Multiple Co-Infecting Caliciviruses in Oral Fluid and Enteric Samples of Swine Detected by a Novel RT-qPCR Assay and a 3′RACE-PCR-NGS Method
Abstract
1. Introduction
2. Materials and Methods
2.1. Design and Setup of the Hydrolysis Probe-Based Triplex RT-qPCR Assay
2.1.1. Production of Virus-Specific RNA Standards
2.1.2. RT-qPCR Reaction Conditions and qPCR Data Analyses
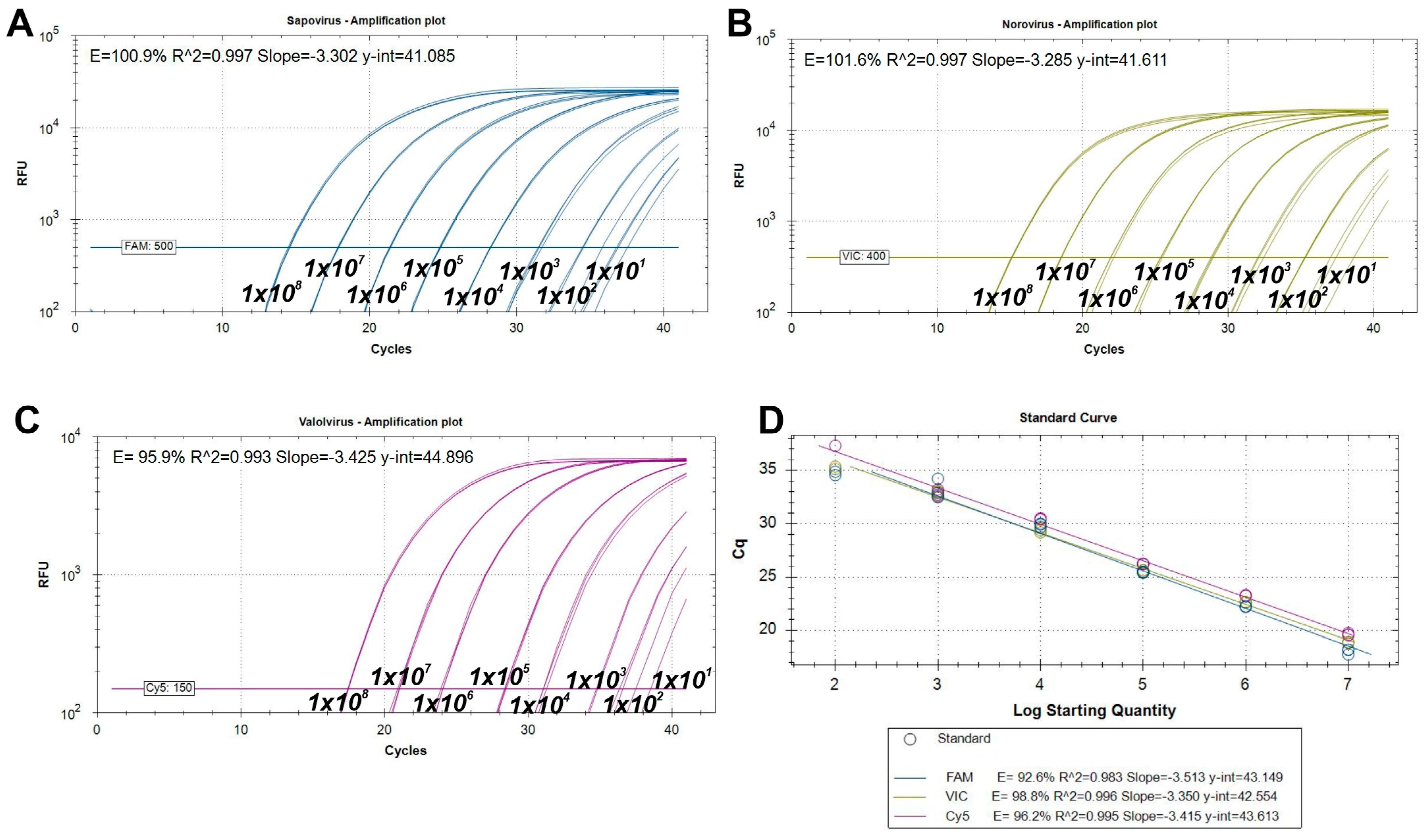
2.1.3. Analytical Performance Tests of the RT-qPCR Assay
2.2. Background Information on Enteric and Oral Fluid Samples, Animals and Farms
2.3. Nucleic Acid Isolation from Enteric and Oral Fluid Samples
2.4. 3′RACE Semi-Nested RT-PCR and Sanger Sequencing
2.5. Next-Generation Sequencing and Data Analysis Pipeline
2.6. Phylogenetic and Sequence Analysis
2.7. Cluster Analysis
2.8. Statistical Analysis
3. Results
3.1. Design and Performance of RT-qPCR Assays
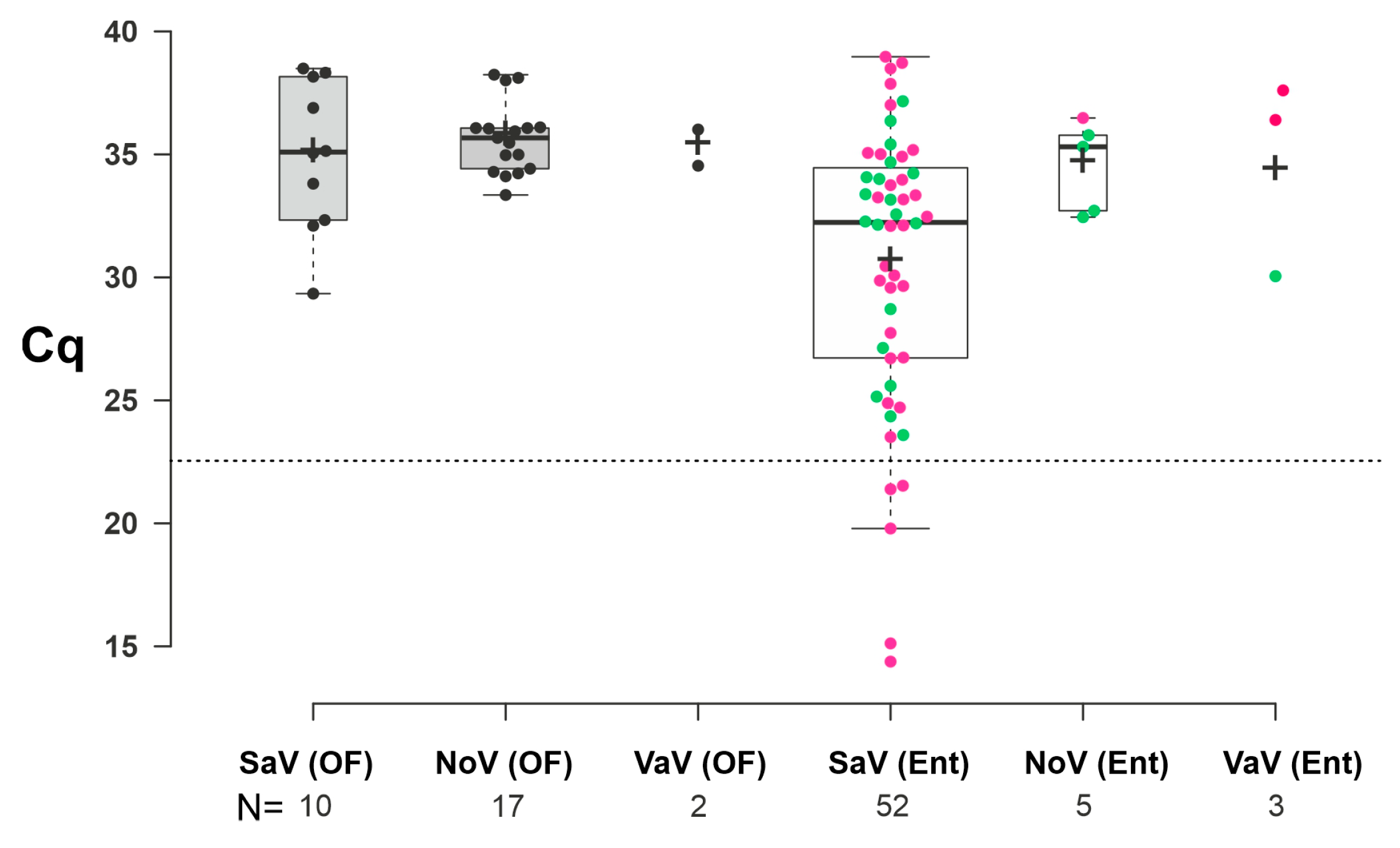
3.2. Epidemiological Investigations of Porcine Enteric Caliciviruses in Enteric and Oral Fluid Samples
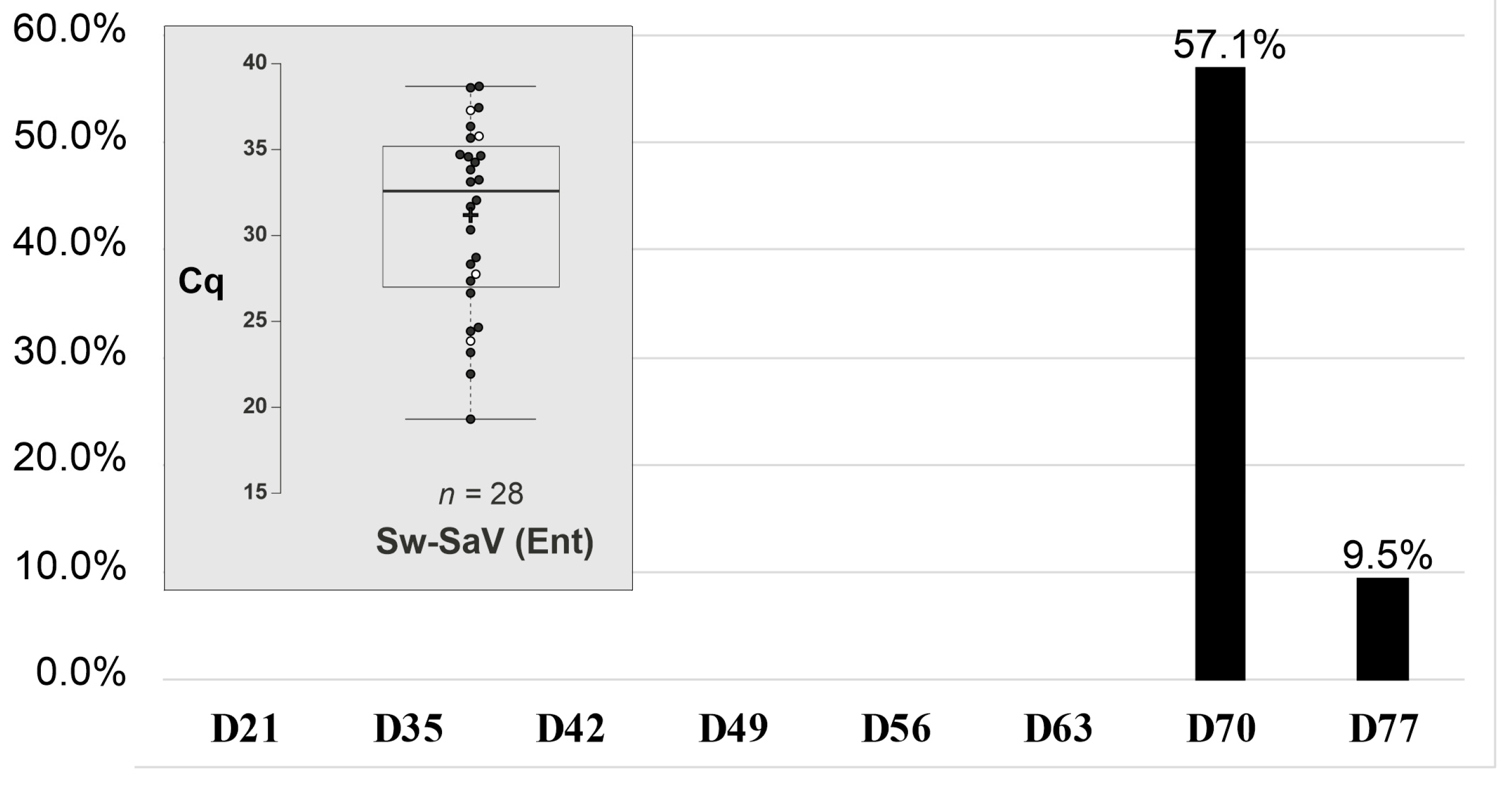
3.3. Retrospective Follow-Up Investigation of Porcine Enteric Caliciviruses in a Newly Established Swine Housing Unit
3.4. Summary of NGS Data Analyses of 3′RACE-snPCR Products
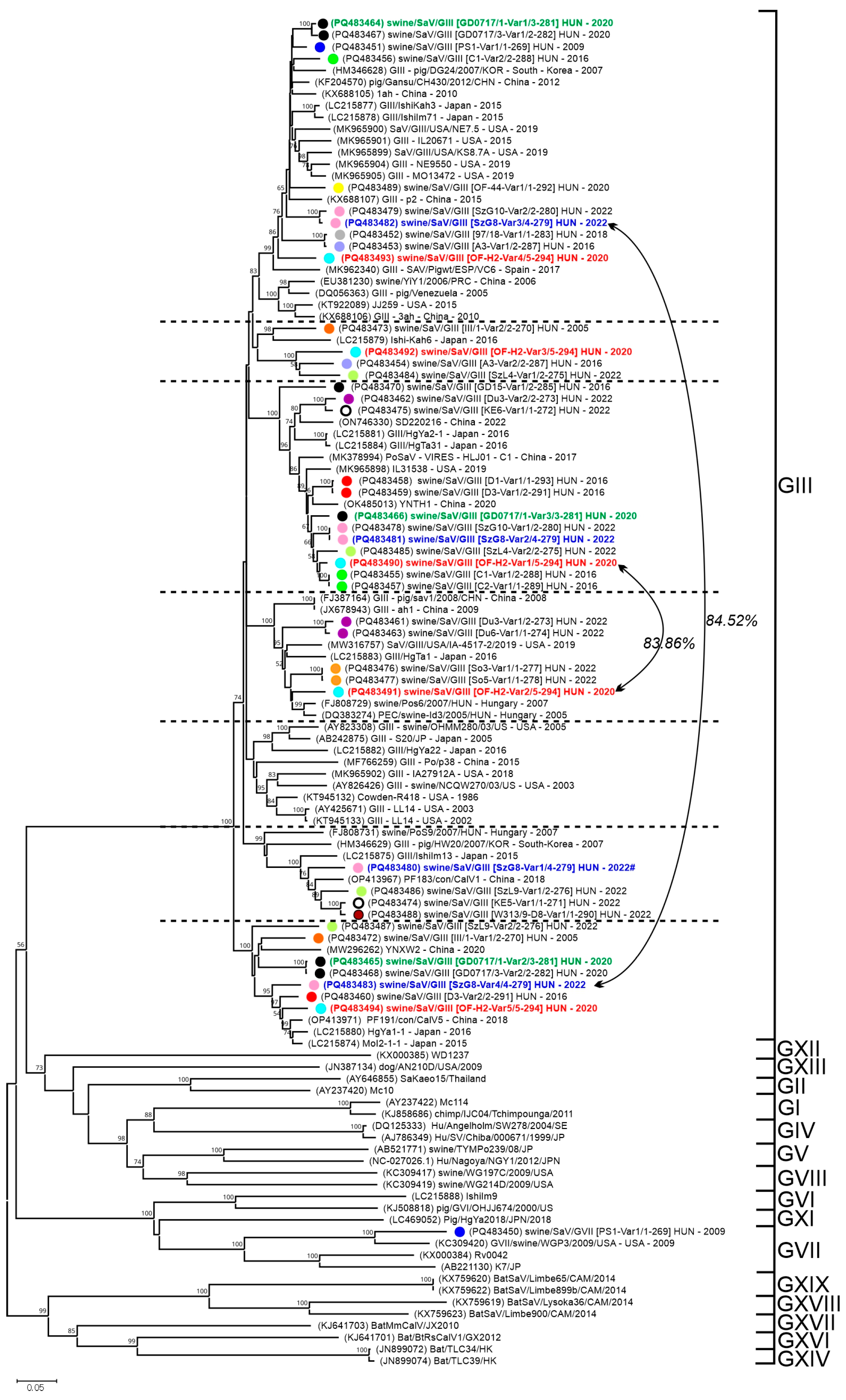
3.4.1. Sequence and VP1-Based Phylogenetic Analyses of Sapovirus Sequences
3.4.2. Sequence and VP1-Based Phylogenetic Analyses of Norovirus Sequences
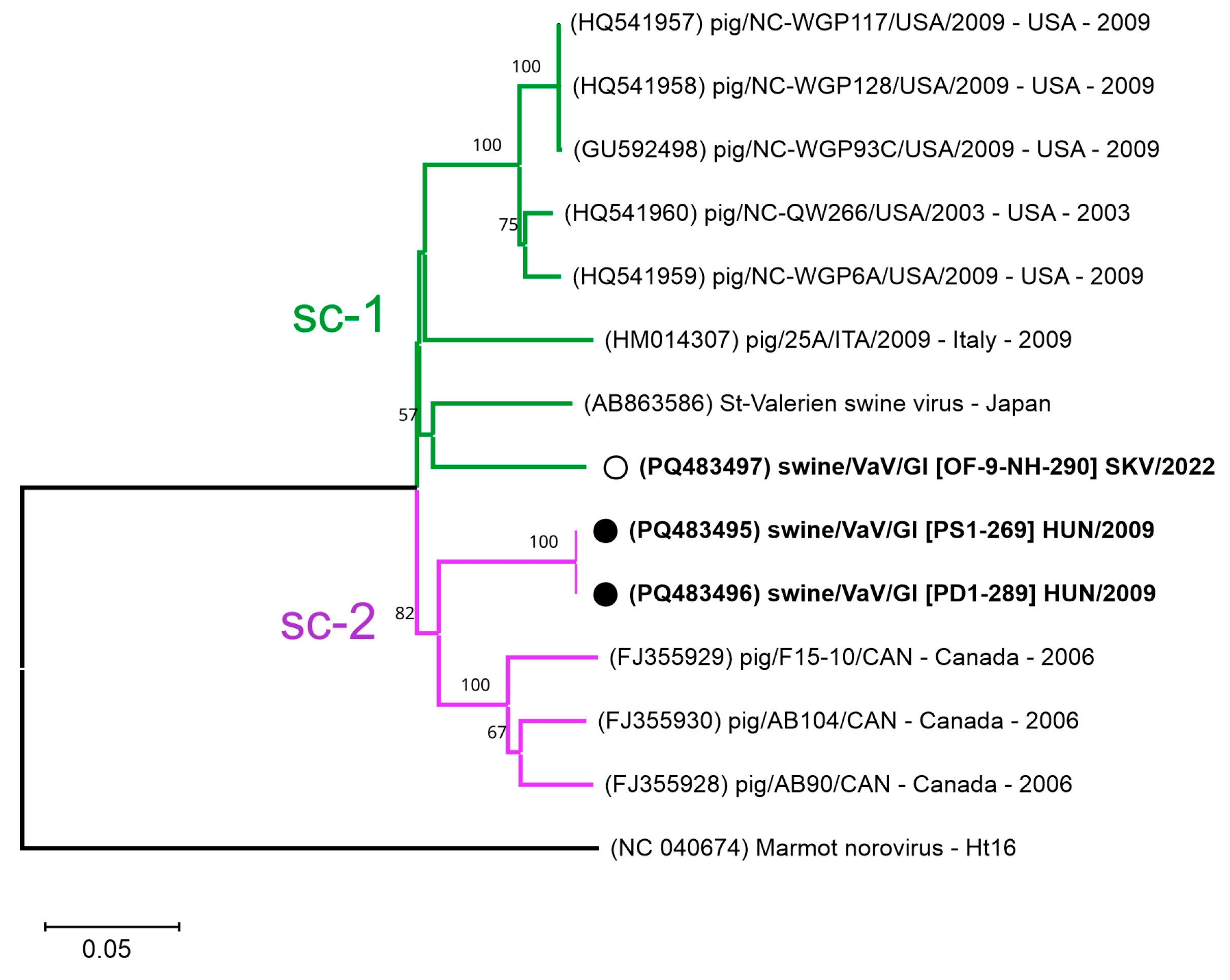
3.4.3. Sequence and VP1-Based Phylogenetic Analyses of Valovirus Sequences
4. Discussion
4.1. Evaluating the Analytical Sensitivity and Efficiency of a Novel Triplex RT-qPCR Assay
4.2. Age-Dependent Prevalence and Mixed Infections of Porcine Enteric Caliciviruses in Enteric Samples
Transient Shedding and High Initial Prevalence of Swine Sapovirus in an Asymptomatic Outbreak
4.3. Age-Dependent Prevalence and Mixed Infections of Porcine Enteric Caliciviruses in Oral Fluid Samples
4.4. Exploring Hidden Variant Diversity of Porcine Enteric Caliciviruses Using Combined 3′RACE-snPCR and NGS Approaches
4.4.1. High Genotypic Diversity and Co-Infection Rates of Sapoviruses
4.4.2. Novel Sub-Clusters and Co-Infections of Norovirus GII.11 and GII.18
