. [Google Scholar] [CrossRef] [PubMed]
- Dimonte, S. Different HIV-1 env frames: gp120 and ASP (antisense protein) biosynthesis, and theirs co-variation tropic amino acid signatures in X4- and R5-viruses. J. Med. Virol. 2017, 89, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, Y.; Gao, Y.; Li, G.; Huang, J. Integrated analysis of residue coevolution and protein structures capture key protein sectors in HIV-1 proteins. PLoS ONE 2015, 10, e0117506. [Google Scholar] [CrossRef] [PubMed]
- Arachchilage, M.H.; Piontkivska, H. Coevolutionary Analysis Identifies Protein-Protein Interaction Sites between HIV-1 Reverse Transcriptase and Integrase. Virus Evol. 2016, 2, vew002. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Theys, K.; Verheyen, J.; Pineda-Pena, A.C.; Khouri, R.; Piampongsant, S.; Eusebio, M.; Ramon, J.; Vandamme, A.M. A new ensemble coevolution system for detecting HIV-1 protein coevolution. Biol. Direct 2015, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.H.; Rocco, C.A.; Mangano, A.; Sen, L.; Aulicino, P.C. Sequence variability in p6 gag protein and gag/pol coevolution in human immunodeficiency type 1 subtype F genomes. AIDS Res. Hum. Retroviruses 2013, 29, 1056–1060. [Google Scholar] [CrossRef]
- Travers, S.A.; Tully, D.C.; McCormack, G.P.; Fares, M.A. A study of the coevolutionary patterns operating within the env gene of the HIV-1 group M subtypes. Mol. Biol. Evol. 2007, 24, 2787–2801. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Julien, J.P.; Cupo, A.; Sok, D.; Stanfield, R.L.; Lyumkis, D.; Deller, M.C.; Klasse, P.J.; Burton, D.R.; Sanders, R.W.; Moore, J.P.; et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 2013, 342, 1477–1483. [Google Scholar] [CrossRef]
- Lyumkis, D.; Julien, J.P.; de Val, N.; Cupo, A.; Potter, C.S.; Klasse, P.J.; Burton, D.R.; Sanders, R.W.; Moore, J.P.; Carragher, B.; et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 2013, 342, 1484–1490. [Google Scholar] [CrossRef] [PubMed]