4.4.3. Genetic Divergence and Novel Sub-Clusters of Swine Valoviruses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vinjé, J.; Estes, M.K.; Esteves, P.; Green, K.Y.; Katayama, K.; Knowles, N.J.; L’Homme, Y.; Martella, V.; Vennema, H.; White, P.A. ICTV Virus Taxonomy Profile: Caliciviridae. J. Gen. Virol. 2019, 100, 1469–1470. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.H.; Costantini, V.; Saif, L.J. Porcine enteric caliciviruses: Genetic and antigenic relatedness to human caliciviruses, diagnosis and epidemiology. Vaccine 2007, 25, 5453–5466. [Google Scholar] [CrossRef] [PubMed]
- Desselberger, U. Caliciviridae Other Than Noroviruses. Viruses 2019, 11, 286. [Google Scholar] [CrossRef]
- Lucero, Y.; Matson, D.O.; Ashkenazi, S.; George, S.; O’Ryan, M. Norovirus: Facts and Reflections from Past, Present, and Future. Viruses 2021, 13, 2399. [Google Scholar] [CrossRef] [PubMed]
- Chhabra, P.; de Graaf, M.; Parra, G.I.; Chan, M.C.; Green, K.; Martella, V.; Wang, Q.; White, P.A.; Katayama, K.; Vennema, H.; et al. Updated classification of norovirus genogroups and genotypes. J. Gen. Virol. 2019, 100, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Cavicchio, L.; Laconi, A.; Piccirillo, A.; Beato, M.S. Swine Norovirus: Past, Present, and Future. Viruses 2022, 14, 537. [Google Scholar] [CrossRef]
- Winder, N.; Gohar, S.; Muthana, M. Norovirus: An Overview of Virology and Preventative Measures. Viruses 2022, 14, 2811. [Google Scholar] [CrossRef]
- Saito, M.; Goel-Apaza, S.; Espetia, S.; Velasquez, D.; Cabrera, L.; Loli, S.; Crabtree, J.E.; Black, R.E.; Kosek, M.; Checkley, W.; et al. Norovirus Working Group in Peru. Multiple norovirus infections in a birth cohort in a Peruvian Periurban community. Clin. Infect. Dis. 2014, 58, 483–491. [Google Scholar] [CrossRef]
- Sun, Y.; Yuan, Y.; Mao, H.; Su, L.; Ge, Q.; Gao, J.; Xu, C.; Gong, L. Molecular Epidemiology of Human Norovirus Variants from Outbreaks in Zhejiang Province, China, during 2021. Adv. Virol. 2024, 2024, 7972494. [Google Scholar] [CrossRef]
- Davidson, I.; Stamelou, E.; Giantsis, I.A.; Papageorgiou, K.V.; Petridou, E.; Kritas, S.K. The Complexity of Swine Caliciviruses. A Mini Review on Genomic Diversity, Infection Diagnostics, World Prevalence and Pathogenicity. Pathogens 2022, 11, 413. [Google Scholar] [CrossRef]
- Parra, G.I. Emergence of norovirus strains: A tale of two genes. Virus Evol. 2019, 5, vez048. [Google Scholar] [CrossRef] [PubMed]
- Cavicchio, L.; Tassoni, L.; Laconi, A.; Cunial, G.; Gagliazzo, L.; Milani, A.; Campalto, M.; Di Martino, G.; Forzan, M.; Monne, I.; et al. Unrevealed genetic diversity of GII Norovirus in the swine population of North East Italy. Sci. Rep. 2020, 10, 9217. [Google Scholar] [CrossRef] [PubMed]
- Villabruna, N.; Koopmans, M.P.G.; de Graaf, M. Animals as Reservoir for Human Norovirus. Viruses 2019, 11, 478. [Google Scholar] [CrossRef]
- Cunha, J.B.; de Mendonça, M.C.; Miagostovich, M.P.; Leite, J.P. Genetic diversity of porcine enteric caliciviruses in pigs raised in Rio de Janeiro State, Brazil. Arch. Virol. 2010, 155, 1301–1305. [Google Scholar] [CrossRef]
- Oka, T.; Wang, Q.; Katayama, K.; Saif, L.J. Comprehensive review of human sapoviruses. Clin. Microbiol. Rev. 2015, 28, 32–53. [Google Scholar] [CrossRef]
- Nagai, M.; Wang, Q.; Oka, T.; Saif, L.J. Porcine sapoviruses: Pathogenesis, epidemiology, genetic diversity, and diagnosis. Virus Res. 2020, 286, 198025. [Google Scholar] [CrossRef]
- Wang, Q.H.; Souza, M.; Funk, J.A.; Zhang, W.; Saif, L.J. Prevalence of noroviruses and sapoviruses in swine of various ages determined by reverse transcription-PCR and microwell hybridization assays. J. Clin. Microbiol. 2006, 44, 2057–2062. [Google Scholar] [CrossRef]
- Reuter, G.; Zimsek-Mijovski, J.; Poljsak-Prijatelj, M.; Di Bartolo, I.; Ruggeri, F.M.; Kantala, T.; Maunula, L.; Kiss, I.; Kecskeméti, S.; Halaihel, N.; et al. Incidence, diversity, and molecular epidemiology of sapoviruses in swine across Europe. J. Clin. Microbiol. 2010, 48, 363–368. [Google Scholar] [CrossRef]
- L’Homme, Y.; Sansregret, R.; Plante-Fortier, E.; Lamontagne, A.M.; Ouardani, M.; Lacroix, G.; Simard, C. Genomic characterization of swine caliciviruses representing a new genus of Caliciviridae. Virus Genes 2009, 39, 66–75. [Google Scholar] [CrossRef]
- Di Martino, B.; Martella, V.; Di Profio, F.; Ceci, C.; Marsilio, F. Detection of St-Valerien-like viruses in swine, Italy. Vet. Microbiol. 2011, 149, 221–224. [Google Scholar] [CrossRef]
- Wang, Q.; Scheuer, K.; Ahang, Z.; Gebreyes, W.A.; Molla, B.Z.; Hoet, A.E.; Saif, L.J. Characterization and prevalence of a new porcine Calicivirus in Swine, United States. Emerg. Infect. Dis. 2011, 17, 1103–1106. [Google Scholar] [CrossRef]