- Caetano, D.G.; Napoleao-Pego, P.; Villela, L.M.; Cortes, F.H.; Cardoso, S.W.; Hoagland, B.; Grinsztejn, B.; Veloso, V.G.; De-Simone, S.G.; Guimaraes, M.L. Patterns of Diversity and Humoral Immunogenicity for HIV-1 Antisense Protein (ASP). Vaccines 2024, 12, 771. [Google Scholar] [CrossRef] [PubMed]
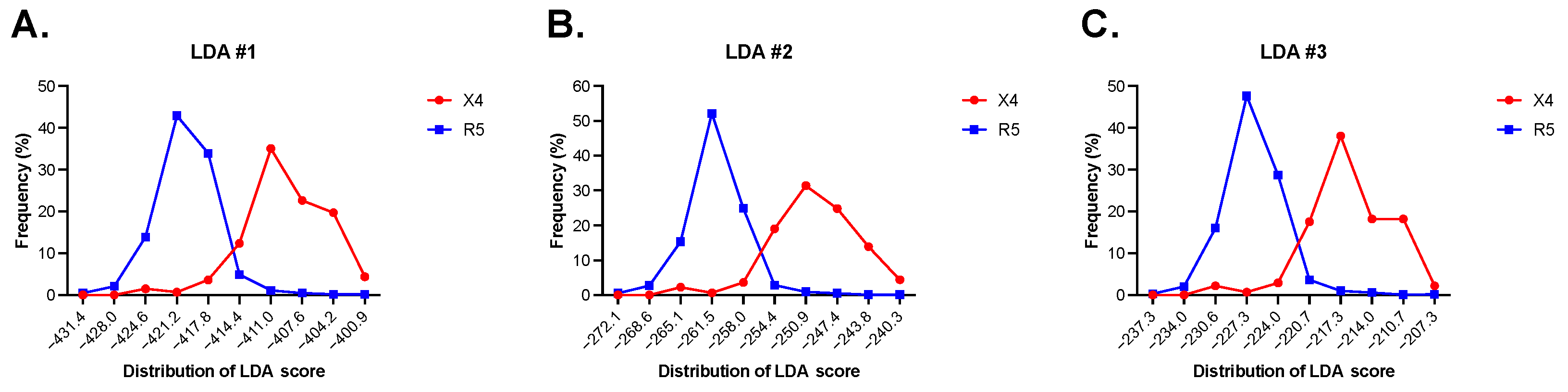
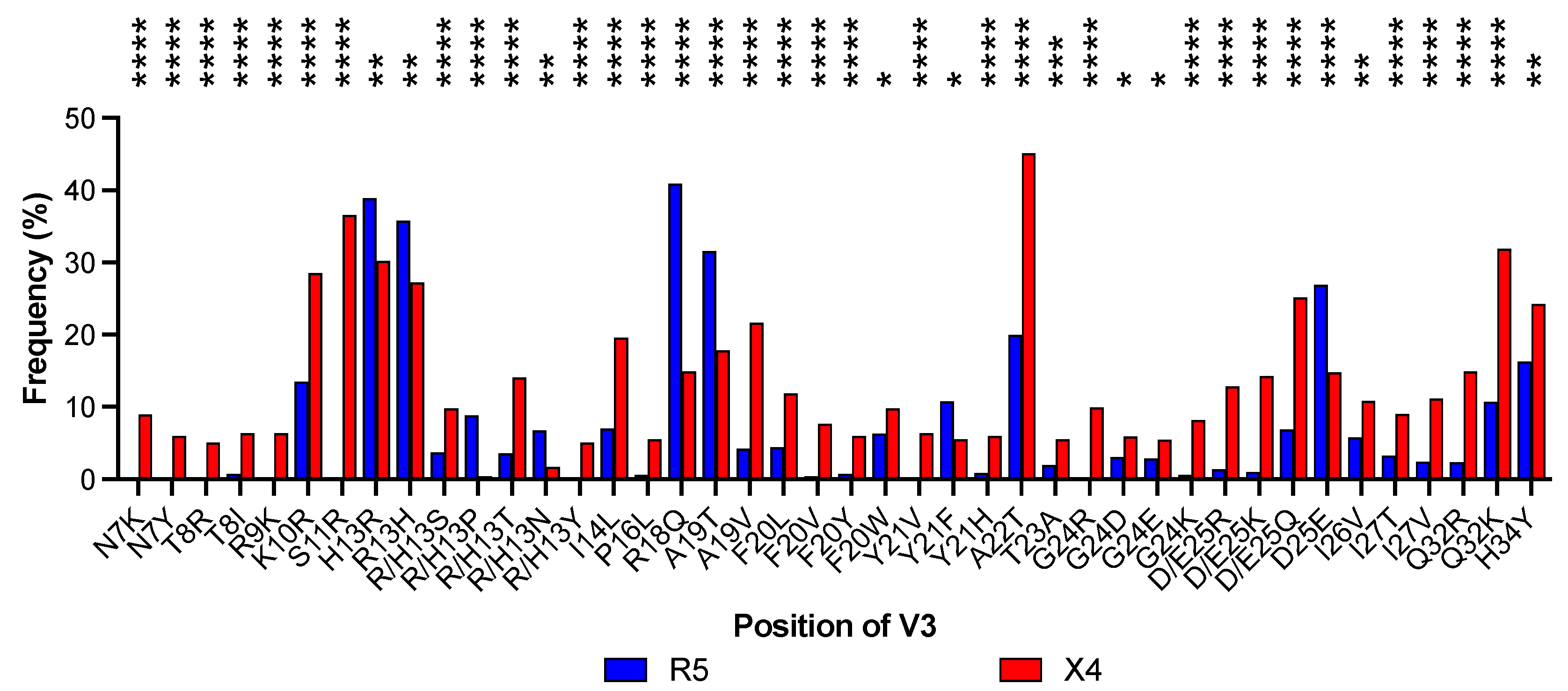
| LDA | Sensitivity 1 | Specificity 2 | Accuracy 3 | MCC 4 | Mean Sensitivity by First Cross-Validation Test | Mean Specificity by First Cross-Validation Test | Mean Accuracy by First Cross-Validation Test | Mean Sensitivity by Second Cross-Validation Test | Mean Specificity by Second Cross-Validation Test | Mean Accuracy by Second Cross-Validation Test |
|---|---|---|---|---|---|---|---|---|---|---|
| LDA#1 | 94.2 | 94.5 | 94.5 | 0.71 | 93.7 | 94.5 | 94.1 | 92.3 | 93.0 | 92.6 |
| LDA#2 | 94.2 | 94.2 | 94.2 | 0.71 | 93.8 | 94.3 | 94.0 | 92.8 | 92.7 | 92.7 |
| LDA#3 | 94.2 | 94.7 | 94.6 | 0.72 | 93.3 | 94.3 | 93.8 | 92.3 | 93.1 | 92.7 |
| ASP Substitution | Location in ASP | V3 Substitution | Prevalent Tropism | φ Binomial Correlation Coefficient 1 | Subtype Prevalence (%) |
|---|---|---|---|---|---|
| K106N | Ectodomain | H13R | R5 | 0.85 (***) | C (93%) |
| R18Q | R5 | 0.87 (***) | C (90%) | ||
| A19T | R5 | 0.56 (**) | C (63%) | ||
| L119R | Ectodomain | H13R | R5 | 0.83 (***) | C (86%) |
| R18Q | R5 | 0.85 (***) | C (87%) | ||
| A19T | R5 | 0.56 (**) | C (60%) | ||
| I161M | C-terminal transmembrane domain | H13R | R5 | 0.56 (**) | C (67%) |
| R18Q | R5 | 0.55 (**) | C (68%) | ||
| A19T | R5 | 0.37 (*) | C (47%) | ||
| L20F | N-terminal intracellular domain | I14L | X4 | 0.45 (*) | B (4.4%) |
| F20W | X4 | 0.67 (**) | B (5.7%) | ||
| D/E25Q | X4 | 0.37 (*) | B (3.9%) | ||
| H157Q | C-terminal transmembrane domain | I14L | X4 | 0.34 (*) | B (5.6%) |
| F20W | X4 | 0.51 (**) | B (5.8%) |