- Sato, G.; Ido, H.; Kiuchi, M.; Kataoka, M.; Katayama, K.; Tohya, Y. Characterization of St-Valerien-like virus genome detected in Japan. J. Vet. Med. Sci. 2014, 76, 1045–1050. [Google Scholar] [CrossRef]
- Bjustrom-Kraft, J.; Christopher-Hennings, J.; Daly, R.; Main, R.; Torrison, J.; Thurn, M.; Zimmerman, J. The use of oral fluid diagnostics in swine medicine. J. Swine Health Prod. 2018, 26, 262–269. [Google Scholar] [CrossRef]
- Igriczi, B.; Dénes, L.; Biksi, I.; Albert, E.; Révész, T.; Balka, G. High Prevalence of Porcine Circovirus 3 in Hungarian Pig Herds: Results of a Systematic Sampling Protocol. Viruses 2022, 14, 1219. [Google Scholar] [CrossRef]
- Igriczi, B.; Dénes, L.; Czétényi, A.; Révész, T.; Somogyi, Z.; Balka, G. Prevalence Estimation and Genetic Characterization of Porcine Parainfluenza Virus 1 (PPIV-1) in Hungary and the First Report of the Virus in Slovakia. Transbound. Emerg. Dis. 2024, 2024, 5117884. [Google Scholar] [CrossRef]
- Tarasiuk, G.; Remmenga, M.D.; O’Hara, K.C.; Talbert, M.K.; Rotolo, M.L.; Zaabel, P.; Zhang, D.; Giménez-Lirola, L.G.; Zimmerman, J.J. Pen-Based Swine Oral Fluid Samples Contain Both Environmental and Pig-Derived Targets. Animals 2024, 14, 766. [Google Scholar] [CrossRef]
- Jiang, X.; Huang, P.W.; Zhong, W.M.; Farkas, T.; Cubitt, D.W.; Matson, D.O. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J. Virol. Methods 1999, 83, 145–154. [Google Scholar] [CrossRef]
- Vennema, H.; de Bruin, E.; Koopmans, M. Rational optimization of generic primers used for Norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J. Clin. Virol. 2002, 25, 233–235. [Google Scholar] [CrossRef]
- Stals, A.; Mathijs, E.; Baert, L.; Botteldoorn, N.; Denayer, S.; Mauroy, A.; Scipioni, A.; Daube, G.; Dierick, K.; Herman, L.; et al. Molecular detection and genotyping of noroviruses. Food Environ. Virol. 2012, 4, 153–167. [Google Scholar] [CrossRef]
- Tohma, K.; Lepore, C.J.; Martinez, M.; Degiuseppe, J.I.; Khamrin, P.; Saito, M.; Mayta, H.; Nwaba, A.U.A.; Ford-Siltz, L.A.; Green, K.Y.; et al. Genome-wide analyses of human noroviruses provide insights on evolutionary dynamics and evidence of coexisting viral populations evolving under recombination constraints. PLoS Pathog. 2021, 17, e1009744. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, M.; Wildenhain, J.; Rappsilber, J.; Tyers, M. BoxPlotR: A web tool for generation of box plots. Nat. Methods 2014, 11, 121–122. [Google Scholar] [CrossRef] [PubMed]
- Boros, Á.; Pankovics, P.; Simmonds, P.; Reuter, G. Novel positive-sense, single-stranded RNA (+ssRNA) virus with di-cistronic genome from intestinal content of freshwater carp (Cyprinus carpio). PLoS ONE 2011, 6, e29145. [Google Scholar] [CrossRef] [PubMed]
- Boros, Á.; Pankovics, P.; Knowles, N.J.; Reuter, G. Natural interspecies recombinant bovine/porcine enterovirus in sheep. J. Gen. Virol. 2012, 93 Pt 9, 1941–1951. [Google Scholar] [CrossRef]
- László, Z.; Pankovics, P.; Reuter, G.; Cságola, A.; Bálint, Á.; Albert, M.; Boros, Á. Multiple Types of Novel Enteric Bopiviruses (Picornaviridae) with the Possibility of Interspecies Transmission Identified from Cloven-Hoofed Domestic Livestock (Ovine, Caprine and Bovine) in Hungary. Viruses 2021, 13, 66. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kroneman, A.; Vennema, H.; Deforche, K.; Avoort, H.v.d.; Peñaranda, S.; Oberste, M.S.; Vinjé, J.; Koopmans, M. An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 2011, 51, 121–125. [Google Scholar] [CrossRef]
- Rousseeuw, P.J. Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Lloyd, S. Least squares quantization in PCM. IEEE Trans. Inf. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- Tibshirani, R.; Walther, G.; Hastie, T. Estimating the number of clusters in a data set via the gap statistic. J. R. Stat. Soc. Ser. B Stat. Methodol. 2001, 63, 411–423. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing; R Core Team: Vienna, Austria, 2021; Available online: https://www-R-project.org/ (accessed on 23 June 2022).
- Bodenhofer, U.; Bonatesta, E.; Horejš-Kainrath, C.; Hochreiter, S. msa: An R package for multiple sequence alignment. Bioinformatics 2015, 31, 3997–3999. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Claude, J.; Strimmer, K. Phylogenetics and Evolution in R language. Bioinformatics 2004, 20, 289–290. [Google Scholar] [CrossRef] [PubMed]
- Kassambara, A. Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package Version 1. 2016. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 23 June 2022).