| ASP Substitution | Location in ASP | V1/V2 Substitution | Prevalent Tropism | φ Binomial Correlation Coefficient 1 | Subtype Prevalence (%) |
|---|---|---|---|---|---|
| K106N | Ectodomain | I/L72M | X4 | 0.78 (***) | C (82%) |
| S83E | R5 | 0.60 (**) | C (54%) | ||
| L119R | Ectodomain | I/L72M | X4 | 0.78 (***) | C (78%) |
| S83E | R5 | 0.57 (**) | C (50%) | ||
| I161M | C-terminal transmembrane domain | I/L72M | X4 | 0.51 (**) | B (65%) |
| S83E | R5 | 0.36 (*) | B (39%) | ||
| L20F | N-terminal intracellular domain | D55T | X4 | 0.88 (***) | C (5.2%) |
| H157Q | C-terminal transmembrane domain | D55T | X4 | 0.68 (***) | C (5.3%) |
| ASP Substitution | Region of gp120 | Substitution | Prevalent Tropism | φ Binomial Correlation Coefficient 1 | Subtype Prevalence (%) |
|---|---|---|---|---|---|
| K106N | Upstream of V1/V2 | A1V | R5 | 0.63 (**) | C (64%) |
| T36K | R5 | 0.86 (***) | C (89%) | ||
| T51R | R5 | 0.37 (*) | C (26%) | ||
| L119R | Upstream of V1/V2 | A1V | R5 | 0.66 (**) | C (63%) |
| T36K | R5 | 0.84 (***) | C (85%) | ||
| T51R | R5 | 0.36 (*) | C (25%) | ||
| I161M | Upstream of V1/V2 | A1V | R5 | 0.42 (*) | C (48%) |
| T36K | R5 | 0.57 (**) | C (67%) | ||
| K106N | Between V1/V2 and V3 | V/T4A | R5 | 0.40 (*) | C (30%) |
| F28Y | R5 | 0.82 (***) | C (86%) | ||
| F99L | R5 | 0.67 (**) | C (79%) | ||
| N128V | R5 | 0.56 (**) | C (45%) | ||
| L119R | Between V1/V2 and V3 | V/T4A | R5 | 0.41 (*) | C (41%) |
| F28Y | R5 | 0.81 (***) | C (82%) | ||
| F99L | R5 | 0.69 (**) | C (76%) | ||
| N128V | R5 | 0.57 (**) | C (44%) | ||
| I161M | Between V1/V2 and V3 | V/T4A | R5 | 0.30 (*) | C (24%) |
| F28Y | R5 | 0.55 (**) | C (65%) | ||
| F99L | R5 | 0.45 (*) | C (59%) | ||
| N128V | R5 | 0.38 (*) | C (34%) | ||
| K106N | Downstream of V3 | A/V17S | R5 | 0.33 (*) | C (22%) |
| A/V17G | R5 | 0.34 (*) | C (20%) | ||
| Q28H | R5 | 0.65 (**) | C (67%) | ||
| V/I48K | R5 | 0.36 (*) | C (27%) | ||
| G70R | R5 | 0.63 (**) | C (69%) | ||