- Fisher, R.A. On the interpretation of χ2 from contingency tables, and the calculation of P. J. R. Stat. Soc. 1922, 85, 87–94. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Kralik, P.; Ricchi, M. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef]
- Mijovski, J.Z.; Poljsak-Prijatelj, M.; Steyer, A.; Barlic-Maganja, D.; Koren, S. Detection and molecular characterisation of noroviruses and sapoviruses in asymptomatic swine and cattle in Slovenian farms. Infect. Genet. Evol. 2010, 10, 413–420. [Google Scholar] [CrossRef]
- Di Bartolo, I.; Tofani, S.; Angeloni, G.; Ponterio, E.; Ostanello, F.; Ruggeri, F.M. Detection and characterization of porcine caliciviruses in Italy. Arch. Virol. 2014, 159, 2479–2484. [Google Scholar] [CrossRef]
- Reuter, G.; Bíró, H.; Szucs, G. Enteric caliciviruses in domestic pigs in Hungary. Arch. Virol. 2007, 152, 611–614. [Google Scholar] [CrossRef]
- Li, J.; Shen, Q.; Zhang, W.; Zhao, T.; Li, Y.; Jiang, J.; Yu, X.; Guo, Z.; Cui, L.; Hua, X. Genomic organization and recombination analysis of a porcine sapovirus identified from a piglet with diarrhea in China. Virol. J. 2017, 14, 57. [Google Scholar] [CrossRef]
- Shen, H.; Zhang, J.; Gauger, P.C.; Burrough, E.R.; Zhang, J.; Harmon, K.; Wang, L.; Zheng, Y.; Petznick, T.; Li, G. Genetic characterization of porcine sapoviruses identified from pigs during a diarrhoea outbreak in Iowa, 2019. Transbound. Emerg. Dis. 2022, 69, 1246–1255. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.V.; Truong, T.H.; Tran, G.T.H.; Rapichai, W.; Rattanasrisomporn, A.; Choowongkomon, K.; Rattanasrisomporn, J. Porcine Sapovirus in Northern Vietnam: Genetic Detection and Characterization Reveals Co-Circulation of Multiple Genotypes. Vet. Sci. 2023, 10, 430. [Google Scholar] [CrossRef] [PubMed]
- Collins, P.J.; Martella, V.; Buonavoglia, C.; O’Shea, H. Detection and characterization of porcine sapoviruses from asymptomatic animals in Irish farms. Vet. Microbiol. 2009, 139, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Lauritsen, K.T.; Hansen, M.S.; Johnsen, C.K.; Jungersen, G.; Böttiger, B. Repeated examination of natural sapovirus infections in pig litters raised under experimental conditions. Acta Vet. Scand. 2015, 57, 60. [Google Scholar] [CrossRef]
- Guo, M.; Hayes, J.; Cho, K.O.; Parwani, A.V.; Lucas, L.M.; Saif, L.J. Comparative pathogenesis of tissue culture-adapted and wild-type Cowden porcine enteric calicivirus (PEC) in gnotobiotic pigs and induction of diarrhea by intravenous inoculation of wild-type PEC. J. Virol. 2001, 75, 9239–9251. [Google Scholar] [CrossRef]
- Kirby, A.; Dove, W.; Ashton, L.; Hopkins, M.; Cunliffe, N.A. Detection of norovirus in mouthwash samples from patients with acute gastroenteritis. J. Clin. Virol. 2010, 48, 285–287. [Google Scholar] [CrossRef]
- Ghosh, S.; Kumar, M.; Santiana, M.; Mishra, A.; Zhang, M.; Labayo, H.; Chibly, A.M.; Nakamura, H.; Tanaka, T.; Henderson, W.; et al. Enteric viruses replicate in salivary glands and infect through saliva. Nature 2022, 607, 345–350. [Google Scholar] [CrossRef]
- Anfruns-Estrada, E.; Sabrià, A.; Fuentes, C.; Sabaté, S.; Razquin, E.; Cornejo, T.; Bartolomé, R.; Torner, N.; Izquierdo, C.; Soldevila, N.; et al. Detection of Norovirus in Saliva Samples from Acute Gastroenteritis Cases and Asymptomatic Subjects: Association with Age and Higher Shedding in Stool. Viruses 2020, 12, 1369. [Google Scholar] [CrossRef]
- Wang, Q.H.; Han, M.G.; Cheetham, S.; Souza, M.; Funk, J.A.; Saif, L.J. Porcine noroviruses related to human noroviruses. Emerg. Infect. Dis. 2005, 11, 1874–1881. [Google Scholar] [CrossRef]