| L119R | Downstream of V3 | A/V17S | R5 | 0.32 (*) | C (21%) |
| A/V17G | R5 | 0.34 (*) | C (20%) | ||
| Q28H | R5 | 0.65 (**) | C (63%) | ||
| V/I48K | R5 | 0.34 (*) | C (25%) | ||
| G70R | R5 | 0.61 (**) | C (66%) | ||
| I161M | Downstream of V3 | Q28H | R5 | 0.45 (*) | C (51%) |
| G70R | R5 | 0.39 (*) | C (51%) |
| Region of the ENV Glycoprotein gp120 | Positions Examined (gaps < 5%) | Positions Associated to Tropism (%) | Amino Acid Substitutions for Covariation Test | ASP Substitutions Significantly Correlated | Mean Value of φ Correlation Coefficient |
|---|---|---|---|---|---|
| V3 loop | 35 | 20 (57.1) | 42 | 5 | 0.60 |
| V1/V2 loops | 65 | 10 (15.4) | 13 | 5 | 0.65 |
| Upstream of V1/V2 | 88 | 3 (3.4) | 3 | 3 | 0.59 |
| Between V1/V2 and V3 | 99 | 7 (7.1) | 8 | 3 | 0.55 |
| Downstream of V3 | 49 | 6 (12.2) | 12 | 3 | 0.46 |
| Group 1 1 | 100 | 30 (30) | 55 | 5 | 0.61 3 |
| Group 2 2 | 236 | 16 (6.7) | 23 | 3 | 0.52 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavesi, A.; Romerio, F. Covariation of Amino Acid Substitutions in the HIV-1 Envelope Glycoprotein gp120 and the Antisense Protein ASP Associated with Coreceptor Usage. Viruses 2025, 17, 323. https://doi.org/10.3390/v17030323
Pavesi A, Romerio F. Covariation of Amino Acid Substitutions in the HIV-1 Envelope Glycoprotein gp120 and the Antisense Protein ASP Associated with Coreceptor Usage. Viruses. 2025; 17(3):323. https://doi.org/10.3390/v17030323
Chicago/Turabian StylePavesi, Angelo, and Fabio Romerio. 2025. "Covariation of Amino Acid Substitutions in the HIV-1 Envelope Glycoprotein gp120 and the Antisense Protein ASP Associated with Coreceptor Usage" Viruses 17, no. 3: 323. https://doi.org/10.3390/v17030323
APA StylePavesi, A., & Romerio, F. (2025). Covariation of Amino Acid Substitutions in the HIV-1 Envelope Glycoprotein gp120 and the Antisense Protein ASP Associated with Coreceptor Usage. Viruses, 17(3), 323. https://doi.org/10.3390/v17030323