- Medici, M.C.; Tummolo, F.; Grazia, S.; Calderaro, A.; Conto, F.; Terio, V.; Chironna, M.; Bonura, F.; Pucci, M.; Bányai, K.; et al. Epidemiological dynamics of norovirus GII.4 variant New Orleans 2009. J. Gen. Virol. 2015, 96, 2919–2927. [Google Scholar] [CrossRef]
- Kuroda, M.; Masuda, T.; Ito, M.; Naoi, Y.; Doan, Y.H.; Haga, K.; Tsuchiaka, S.; Kishimoto, M.; Sano, K.; Omatsu, T.; et al. Genetic diversity and intergenogroup recombination events of sapoviruses detected from feces of pigs in Japan. Infect. Genet. Evol. 2017, 55, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Xiong, K.; Fang, R.; Li, M. Weaning stress and intestinal health of piglets: A review. Front. Immunol. 2022, 13, 1042778. [Google Scholar] [CrossRef] [PubMed]
- Mauroy, A.; Scipioni, A.; Mathijs, E.; Miry, C.; Ziant, D.; Thys, C.; Thiry, E. Noroviruses and sapoviruses in pigs in Belgium. Arch. Virol. 2008, 153, 1927–1931. [Google Scholar] [CrossRef] [PubMed]
- Bull, R.A.; Eden, J.S.; Rawlinson, W.D.; White, P.A. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathog. 2010, 6, e1000831. [Google Scholar] [CrossRef]
- Parra, G.I.; Tohma, K.; Ford-Siltz, L.A.; Eguino, P.; Kendra, J.A.; Pilewski, K.A.; Gao, Y. Antigenic Evolution after a Decade of Norovirus GII.4 Sydney_2012 Circulation in Humans. J. Virol. 2023, 97, e0171622. [Google Scholar] [CrossRef]
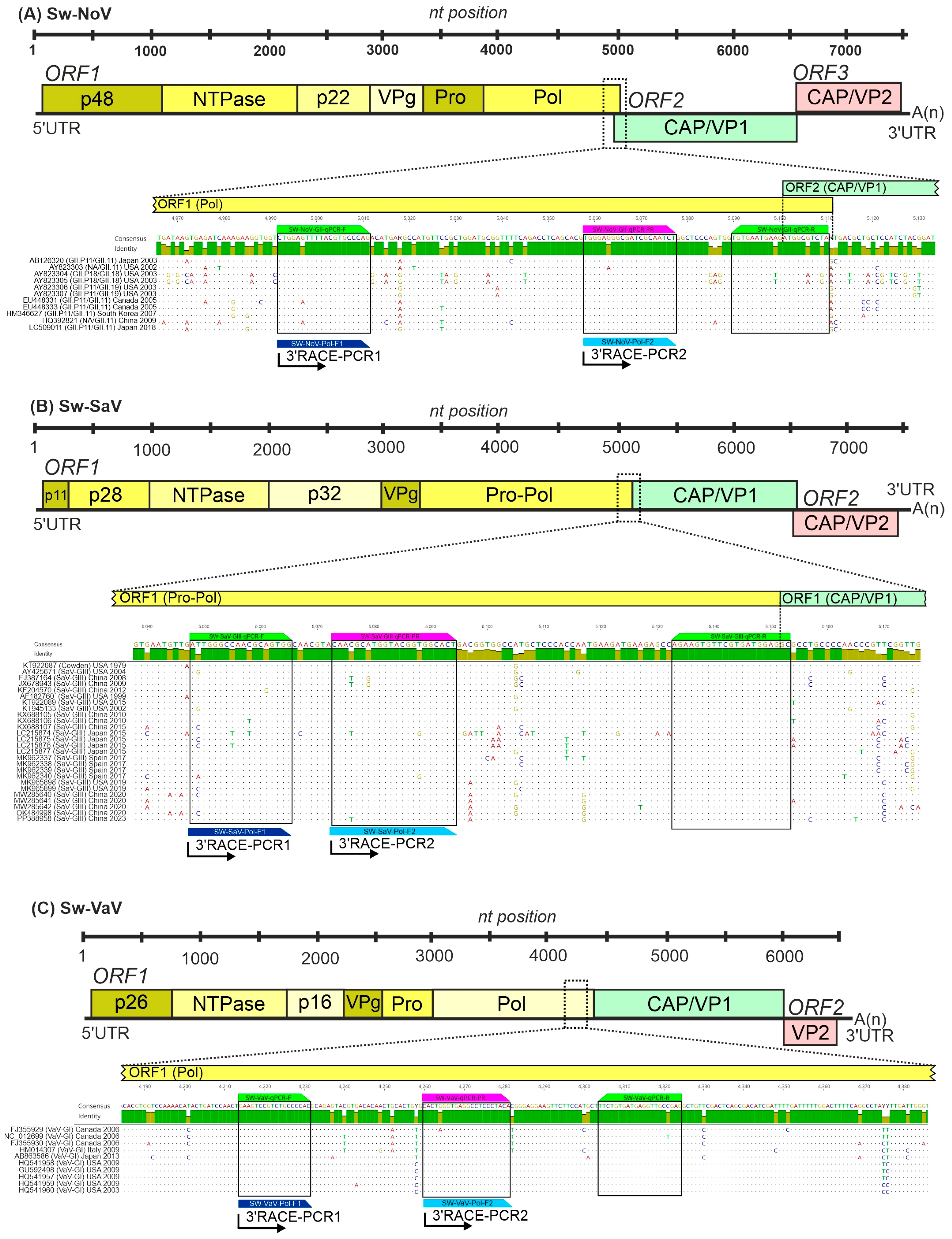

| Target (Region) | Reaction Type | Oligonucleotide Primer ID | Sequence (5′–3′) | Length (nt) | Tm (°C) | PCR Product Size |
|---|---|---|---|---|---|---|
| Swine norovirus GII (Pol-Cap junction) | RT-qPCR | SW-NoV-GII-qPCR-F | CTG GAG TTT TAC GTG CCC AG | 20 | 63 | 120 bp |
| SW-NoV-GII-qPCR-PR | /SUN/TGG GAG GGC/ZEN/GAT CGC AAT CT/IABkFQ/ | 20 | 69 | |||
| SW-NoV-GII-qPCR-R | TAG ACG CCA TCT TCA TTC ACA | 21 | 63 | |||
| 3′RACE-snPCR1 | SW-NoV-Pol-F1 | CTG GAG TTT TAC GTG CCC AG | 20 | 63 | ≈2570 bp * | |
| 3′RACE-snPCR2 | SW-NoV-Pol-F2 | TGG GAG GGC GAT CGC AAT CT | 20 | 69 | ≈2500 bp * | |
| Swine sapovirus GIII (Pol-Cap junction) | RT-qPCR | SW-SaV-GIII-qPCR-F | ATT GGG CCA ACG CAG TGG | 18 | 64 | 106 bp |
| SW-SaV-GIII-qPCR-PR | /6-FAM/CAA CGC ATG/ZEN/GTA CGG TGG CAC T/IABkFQ/ | 22 | 69 | |||
| SW-SaV-GIII-qPCR-R | GCC TCC ATC ACG AAC ACT TCT | 21 | 64 | |||
| 3′RACE-snPCR1 | SW-SaV-Pol-F1 | ATT GGG CCA ACG CAG TGG | 18 | 64 | ≈2300 bp * | |
| 3′RACE-snPCR2 | SW-SaV-Pol-F2 | CAA CGC ATG GTA CGG TGG CAC T | 22 | 69 | ≈2270 bp * | |
| Swine valovirus GI (Pol) | RT-qPCR | SW-VaV-qPCR-F | GAA GTC CGT CTG CCC CAC | 18 | 64 | 112 bp |
| SW-VaV-qPCR-PR | /Cy5/CAC TGG GTG/TAO/AGG CCT CCC TAC A/IAbRQSp/ | 22 | 69 | |||
| SW-VaV-qPCR-R | CTC GGC AAC CTC ATC ACA GAA | 21 | 64 | |||
| 3′RACE-snPCR1 | SW-VaV-Pol-F1 | GAA GTC CGT CTG CCC CAC | 18 | 64 | ≈2200 bp * | |
| 3′RACE-snPCR2 | SW-VaV-Pol-F2 | CAC TGG GTG AGG CCT CCC TAC A | 22 | 69 | ≈2150 bp * | |
| 3′ Poly(A)-tail | 3′RACE-RT | Oligo dT-Anchor-Adapter # | GGC CGC GCC ACC AAT TTA AA T(15)V | 36 | >42 | |
| 3′RACE-snPCR1/2 | Adapter-1 * | GGC CGC GCC ACC AAT TTA AAT | 21 | 66 |
| Copies/ Reaction | Mean Cq (SD) | Detected/Tested (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Singleplex | Triplex | Singleplex | Triplex | |||||||||
| Sw-SaV | Sw-NoV | Sw-VaV | Sw-SaV | Sw-NoV | Sw-VaV | Sw-SaV | Sw-NoV | Sw-VaV | Sw-SaV | Sw-NoV | Sw-VaV | |
| (1 × 101) 10 | 37.05 (0.71) | 37.53 (0.09) | 38.46 (0.01) | - (-) | 38.30 (0.64) | 38.87 (0.88) | 3/10 (30) | 4/10 (40) | 2/10 (20) | 0/10 (0) | 5/10 (50) | 2/10 (20) |
| (1 × 102) 100 | 34.99 (0.28) | 35.18 (0.42) | 36.16 (0.61) | 37.82 (0.53) | 35.46 (0.59) | 36.41 (0.99) | 10/10 (100) | 10/10 (100) | 10/10 (100) | 4/10 (40) | 10/10 (100) | 10/10 (100) |
| (1 × 103) 1000 | 31.79 (0.24) | 32.16 (0.12) | 32.81 (0.25) | 33.00 (0.27) | 31.91 (0.20) | 32.66 (0.30) | 10/10 (100) | 10/10 (100) | 10/10 (100) | 10/10 (100) | 10/10 (100) | 10/10 (100) |
| Age Groups | Disease Status | Sw-SaV | Sw-NoV | Sw-VaV |
|---|---|---|---|---|
| Suckling pigs | diarrheic | 1/8 (12.50%) | 0/8 | 0/8 |
| (1–20 days) | non-diarrheic | 0/9 (0%) | 0/9 | 0/9 |
| overall | 1/17 (5.88%) | 0/17 (0%) | 0/17 (0%) | |
| Nursery pigs | diarrheic | 32/55 (58.18%) | 1/55 (1.82%) | 2/55 (3.64%) |
| (21–77 days) | non-diarrheic | 17/72 (26.61%) | 4/72 (5.56%) | 1/72 (1.39%) |
| overall | 49/127 (38.58%) | 5/127 (3.94%) | 3/127 (2.36%) | |
| Fattening pigs | diarrheic | 0/0 | 0/0 | 0/0 |
| (78–140 days) | non-diarrheic | 1/39 (2.56%) | 0/39 | 0/39 |
| overall | 1/39 (2.56%) | 0/39 (0%) | 0/39 (0%) | |
| Sows | diarrheic | 0/0 | 0/0 | 0/0 |
| (≥141 days) | non-diarrheic | 1/15 (6.66%) | 0/15 | 0/15 |
| overall | 1/15 (6.66%) | 0/15 (0%) | 0/15 (0%) | |
| ∑ | 52/198 (26.26%) | 5/198 (2.53%) | 3/198 (1.52%) |
| Age Groups | No. of Samples | Sw-SaV | Sw-NoV | Sw-VaV |
|---|---|---|---|---|
| Suckling pigs | 1 | 0 | 0 | 0 |
| (1–20 days) | ||||
| Nursery pigs | 107 | 7 (6.54%) | 1 (0.94%) | 1 (0.94%) |
| (21–77 days) | ||||
| Fattening pigs | 116 | 3 (2.58%) | 16 (13.79%) | 1 (0.86%) |
| (78–140 days) | ||||
| Sows | 4 | 0 | 0 | 0 |
| (≥141 days) | ||||
| Overall | 228 | 10 (4.39%) | 17 (7.46%) | 2 (0.88%) |
| Virus | Sample/Farm Type | Overall Pos. Farms | Low Prevalence Farms | Medium Prevalence Farms | High Prevalence Farms |
|---|---|---|---|---|---|
| Sw-SaV | “enteric” | 17/25 (68.0%) | 6/17 (35.3%) | 7/17 (41.2%) | 4/17 (23.5%) |
| “oral fluid” | 6/24 (25.0%) | 5/6 (83.3%) | 1/6 (16.7%) | 0/6 | |
| Sw-NoV | “enteric” | 3/25 (12.0%) | 2/3 (66.7%) | 0/3 | 1/3 (33.3%) |
| “oral fluid” | 10/24 (41.7%) | 9/10 (90.0%) | 1/10 (10%) | 0/10 | |
| Sw-VaV | “enteric” | 2/25 (8.0%) | 1/2 (50.0%) | 0/2 | 1/2 (50.0%) |
| “oral fluid” | 2/24 (8.3%) | 2/2 (100%) | 0/2 | 0/2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
László, Z.; Pankovics, P.; Urbán, P.; Herczeg, R.; Balka, G.; Igriczi, B.; Cságola, A.; Albert, M.; Tóth, F.; Reuter, G.; et al. Multiple Co-Infecting Caliciviruses in Oral Fluid and Enteric Samples of Swine Detected by a Novel RT-qPCR Assay and a 3′RACE-PCR-NGS Method. Viruses 2025, 17, 193. https://doi.org/10.3390/v17020193
László Z, Pankovics P, Urbán P, Herczeg R, Balka G, Igriczi B, Cságola A, Albert M, Tóth F, Reuter G, et al. Multiple Co-Infecting Caliciviruses in Oral Fluid and Enteric Samples of Swine Detected by a Novel RT-qPCR Assay and a 3′RACE-PCR-NGS Method. Viruses. 2025; 17(2):193. https://doi.org/10.3390/v17020193
Chicago/Turabian StyleLászló, Zoltán, Péter Pankovics, Péter Urbán, Róbert Herczeg, Gyula Balka, Barbara Igriczi, Attila Cságola, Mihály Albert, Fruzsina Tóth, Gábor Reuter, and et al. 2025. "Multiple Co-Infecting Caliciviruses in Oral Fluid and Enteric Samples of Swine Detected by a Novel RT-qPCR Assay and a 3′RACE-PCR-NGS Method" Viruses 17, no. 2: 193. https://doi.org/10.3390/v17020193
APA StyleLászló, Z., Pankovics, P., Urbán, P., Herczeg, R., Balka, G., Igriczi, B., Cságola, A., Albert, M., Tóth, F., Reuter, G., & Boros, Á. (2025). Multiple Co-Infecting Caliciviruses in Oral Fluid and Enteric Samples of Swine Detected by a Novel RT-qPCR Assay and a 3′RACE-PCR-NGS Method. Viruses, 17(2), 193. https://doi.org/10.3390/v17020193








